Effect of Allergen-Specific Immunotherapy on Transcriptomic Changes in Canine Atopic Dermatitis
Abstract
1. Introduction
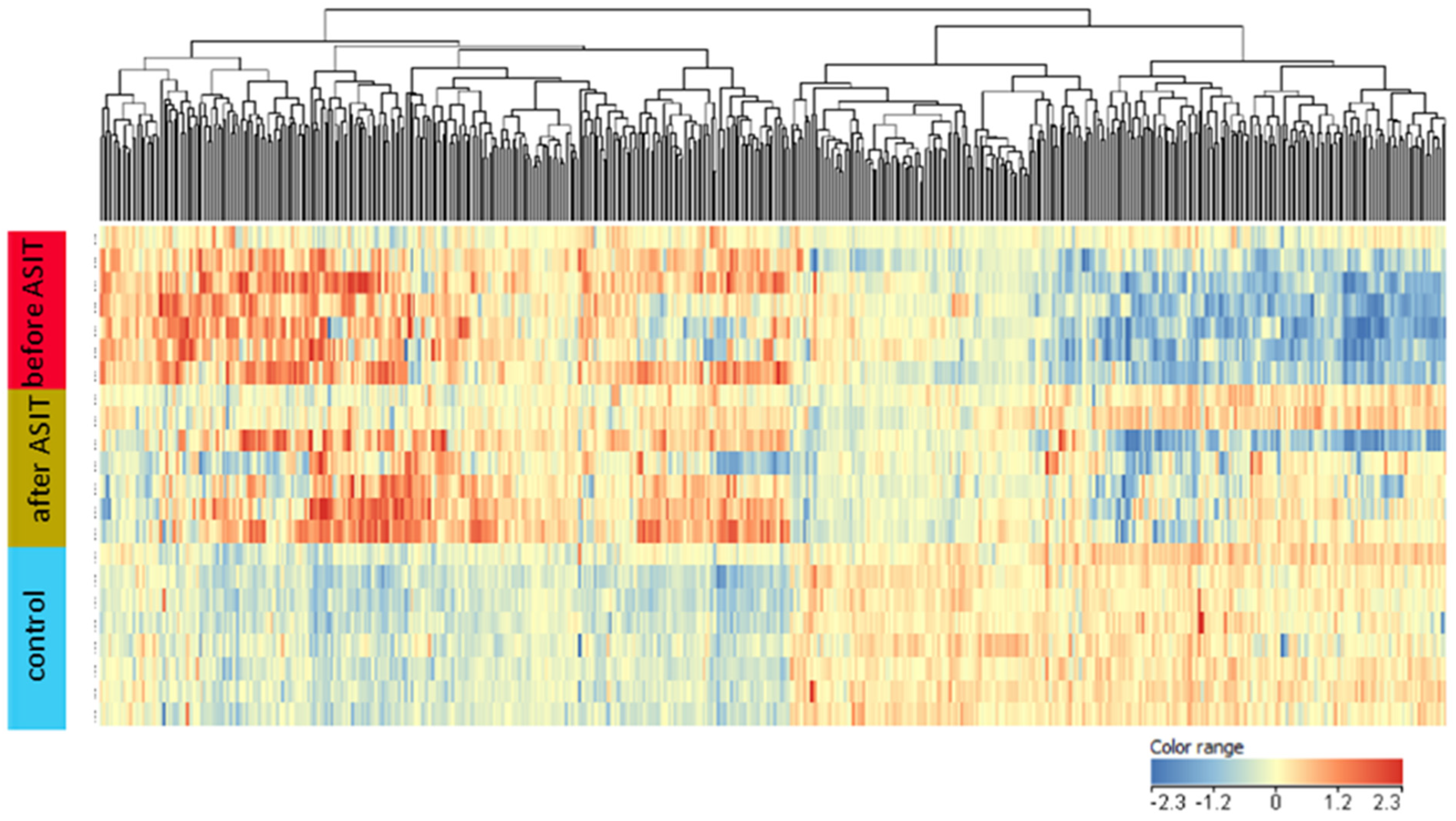
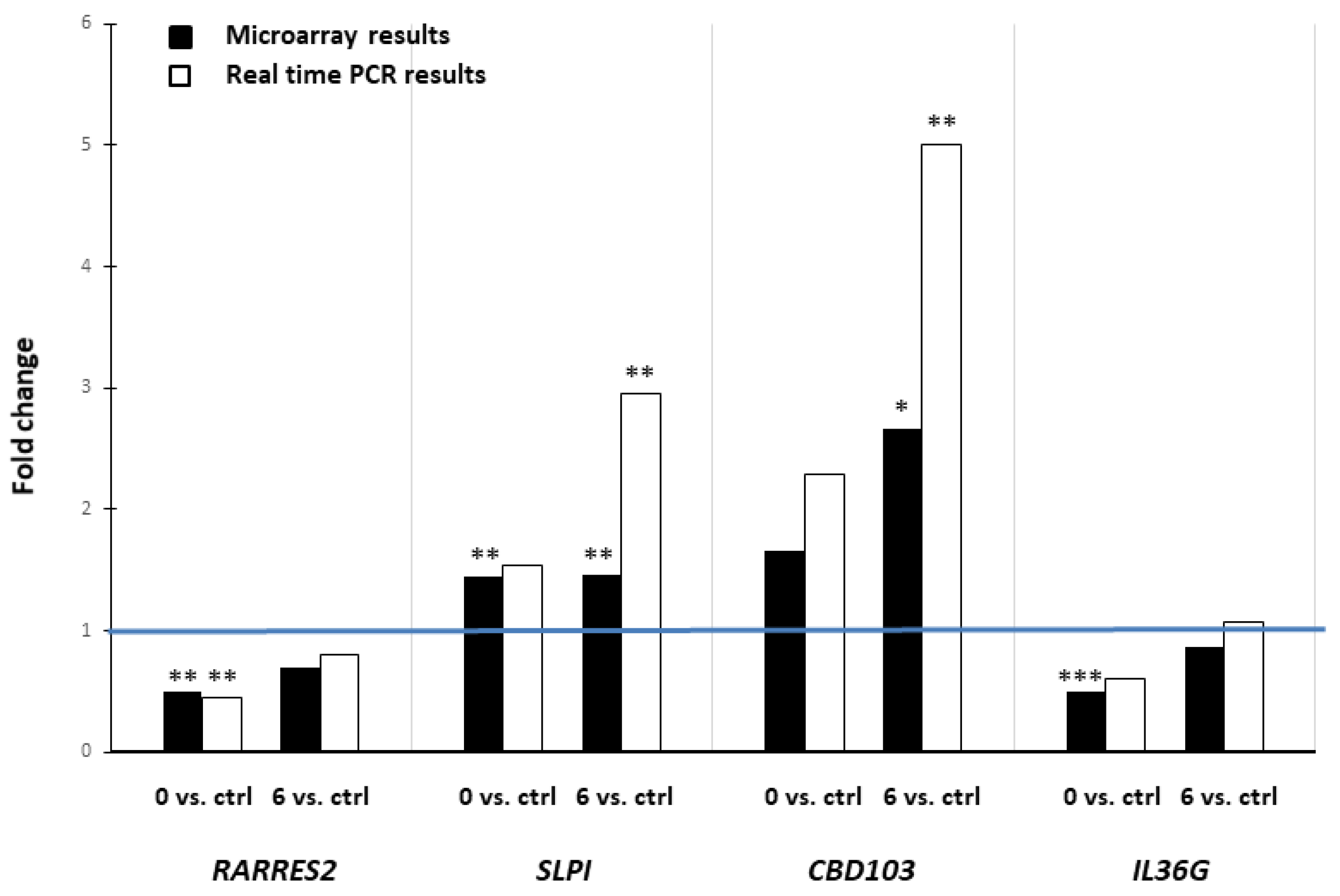
2. Results
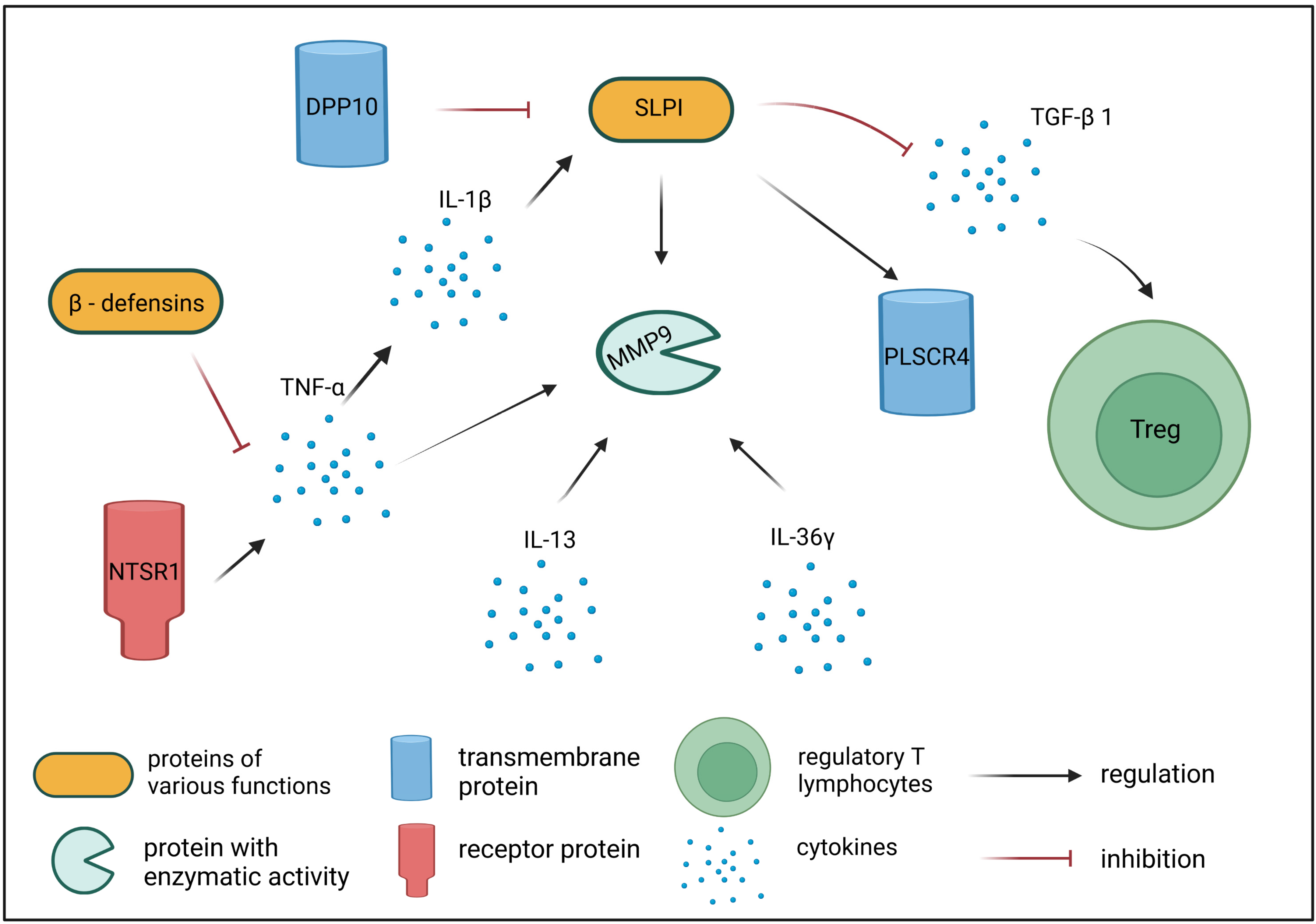
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. cAD Diagnosis and Sample Collection
4.3. Allergen-Specific Immunotherapy (ASIT)
4.4. Gene Expression Microarray Analysis
4.5. Real-Time RT-PCR
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hensel, P.; Santoro, D.; Favrot, C.; Hill, P.; Griffin, C. Canine atopic dermatitis: Detailed guidelines for diagnosis and allergen identification. BMC Vet. Res. 2015, 11, 196. [Google Scholar] [CrossRef]
- Favrot, C. Clinical signs and diagnosis of canine atopic dermatitis. In Proceedings of the 3. Congresso Latinoamericano de Dermatologia Veterinaria, Buenos Aires, Argentina, 26–27 November 2015. [Google Scholar]
- Olivry, T.; Banovic, F. Treatment of canine atopic dermatitis: Time to revise our strategy? Vet. Dermatol. 2019, 30, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Arasi, S.; Corsello, G.; Villani, A.; Pajno, G.B. The future outlook on allergen immunotherapy in children: 2018 and beyond. Ital. J. Pediatr. 2018, 44, 80. [Google Scholar] [CrossRef] [PubMed]
- Fennis, E.E.M.; van Damme, C.M.M.; Schlotter, Y.M.; Sinke, J.D.; Leistra, M.H.G.; Bartels, R.T.; Broere, F. Efficacy of subcutaneous allergen immunotherapy in atopic dogs: A retrospective study of 664 cases. Vet. Dermatol. 2022, 33, 321-e75. [Google Scholar] [CrossRef] [PubMed]
- Gedon, N.K.Y.; Mueller, R.S. Atopic dermatitis in cats and dogs: A difficult disease for animals and owners. Clin. Transl. Allergy 2018, 8, 41. [Google Scholar] [CrossRef]
- Fischer, N.M.; Müller, R.S. Allergen Specific Immunotherapy in Canine Atopic Dermatitis: An Update. Curr. Dermatol. Rep. 2019, 8, 297–302. [Google Scholar] [CrossRef]
- Valenta, R.; Campana, R.; Marth, K.; van Hage, M. Allergen-specific immunotherapy: From therapeutic vaccines to prophylactic approaches. J. Intern. Med. 2012, 272, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Alvaro-Lozano, M.; Akdis, C.A.; Akdis, M.; Alviani, C.; Angier, E.; Arasi, S.; Arzt-Gradwohl, L.; Barber, D.; Bazire, R.; Cavkaytar, O.; et al. EAACI Allergen Immunotherapy User’s Guide. Pediatr. Allergy Immunol. 2020, 31, 1–101. [Google Scholar] [CrossRef]
- Kucuksezer, U.C.; Ozdemir, C.; Cevhertas, L.; Ogulur, I.; Akdis, M.; Akdis, C.A. Mechanisms of allergen-specific immunotherapy and allergen tolerance. Allergol. Int. 2020, 69, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Czerwińska, E.; Nittner-Marszalska, M.; Mastalerz-Migas, A.; Szenborn, L. Prophylactic vaccinations management in patients undergoing allergen immunotherapy—A review. Fam. Med. Prim. Care Rev. 2022, 24, 163–167. [Google Scholar] [CrossRef]
- Shamji, M.H.; Sharif, H.; Layhadi, J.A.; Zhu, R.; Kishore, U.; Renz, H. Diverse immune mechanisms of allergen immunotherapy for allergic rhinitis with and without asthma. J. Allergy Clin. Immunol. 2022, 149, 791–801. [Google Scholar] [CrossRef]
- Majewska, A.; Dembele, K.; Dzieńdzikowska, K.; Prostek, A.; Gajewska, M. Cytokine and Lymphocyte Profiles in Dogs with Atopic Dermatitis after Allergen-Specific Immunotherapy. Vaccines 2022, 10, 1037. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.C.; Griffin, C.E.; Hill, P.B. Dermatophagoides farinae-specific IgG responses in atopic dogs undergoing allergen-specific immunotherapy with aqueous vaccines. Vet. Dermatol. 2008, 19, 215–220. [Google Scholar] [CrossRef] [PubMed]
- DeBoer, D.J.; Verbrugge, M.; Morris, M. Clinical and immunological responses of dust mite sensitive, atopic dogs to treatment with sublingual immunotherapy (SLIT). Vet. Dermatol. 2016, 27, 82-e23. [Google Scholar] [CrossRef]
- Keppel, K.E.; Campbell, K.L.; Zuckermann, F.A.; Greeley, E.A.; Schaeffer, D.J.; Husmann, R.J. Quantitation of canine regulatory T cell populations, serum interleukin-10 and allergen-specific IgE concentrations in healthy control dogs and canine atopic dermatitis patients receiving allergen-specific immunotherapy. Vet. Immunol. Immunopathol. 2008, 123, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Martini, F.; Rostaher, A.; Favrot, C.; Fischer, N. Interleukin 10 and transforming growth factor-beta 1 plasma levels in atopic dogs before and during immunotherapy. Vet. Rec. 2022, 190, e1270. [Google Scholar] [CrossRef]
- Beccati, M.; Martini, V.; Comazzi, S.; Fanton, N.; Cornegliani, L. Lymphocyte subpopulations and Treg cells in dogs with atopic dermatitis receiving ciclosporin therapy: A prospective study. Vet. Dermatol. 2016, 2, 17-e5. [Google Scholar] [CrossRef]
- Hauck, V.; Hügli, P.; Meli, M.L.; Rostaher, A.; Fischer, N.; Hofmann-Lehmann, R.; Favrot, C. Increased numbers of FoxP3-expressing CD4+ CD25+ regulatory T cells in peripheral blood from dogs with atopic dermatitis and its correlation with disease severity. Vet. Dermatol. 2016, 27, 26-e9. [Google Scholar] [CrossRef] [PubMed]
- Majewska, A.; Gajewska, M.; Dembele, K.; Maciejewski, H.; Prostek, A.; Jank, M. Lymphocytic, cytokine and transcriptomic profiles in peripheral blood of dogs with atopic dermatitis. BMC Vet. Res. 2016, 12, 174. [Google Scholar] [CrossRef]
- Zheng, Y.; Luo, S.; Wang, G.; Peng, Z.; Zeng, W.; Tan, S.; Xi, Y.; Fan, J. Downregulation of tazarotene induced gene-2 (TIG2) in skin squamous cell carcinoma. Eur. J. Dermatol. 2008, 18, 638–641. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yang, W.; Yang, X.; Lin, Y.; Lv, J.; Dou, X.; Luo, Q.; Dong, J.; Chen, Z.; Chu, Y.; et al. Chemerin suppresses murine allergic asthma by inhibiting CCL2 production and subsequent airway recruitment of inflammatory dendritic cells. Allergy 2014, 69, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Saurat, J.H. Retinoids and psoriasis: Novel issues in retinoid pharmacology and implications for psoriasis treatment. J. Am. Acad. Dermatol. 1999, 41, S2–S6. [Google Scholar] [CrossRef] [PubMed]
- Mihály, J.; Gamlieli, A.; Worm, M.; Rühl, R. Decreased retinoid concentration and retinoid signalling pathways in human atopic dermatitis. Exp. Dermatol. 2011, 20, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Poobalasingam, T.; Yates, L.L.; Walker, S.A.; Taylor, M.S.; Chessum, L.; Harrison, J.; Tsaprouni, L.; Adcock, I.M.; Lloyd, C.M.; et al. Manipulation of dipeptidylpeptidase 10 in mouse and human in vivo and in vitro models indicates a protective role in asthma. Dis. Model. Mech. 2018, 11, dmm031369. [Google Scholar] [CrossRef]
- Zagha, E.; Ozaita, A.; Chang, S.Y.; Nadal, M.S.; Lin, U.; Saganich, M.J.; McCormack, T.; Akinsanya, K.O.; Qi, S.Y.; Rudy, B. DPP10 modulates Kv4-mediated A-type potassium channels. J. Biol. Chem. 2005, 280, 18853–18861. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.M.; Jun, E.; Conlon, H.; Sadiq, S.A. Inhibition of SLPI ameliorates disease activity in experimental autoimmune encephalomyelitis. BMC Neurosci. 2012, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Tateosian, N.L.; Reiteri, R.M.; Amiano, N.O.; Costa, M.J.; Villalonga, X.; Guerrieri, D.; Maffía, P.C. Neutrophil elastase treated dendritic cells promote the generation of CD4(+)FOXP3(+) regulatory T cells in vitro. Cell. Immunol. 2011, 269, 128–134. [Google Scholar] [CrossRef]
- Py, B.; Basmaciogullari, S.; Bouchet, J.; Zarka, M.; Moura, I.C.; Benhamou, M.; Monteiro, R.C.; Hocini, H.; Madrid, R.; Benichou, S. The phospholipid scramblases 1 and 4 are cellular receptors for the secretory leukocyte protease inhibitor and interact with CD4 at the plasma membrane. PLoS ONE 2009, 4, e5006. [Google Scholar] [CrossRef]
- Ram, M.; Sherer, Y.; Shoenfeld, Y. Matrix metalloproteinase-9 and autoimmune diseases. J. Clin. Immunol. 2006, 26, 299–307. [Google Scholar] [CrossRef]
- Ingram, J.; Kraft, M. Metalloproteinases as modulators of allergic asthma: Therapeutic perspectives. Met. Med. 2015, 2, 61–74. [Google Scholar] [CrossRef]
- Puxeddu, I.; Petrelli, F.; Angelotti, F.; Croia, C.; Migliorini, P. Biomarkers In Chronic Spontaneous Urticaria: Current Targets And Clinical Implications. J. Asthma Allergy 2019, 12, 285–295. [Google Scholar] [CrossRef]
- Zhang, D.; Simmen, R.C.; Michel, F.J.; Zhao, G.; Vale-Cruz, D.; Simmen, F.A. Secretory leukocyte protease inhibitor mediates proliferation of human endometrial epithelial cells by positive and negative regulation of growth-associated genes. J. Biol. Chem. 2002, 277, 29999–30009. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.; Shapiro, S.; Karni, A.; Weiner, H.L.; Miller, A. Gelatinases (MMP-2 and MMP-9) are preferentially expressed by Th1 vs. Th2 cells. J. Neuroimmunol. 2005, 163, 157–164. [Google Scholar] [CrossRef]
- Harper, J.I.; Godwin, H.; Green, A.; Wilkes, L.E.; Holden, N.J.; Moffatt, M.; Cookson, W.O.; Layton, G.; Chandler, S. A study of matrix metalloproteinase expression and activity in atopic dermatitis using a novel skin wash sampling assay for functional biomarker analysis. Br. J. Dermatol. 2010, 162, 397–403. [Google Scholar] [CrossRef]
- Purwar, R.; Kraus, M.; Werfel, T.; Wittmann, M. Modulation of keratinocyte-derived MMP-9 by IL-13, a possible role for the pathogenesis of epidermal inflammation. J. Investig. Dermatol. 2008, 128, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Schwingshackl, A.; Duszyk, M.; Brown, N.; Moqbel, R. Human eosinophils release matrix metalloproteinase-9 on stimulation with TNF-alpha. J. Allergy Clin. Immunol. 1999, 104, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, E.; Rodriguez-Canales, J.; Hewitt, S.M.; Elmasri, W.; Han, J.; Han, S.; Davidson, B.; Kohn, E.C. Paracrine SLPI secretion upregulates MMP-9 transcription and secretion in ovarian cancer cells. Gynecol. Oncol. 2011, 122, 656–662. [Google Scholar] [CrossRef]
- Poachanukoon, O.; Meesuk, L.; Pattanacharoenchai, N.; Monthanapisut, P.; Dechatiwongse Na Ayudhya, T.; Koontongkaew, S. Zingiber cassumunar ROXb. and its active constituent inhibit MMP-9 direct activation by house dust mite allergens and MMP-9 expression in PMA-stimulated human airway epithelial cells. Asian Pac. J. Allergy Immunol. 2015, 33, 42–51. [Google Scholar] [CrossRef]
- St-Gelais, F.; Jomphe, C.; Trudeau, L.E. The role of neurotensin in central nervous system pathophysiology: What is the evidence? J. Psychiatry Neurosci. 2006, 31, 229–245. [Google Scholar]
- Ferris, C.F.; Carraway, R.E.; Hammer, R.A.; Leeman, S.E. Release and degradation of neurotensin during perfusion of rat small intestine with lipid. Regul. Pept. 1985, 12, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Pellino, G.; Fiorentino, F.; Rasheed, S.; Darzi, A.; Tekkis, P.; Kontovounisios, C. A Review of the Role of Neurotensin and Its Receptors in Colorectal Cancer. Gastroenterol. Res. Pract. 2017, 2017, 6456257. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.; Neves, B.M.; Moura, L.; Cruz, M.T.; Carvalho, E. Neurotensin downregulates the pro-inflammatory properties of skin dendritic cells and increases epidermal growth factor expression. Biochim. Biophys. Acta 2011, 1813, 1863–1871. [Google Scholar] [CrossRef] [PubMed]
- Ostlere, L.S.; Cowen, T.; Rustin, M.H. Neuropeptides in the skin of patients with atopic dermatitis. Clin. Exp. Dermatol. 1995, 20, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Singh, L.K.; Boucher, W.; Pang, X.; Letourneau, R.; Seretakis, D.; Green, M.; Theoharides, T.C. Potent mast cell degranulation and vascular permeability triggered by urocortin through activation of corticotropin-releasing hormone receptors. J. Pharmacol. Exp. Ther. 1999, 288, 1349–1356. [Google Scholar] [PubMed]
- Donelan, J.; Boucher, W.; Papadopoulou, N.; Lytinas, M.; Papaliodis, D.; Dobner, P.; Theoharides, T.C. Corticotropin-releasing hormone induces skin vascular permeability through a neurotensin-dependent process. Proc. Natl. Acad. Sci. USA 2006, 103, 7759–7764. [Google Scholar] [CrossRef]
- Carraway, R.; Cochrane, D.E.; Lansman, J.B.; Leeman, S.E.; Paterson, B.M.; Welch, H.J. Neurotensin stimulates exocytotic histamine secretion from rat mast cells and elevates plasma histamine levels. J. Physiol. 1982, 323, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Feldberg, R.S.; Cochrane, D.E.; Carraway, R.E.; Brown, E.; Sawyer, R.; Hartunian, M.; Wentworth, D. Evidence for a neurotensin receptor in rat serosal mast cells. Inflamm. Res. 1998, 47, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Alysandratos, K.D.; Asadi, S.; Angelidou, A.; Zhang, B.; Sismanopoulos, N.; Yang, H.; Critchfield, A.; Theoharides, T.C. Neurotensin and CRH interactions augment human mast cell activation. PLoS ONE 2012, 7, e48934. [Google Scholar] [CrossRef]
- Koczy-Baron, E.; Jochem, J.; Kasperska-Zajac, A. Increased plasma concentration of vascular endothelial growth factor in patients with atopic dermatitis and its relation to disease severity and platelet activation. Inflamm. Res. 2012, 61, 1405–1409. [Google Scholar] [CrossRef] [PubMed]
- Samochocki, Z.; Bogaczewicz, J.; Sysa-Jędrzejowska, A.; McCauliffe, D.P.; Kontny, E.; Wozniacka, A. Expression of vascular endothelial growth factor and other cytokines in atopic dermatitis, and correlation with clinical features. Int. J. Dermatol. 2016, 55, e141–e146. [Google Scholar] [CrossRef]
- Vasiadi, M.; Mondolfi, A.P.; Alysandratos, K.D.; Therianou, A.; Katsarou-Katsari, A.; Petrakopoulou, T.; Theoharidis, A.; Miniati, A.; Theoharides, T.C. Neurotensin serum levels and skin gene expression are increased in atopic dermatitis. Br. J. Dermatol. 2013, 169, 695–699. [Google Scholar] [CrossRef]
- Semple, F.; Dorin, J.R. β-Defensins: Multifunctional modulators of infection, inflammation and more? J. Innate Immun. 2012, 4, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.R.; Davidson, D.J.; Dorin, J.R. The Dichotomous Responses Driven by β-Defensins. Front. Immunol. 2020, 11, 1176. [Google Scholar] [CrossRef] [PubMed]
- Toke, O. Antimicrobial peptides: New candidates in the fight against bacterial infections. Biopolymers 2005, 80, 717–735. [Google Scholar] [CrossRef] [PubMed]
- van Damme, C.M.; Willemse, T.; van Dijk, A.; Haagsman, H.P.; Veldhuizen, E.J. Altered cutaneous expression of beta-defensins in dogs with atopic dermatitis. Mol. Immunol. 2009, 46, 2449–2455. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chertov, O.; Bykovskaia, S.N.; Chen, Q.; Buffo, M.J.; Shogan, J.; Anderson, M.; Schröder, J.M.; Wang, J.M.; Howard, O.M.; et al. Beta-defensins: Linking innate and adaptive immunity through dendritic and T cell CCR6. Science 1999, 286, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Lancto, C.; Torres, S.M.; Hendrickson, J.A.; Martins, K.V.; Rutherford, M.S. Altered expression of antimicrobial peptide genes in the skin of dogs with atopic dermatitis and other inflammatory skin conditions. Vet. Dermatol. 2013, 24, 414–421.e90. [Google Scholar] [CrossRef]
- Leonard, B.C.; Marks, S.L.; Outerbridge, C.A.; Affolter, V.K.; Kananurak, A.; Young, A.; Moore, P.F.; Bannasch, D.L.; Bevins, C.L. Activity, expression and genetic variation of canine β-defensin 103, a multifunctional antimicrobial peptide in the skin of domestic dogs. J. Innate Immun. 2012, 4, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Towne, J.E.; Garka, K.E.; Renshaw, B.R.; Virca, G.D.; Sims, J.E. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J. Biol. Chem. 2004, 279, 13677–13688. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.C.; Xu, W.D.; Liu, X.Y.; Liu, X.Y.; Huang, A.F.; Su, L.C. Biology of IL-36 Signaling and Its Role in Systemic Inflammatory Diseases. Front. Immunol. 2019, 10, 2532. [Google Scholar] [CrossRef] [PubMed]
- Gresnigt, M.S.; van de Veerdonk, F.L. Biology of IL-36 cytokines and their role in disease. Semin. Immunol. 2013, 25, 458–465. [Google Scholar] [CrossRef]
- Costelloe, C.; Watson, M.; Murphy, A.; McQuillan, K.; Loscher, C.; Armstrong, M.E.; Garlanda, C.; Mantovani, A.; O’Neill, L.A.; Mills, K.H.; et al. IL-1F5 mediates anti-inflammatory activity in the brain through induction of IL-4 following interaction with SIGIRR/TIR8. J. Neurochem. 2008, 105, 1960–1969. [Google Scholar] [CrossRef]
- Turtoi, A.; Brown, I.; Schläger, M.; Schneeweiss, F.H. Gene expression profile of human lymphocytes exposed to (211)At alpha particles. Radiat. Res. 2010, 174, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Vigne, S.; Palmer, G.; Lamacchia, C.; Martin, P.; Talabot-Ayer, D.; Rodriguez, E.; Ronchi, F.; Sallusto, F.; Dinh, H.; Sims, J.E.; et al. IL-36R ligands are potent regulators of dendritic and T cells. Blood 2011, 118, 5813–5823. [Google Scholar] [CrossRef]
- Vigne, S.; Palmer, G.; Martin, P.; Lamacchia, C.; Strebel, D.; Rodriguez, E.; Olleros, M.L.; Vesin, D.; Garcia, I.; Ronchi, F.; et al. IL-36 signaling amplifies Th1 responses by enhancing proliferation and Th1 polarization of naive CD4+ T cells. Blood 2012, 120, 3478–3487. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Fariñas, M.; Ungar, B.; Correa da Rosa, J.; Ewald, D.A.; Rozenblit, M.; Gonzalez, J.; Xu, H.; Zheng, X.; Peng, X.; Estrada, Y.D.; et al. RNA sequencing atopic dermatitis transcriptome profiling provides insights into novel disease mechanisms with potential therapeutic implications. J. Allergy Clin. Immunol. 2015, 135, 1218–1227. [Google Scholar] [CrossRef]
- Buhl, A.L.; Wenzel, J. Interleukin-36 in Infectious and Inflammatory Skin Diseases. Front. Immunol. 2019, 10, 1162. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, Y.; Li, C.; Chang, L.; Wang, W.; Wang, Z.; Gao, X.; Ryffel, B.; Wu, Y.; Lai, Y. IL-36γ Induced by the TLR3-SLUG-VDR Axis Promotes Wound Healing via REG3A. J. Investig. Dermatol. 2017, 137, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Rodriguez, E.; Degenhardt, F.; Baurecht, H.; Wehkamp, U.; Volks, N.; Szymczak, S.; Swindell, W.R.; Sarkar, M.K.; Raja, K.; et al. Atopic Dermatitis Is an IL-13-Dominant Disease with Greater Molecular Heterogeneity Compared to Psoriasis. J. Investig. Dermatol. 2019, 139, 1480–1489. [Google Scholar] [CrossRef]
- D’Erme, A.M.; Wilsmann-Theis, D.; Wagenpfeil, J.; Hölzel, M.; Ferring-Schmitt, S.; Sternberg, S.; Wittmann, M.; Peters, B.; Bosio, A.; Bieber, T.; et al. IL-36γ (IL-1F9) is a biomarker for psoriasis skin lesions. J. Investig. Dermatol. 2015, 135, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Quaranta, M.; Knapp, B.; Garzorz, N.; Mattii, M.; Pullabhatla, V.; Pennino, D.; Andres, C.; Traidl-Hoffmann, C.; Cavani, A.; Theis, F.J.; et al. Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci. Transl. Med. 2014, 6, 244ra90. [Google Scholar] [CrossRef]
- Tsang, M.S.M.; Sun, X.; Wong, C.K. The Role of New IL-1 Family Members (IL-36 and IL-38) in Atopic Dermatitis, Allergic Asthma, and Allergic Rhinitis. Curr. Allergy Asthma Rep. 2020, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Tengvall, K.; Bergvall, K.; Olsson, M.; Ardesjö-Lundgren, B.; Farias, F.H.G.; Kierczak, M.; Hedhammar, Å.; Lindblad-Toh, K.; Andersson, G. Transcriptomes from German shepherd dogs reveal differences in immune activity between atopic dermatitis affected and control skin. Immunogenetics 2020, 72, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Brinkhof, B.; Spee, B.; Rothuizen, J.; Penning, L.C. Development and evaluation of canine reference genes for accurate quantification of gene expression. Anal. Biochem. 2006, 356, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.; Garden, O.A.; Werling, D.; Allenspach, K. Gene expression of selected signature cytokines of T cell subsets in duodenal tissues of dogs with and without inflammatory bowel disease. Vet. Immunol. Immunopathol. 2012, 146, 87–91. [Google Scholar] [CrossRef] [PubMed]
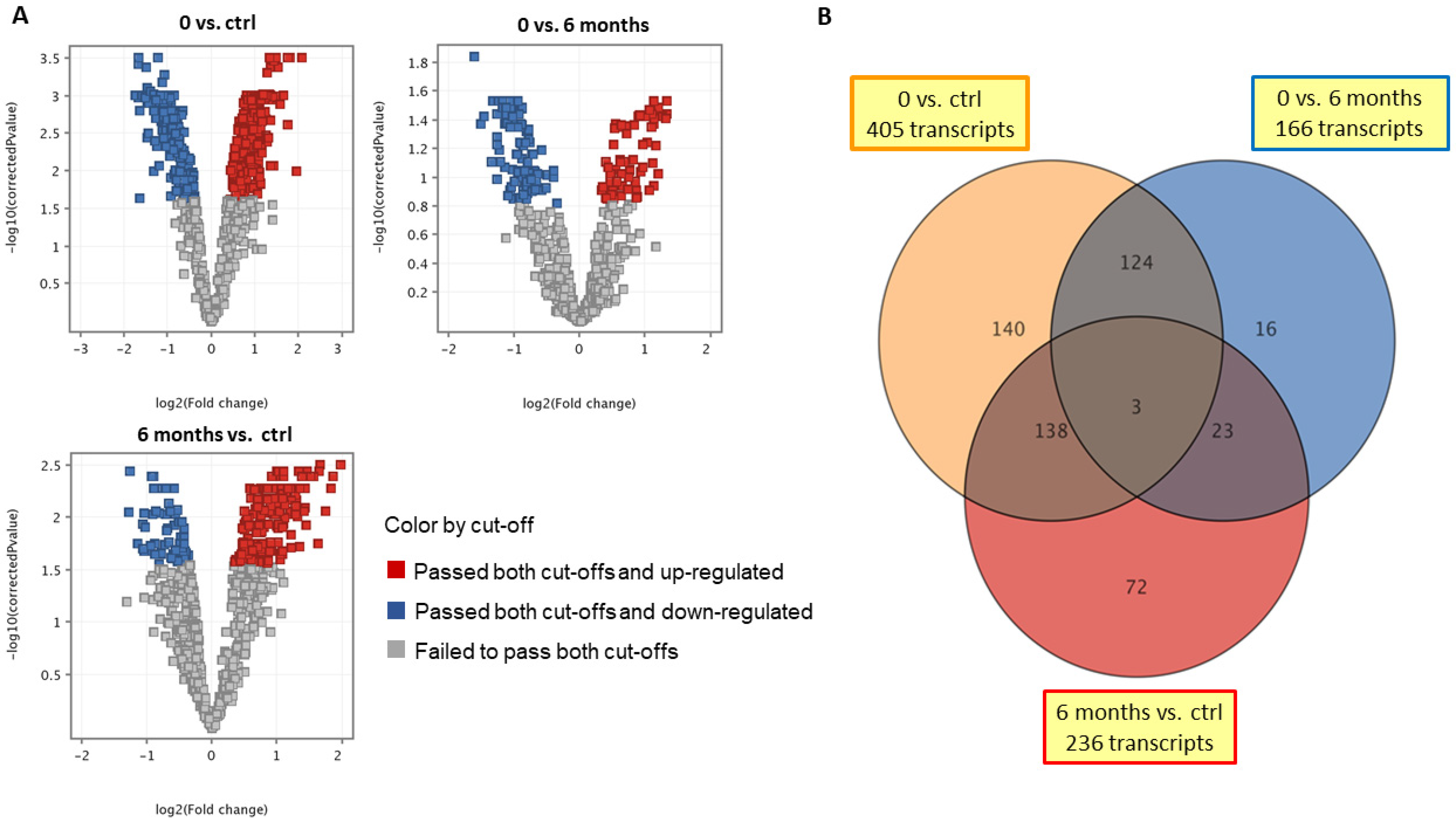
| Gene Symbol | Gene Name | GenBank Accession | FC (0 vs. 6 Months) | p-Values (0 vs. 6 Months) | FC (0 Months vs. Ctrl) | p-Values (0 Months vs. Ctrl) | FC (6 Months vs. Ctrl) | p-Values (6 Months vs. Ctrl) | Corrected p-Values (Ctrl vs. 0 Months vs. 6 Months) |
|---|---|---|---|---|---|---|---|---|---|
| RARRES2 | retinoic acid receptor responder 2 | XM_005629654 | −1.39 down | ns | −2.00 down | 0.0015 | −1.44 down | ns | 0.0089 |
| DPP10 | dipeptidyl peptidase-like 10 | CX005277 | −1.90 down | 0.0006 | −1.69 down | 0.0028 | 1.12 | ns | 0.0045 |
| SLPI | secretory leukocyte peptidase inhibitor | NM_001113171 | −1.01 | ns | 1.45 up | 0.0049 | 1.46 up | 0.00426 | 0.0081 |
| PLSCR4 | phospholipid scramblase 4 | XM_005634546 | 2.28 up | 0.0222 | 2.34 up | 0.0144 | 1.03 | ns | 0.0192 |
| MMP9 | matrix metalloproteinase 9 | NM_001003219 | 1.08 | ns | 3.36 up | 0.0239 | 3.10 up | 0.0357 | 0.0244 |
| NTSR1 | neurotensin receptor 1 | XM_543088 | 2.40 up | 0.0014 | 1.67 up | 0.0492 | −1.44 down | ns | 0.0083 |
| CBD103 | beta-defensin 103 | NM_001129980 | −1.60 down | ns | 1.66 up | ns | 2.66 up | 0.0240 | 0.0431 |
| DEFB122 | beta-defensin 122 | NM_001024641 | −2.14 down | 0.0408 | −2.88 down | 0.0034 | −1.35 down | ns | 0.0123 |
| IL36G | interleukin 36 gamma | XM_005630449 | −1.71 down | 0.0059 | −1.99 down | 0.0005 | −1.16 | ns | 0.0045 |
| Gene Symbol | Gene Name | Summary of Gene Function |
|---|---|---|
| RARRES2 | retinoic acid receptor responder 2 | Encodes a precursor of chemerin that acts via retinoic acid receptor (RAR), causing chemoattraction of dendritic cells and macrophages, linking innate and adaptive immunity [21,22]. RARRES2 gene is downregulated in skin of AD patients [24]. |
| DPP10 | dipeptidyl peptidase-like 10 | Encodes a single-pass type II membrane protein with no detectable protease activity, but with ability to bind other proteins, such as specific voltage-gated potassium channels, altering their expression and biophysical properties. Mutations in this gene have been associated with asthma [25]. Overexpression of DPP10 enhances glucocorticoid receptor (GR) activation even without glucocorticoid treatment. DPP10 protein may influence endogenous anti-inflammatory corticosteroid production [25]. |
| SLPI | secretory leukocyte peptidase inhibitor | Encodes a secreted inhibitor protecting epithelial tissues from endogenous serine proteases. SLPI protein also mediates the suppression of TGF-β expression and interferes with the differentiation of Treg cells [28]. SLPI has an affinity for leukocyte elastase and blocks its activity, which is needed for increased TGF-β expression in dendritic cells, in turn, leading to an increased number of CD4 + FOXP3+ cells [28]. |
| PLSCR4 | phospholipid scramblase 4 | Encodes a cell membrane protein necessary for CD4 receptor binding activity in T lymphocytes [29]. SLPI binding to PLSCR4 induces its translocation from extracellular matrix to cytoplasm and nuclei of monocytes, macrophages, and B and Th lymphocytes [29]. |
| MMP9 | matrix metalloproteinase 9 | Encodes MMP-9—one of the key metalloproteinases involved in the development and course of inflammatory reactions following allergen challenge. MMP-9 is secreted by several inflammatory cells, e.g., eosinophils, neutrophils, T cells, macrophages, and mast cells. MMP-9 promotes migration and activation of immune cells by cleaving pro-inflammatory chemokines and cytokines, therefore contributing to inflammatory processes [30,31,32]. Increased expression of MMP9 gene is observed in acute compared with chronic AD skin lesions [36]. |
| NTSR1 | neurotensin receptor 1 | Encodes a membrane protein belonging to the superfamily of G protein-coupled receptors. NTSR1 is a specific receptor for neurotensin, which, aside from its role as a neurotransmitter, is also known for its pro-inflammatory role, induction of vasodilatation, vascular permeability, activation of mast cell degranulation, and enhancement of directional migration and phagocytosis of neutrophils [43]. Neurotensin can be involved in the pathogenesis of inflammatory skin disorders, including AD, especially those exacerbated by stress, scratching, and sweating [44]. |
| CBD103 | beta-defensin 103 | Encodes a protein belonging to defensins—a family of microbicidal and cytotoxic peptides. CBD103 can be detected in skin and peripheral blood mononuclear cells, but shows decreased expression in cAD patients [56]. |
| DEFB122 | beta-defensin 122 | Encodes a protein belonging to defensins—a family of microbicidal and cytotoxic peptides. Decreased expression of DEFB122 transcript was observed in lesional and non-lesional skin of cAD patients in comparison to the skin of healthy dogs [58]. |
| IL36G | interleukin 36 gamma | Encodes interleukin 36 gamma (IL-36γ) belonging to the IL-1 superfamily. IL-36 is involved in immune cell activation, antigen presentation, and pro-inflammatory factor production [61]. IL-36 cytokine expression can be found in keratinocytes, B-lymphocytes, T-lymphocytes, dendritic cells, and monocytes [63,64,65,66,67,68]. Increased expression of IL36G was shown in lesional skin of AD patients compared to non-lesional skin [67,73,74]. |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | NCBI Accession Number |
|---|---|---|---|
| CBD103 | TGTGTGTCCTGCAACCTTAT | CACCGACCGCTCCTTATTC | NM 001129980.1 |
| IL36G | ATCACTGTTCTCCCATGCAA | CCAGTATCTCCTCCTCCTTTAG | XM_005630449.2 |
| RARRES2 | GGAGACCAGTGTGGACAGA | CATTTCCGCTTCCTCCCATT | XM_022403838.1 |
| SLPI | ATCCCGTTAATGTCTCCAACTC | AATGGCAGGTATCAGGCTTATT | NM_001113171.1 |
| RSP19 | CCTTCCTCAAAAAGTCTGGG | GTTCTCATCGTAGGGAGCAAG | XM_533657.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majewska, A.; Gajewska, M.; Dembele, K. Effect of Allergen-Specific Immunotherapy on Transcriptomic Changes in Canine Atopic Dermatitis. Int. J. Mol. Sci. 2023, 24, 11616. https://doi.org/10.3390/ijms241411616
Majewska A, Gajewska M, Dembele K. Effect of Allergen-Specific Immunotherapy on Transcriptomic Changes in Canine Atopic Dermatitis. International Journal of Molecular Sciences. 2023; 24(14):11616. https://doi.org/10.3390/ijms241411616
Chicago/Turabian StyleMajewska, Alicja, Małgorzata Gajewska, and Kourou Dembele. 2023. "Effect of Allergen-Specific Immunotherapy on Transcriptomic Changes in Canine Atopic Dermatitis" International Journal of Molecular Sciences 24, no. 14: 11616. https://doi.org/10.3390/ijms241411616
APA StyleMajewska, A., Gajewska, M., & Dembele, K. (2023). Effect of Allergen-Specific Immunotherapy on Transcriptomic Changes in Canine Atopic Dermatitis. International Journal of Molecular Sciences, 24(14), 11616. https://doi.org/10.3390/ijms241411616







