Report on the Effect of the Implementation of an Early Detection and Prevention of Cancer Program on Families at High Hereditary Risk—Concentrating on Patients Undergoing Genetic Diagnostics and Counseling in Central Poland
Abstract
1. Introduction
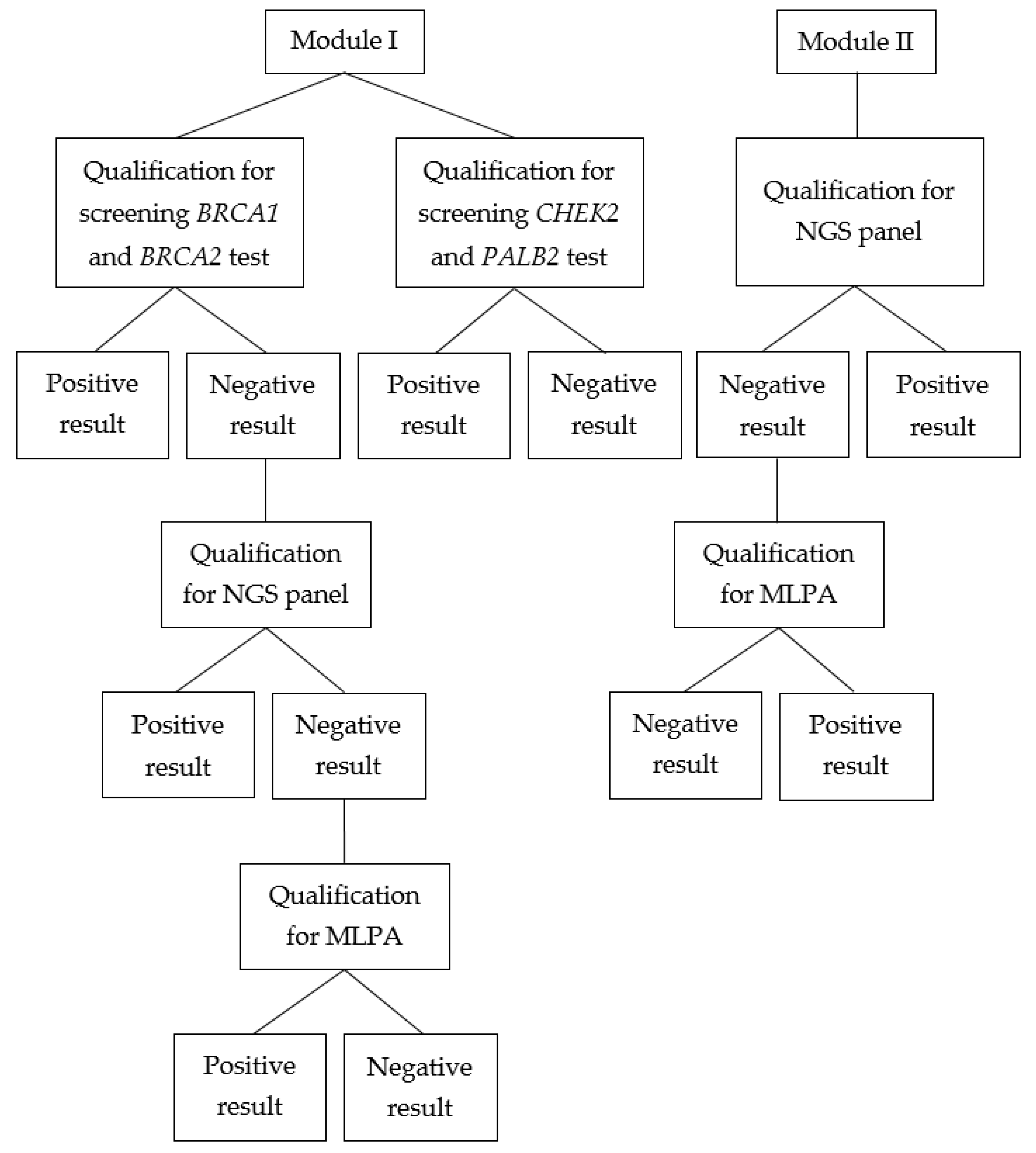
2. Results
2.1. Module I
2.2. Module II
3. Discussion
4. Materials and Methods
4.1. Patient Qualification—Module I
4.1.1. Qualification Criteria for the Highest-Risk Group
- Individuals from families with 3 or more cases of breast cancer and/or ovarian cancer among first- and second-degree relatives (including the proband);
- Individuals identified with a pathogenic mutation in the BRCA1, BRCA2, or PALB2 genes, regardless of their family history.
4.1.2. Qualification Criteria for the High-Risk Group
- Families with 2 cases of breast and/or ovarian cancer among the proband or first- and second-degree relatives (or 2 cases among second- and third-degree relatives on the paternal side), particularly when at least one affected individual had been diagnosed with ovarian cancer and one case of said cancer had occurred before the age of 50.
- Families with bilateral breast cancer diagnosed in first- and second-degree relatives.
- Families with breast cancer diagnosed before the age of 40 in first- and second-degree relatives.
- Families with breast cancer diagnosed in males among first- and second-degree relatives.
4.1.3. Qualification for Genetic Testing
- All individuals diagnosed with ovarian/fallopian tube/peritoneal cancer;
- All individuals diagnosed with breast cancer;
- First- and second-degree relatives of individuals with breast and/or ovarian cancer for whom marker mutations could not be established and diagnostics could not be employed for the affected individual.
- All individuals diagnosed with breast cancer;
- First-degree relatives of individuals with breast cancer from families meeting the criteria for high and the highest risk of breast cancer.
- The affected individual had been diagnosed with breast cancer or ovarian cancer and had at least 2 first- or second-degree relatives with a diagnosis of breast and/or ovarian cancer, where at least one of such cases had occurred before the age of 50;
- The affected individual had been diagnosed with breast cancer before the age of 50 or ovarian cancer at any age and had a first- or second-degree relative who had been diagnosed with breast cancer (breast cancer in males) and/or ovarian cancer;
- The same affected individual had been diagnosed with both breast and ovarian cancer or bilateral breast cancer, including at least one case below the age of 50;
- The affected individual had been diagnosed with ovarian cancer and had at least one relative with breast cancer—which had been diagnosed before the age of 50—or who had been diagnosed with ovarian cancer.
4.2. Patient Qualification—Module II
4.3. Laboratory Methodology
4.4. Editorial Policy and Ethical Considerations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Górski, B.; Jakubowska, A.; Huzarski, T.; Byrski, T.; Gronwald, J.; Grzybowska, E.; Mackiewicz, A.; Stawicka, M.; Bebenek, M.; Sorokin, D.; et al. A High Proportion of Founder BRCA1 Mutations in Polish Breast Cancer Families. Int. J. Cancer 2004, 110, 683–686. [Google Scholar] [CrossRef]
- Górski, B.; Lubiński, J. BRCA1 Testing. Hered. Cancer Clin. Pract. 2008, 6, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Dzidkowska, J.; Wojciechowska, U.; Czaderny, K.; Olasek, P.; Ciuba, A. Nowotwory Złośliwe w Polsce w 2019 Roku; Ministerstwo Zdrowia: Warsaw, Poland, 2021. [Google Scholar]
- American Cancer Society|Cancer Facts & Statistics. Available online: http://cancerstatisticscenter.cancer.org/ (accessed on 23 July 2022).
- Menkiszak, J.; Gronwald, J.; Górski, B.; Jakubowska, A.; Huzarski, T.; Byrski, T.; Foszczyńska-Kłoda, M.; Haus, O.; Janiszewska, H.; Perkowska, M.; et al. Hereditary Ovarian Cancer in Poland. Int. J. Cancer 2003, 106, 942–945. [Google Scholar] [CrossRef] [PubMed]
- Szwiec, M.; Jakubowska, A.; Górski, B.; Huzarski, T.; Tomiczek-Szwiec, J.; Gronwald, J.; Dębniak, T.; Byrski, T.; Kluźniak, W.; Wokołorczyk, D.; et al. Recurrent Mutations of BRCA1 and BRCA2 in Poland: An Update. Clin. Genet. 2015, 87, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, P.; Jasiowka, M.; Strycharz, E.; Sobol, M.; Hodorowicz-Zaniewska, D.; Skotnicki, P.; Byrski, T.; Blecharz, P.; Marczyk, E.; Cedrych, I.; et al. Recurrent Mutations of BRCA1, BRCA2 and PALB2 in the Population of Breast and Ovarian Cancer Patients in Southern Poland. Hered. Cancer Clin. Pract. 2016, 14, 5. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lubiński, J. Genetyka Kliniczna Nowotworów 2018; libroprint.pl: Szczecin, Poland, 2018. [Google Scholar]
- Złowocka-Perłowska, E.; Dębniak, T.; Słojewski, M.; Lemiński, A.; Soczawa, M.; van de Wetering, T.; Trubicka, J.; Kluźniak, W.; Wokołorczyk, D.; Cybulski, C.; et al. Recurrent PALB2 Mutations and the Risk of Cancers of Bladder or Kidney in Polish Population. Hered. Cancer Clin. Pract. 2021, 19, 6. [Google Scholar] [CrossRef] [PubMed]
| Test | Number of Tests | Number of Pathogenic Mutations | Percentage of Pathogenic Mutations |
|---|---|---|---|
| BRCA1 and BRCA2 screening test | 880 | 39 | 4.43% |
| Highest-risk group | 141 | 22 | 15.60% |
| High-risk group | 372 | 11 | 2.96% |
| CHEK2 screening test | 843 | 9 | 1.07% |
| Highest-risk group | 84 | 9 | 10.71% |
| High-risk group | 329 | 0 | 0% |
| PALB2 screening test | 843 | 3 | 0.35% |
| Highest-risk group | 84 | 0 | 0% |
| High-risk group | 329 | 3 | 0.91% |
| MLPA BRCA1 | 70 | 3 | 4.29% |
| MLPA BRCA2 | 62 | 2 | 3.33% |
| Next-generation sequencing | 55 | 10 | 18.18% |
| Gene | Mutation | dbSNP Number | ClinVar Submissions | Internal Classification |
|---|---|---|---|---|
| BRCA1 | NM_007294.4: c.4986+6T>G | rs80358086 | 12 (P), 2 (LP) | Pathogenic |
| BRCA1 | NM_007294.4:c.81-2A>C | rs397509326 | 2 (P), 2 (LP) | Pathogenic |
| BRCA1 | NM_007294.4:c.115T>C | rs80357164 | 15 (P), 1 (VUS) | Pathogenic |
| BRCA1 | NM_007294.4:c.5034_5037del | rs80357580 | 27 (P) | Pathogenic |
| BRCA1 | NM_007294.4:c.2761C>T | rs80357377 | 10 (P) | Pathogenic |
| BRCA2 | NM_000059.4:c.3975_3978dup | rs397515636 | 23 (P) | Pathogenic |
| BRCA2 | NM_000059:4:c.7990_7991del | - | 0 | Likely pathogenic |
| PALB2 | NM_024675.3:c.2962C>T | rs118203999 | 8 (P) | Pathogenic |
| PALB2 | NM_024675.3:c.697del | rs180177090 | 6 (P) | Pathogenic |
| RAD51C | NM_058216.3:c.577C>T | rs200293302 | 13 (P), 1 (LP) | Pathogenic |
| Gene | Mutation | dbSNP Number | ClinVar Submissions | Internal Classification | Number of Patients |
|---|---|---|---|---|---|
| APC | NM_000038.4:c.3927_3931del | rs121913224 | 30 (P), 1 (LP) | Pathogenic | 5 |
| APC | NM_000038.6:c.2626C>T | rs121913333 | 11 (P), 1 (LP) | Pathogenic | 5 |
| APC | NM_000038.6:c.4438C>T | - | 1 (P) | Pathogenic | 1 |
| MLH1 | NM_000249.4:c.83C>T | rs63750792 | 7 (P), 1 (LP) | Pathogenic | 1 |
| MLH1 | NM_000249.4:c.1897-2A>G | rs267607871 | 4 (LP), 5 (VUS) | Pathogenic | 1 |
| MSH6 | NM_000179:c.423del | rs1114167728 | 3 (P), 1 (LP) | Pathogenic | 3 |
| STK11 | NM_000455.5:c.891G>T | rs730881984 | 1 (P) | Pathogenic | 1 |
| BRCA1 | NM_007294.4:c.181T>G | rs28897672 | 56 (P) | Pathogenic | 1 |
| BRCA1 | NM_007294.4:c.5251C>T | rs80357123 | 36 (P) | Pathogenic | 1 |
| BRCA2 | NM_000059.4:c.3076A>T | rs80358552 | 8 (P) | Pathogenic | 1 |
| PALB2 | NM_024675.4:c.759del | rs1060499830 | 2 (P), 1 (LP) | Pathogenic | 1 |
| ATM | NM_000051.4:c.742C>T | rs730881336 | 10 (P), 1 (LP) | Pathogenic | 3 |
| NBN | NM_002485.5:c.657_661del | rs587776650 | 36 (P), 1 (LP) | Pathogenic | 1 |
| Gene | HGVS Variant | The Common Name of Mutation | Comment |
|---|---|---|---|
| BRCA1 | NM_007294.3:c.5266dup | 5382insC | Mutation included in the NCCP |
| BRCA1 | NM_007294.3:c.181T>G | C61G | Mutation included in the NCCP |
| BRCA1 | NM_007294.3:c.4035del | 4153delA | Mutation included in the NCCP |
| BRCA1 | NM_007294.3:c.68_69del | 185delAG | Mutation included in the NCCP |
| BRCA1 | NM_007294.3:c.3700_3704del | 3819del 5 | Mutation included in the NCCP |
| BRCA1 | NM_007294.3:c.3756_3759del | - | Mutation investigated additionally. |
| BRCA1 | NM_007294.3:c.1961del | - | Mutation investigated additionally. |
| BRCA2 | NM_000059.3:c.5946del | - | Mutation investigated additionally. |
| Gene | HGVS Variant | The Common Name of Mutation | Comment |
|---|---|---|---|
| CHEK2 | NM_007194.3:c.444+1G>A | IVS+1G>A | - |
| CHEK2 | NM_007194.3:c.1100del | 1100delC | - |
| CHEK2 | NM_007194.3:c.909-?_1095+?del | del 5395 | - |
| PALB2 | NM_024675.3:c.509_510del | - | - |
| PALB2 | NM_024675.3:c.172_175del | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kałużewski, T.; Kubiak, I.; Bednarek, M.; Sałamunia, J.; Kucharska, D.; Kępczyński, Ł.; Stempień, M.; Kubicki, T.; Trzciński, R.; Gordon-Sönmez, Z.; et al. Report on the Effect of the Implementation of an Early Detection and Prevention of Cancer Program on Families at High Hereditary Risk—Concentrating on Patients Undergoing Genetic Diagnostics and Counseling in Central Poland. Int. J. Mol. Sci. 2023, 24, 13178. https://doi.org/10.3390/ijms241713178
Kałużewski T, Kubiak I, Bednarek M, Sałamunia J, Kucharska D, Kępczyński Ł, Stempień M, Kubicki T, Trzciński R, Gordon-Sönmez Z, et al. Report on the Effect of the Implementation of an Early Detection and Prevention of Cancer Program on Families at High Hereditary Risk—Concentrating on Patients Undergoing Genetic Diagnostics and Counseling in Central Poland. International Journal of Molecular Sciences. 2023; 24(17):13178. https://doi.org/10.3390/ijms241713178
Chicago/Turabian StyleKałużewski, Tadeusz, Izabela Kubiak, Michał Bednarek, Jordan Sałamunia, Dorota Kucharska, Łukasz Kępczyński, Marek Stempień, Tobiasz Kubicki, Radzisław Trzciński, Zofia Gordon-Sönmez, and et al. 2023. "Report on the Effect of the Implementation of an Early Detection and Prevention of Cancer Program on Families at High Hereditary Risk—Concentrating on Patients Undergoing Genetic Diagnostics and Counseling in Central Poland" International Journal of Molecular Sciences 24, no. 17: 13178. https://doi.org/10.3390/ijms241713178
APA StyleKałużewski, T., Kubiak, I., Bednarek, M., Sałamunia, J., Kucharska, D., Kępczyński, Ł., Stempień, M., Kubicki, T., Trzciński, R., Gordon-Sönmez, Z., Bartosińska-Dyc, A., Gach, A., & Kałużewski, B. (2023). Report on the Effect of the Implementation of an Early Detection and Prevention of Cancer Program on Families at High Hereditary Risk—Concentrating on Patients Undergoing Genetic Diagnostics and Counseling in Central Poland. International Journal of Molecular Sciences, 24(17), 13178. https://doi.org/10.3390/ijms241713178








