Bioreactor Technologies for Enhanced Organoid Culture
Abstract
1. Introduction
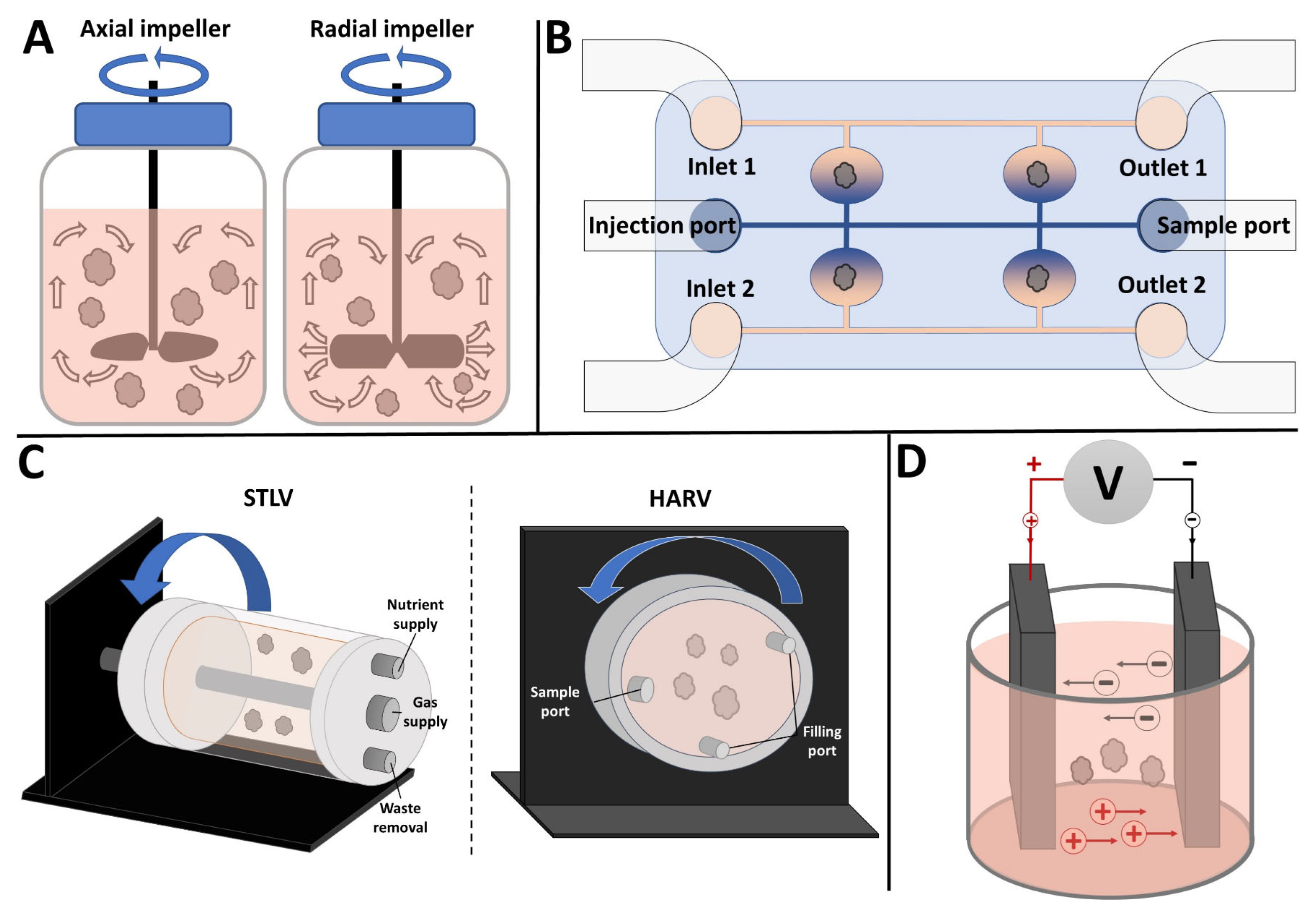
2. Stirred Bioreactors
2.1. Improving Tissue Oxygenation
2.2. Achieving Proper Lineage Progression and 3D Spatial Organization
2.3. Types and Customizability
2.4. Limitations of SBRs
| Reference | Cell Source | Goal | Bioreactor Set-Up | Outcomes |
|---|---|---|---|---|
| Lancaster et al. [25,26]. | hESCs and hiPSC. | Generation of cerebral organoids using stirred bioreactors. | 125 mL spinner flask. | Successful generation of organoids expressing markers for neurons and specific brain regions. |
| Qian et al. [21]. | hiPSCs. | Region-specific brain organoids to model exposure to ZIKA virus. | Custom miniaturized spinning bioreactor (SpinΩ). | Successful generation of organoids specific to brain regions using a bioreactor. |
| Fluri et al. [28]. | Fibroblast to hiPSCs. | Derivation and expansion of iPSCs from somatic cells. | Spinner flasks. | Culture in spinner flasks obviates need for feeder cells, serum, and solid substrate. iPSCs comparable to monolayer. |
| Zur Nieden et al. [30]. | mESCs. | Pluripotent expansion and proliferation. | 100 mL stirred flask. | hESCs in suspension maintain pluripotency and are easily scalable. |
| Ovando-Roche et al. [22]. | hiPSCs. | Preservation of retina-like architecture and enhanced generation of photoreceptors . | 100 mL stirred flask. | Improved laminar stratification and increase in photoreceptor yield. |
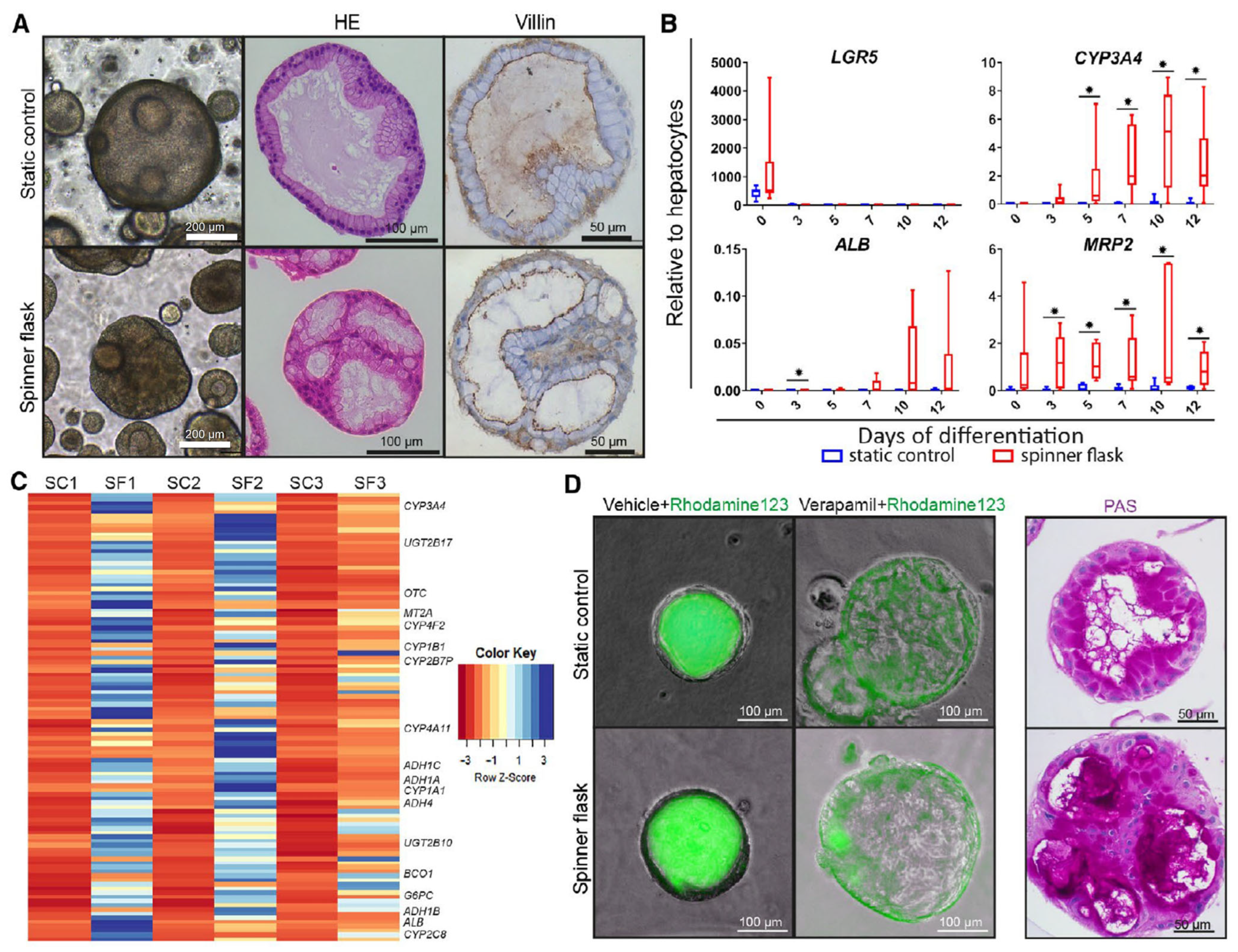
| Schneeberger et al. [31]. | Patient-derived liver biopsies. | Generation of large-scale liver organoids. | 100 mL spinner flask. | Culture in spinner flask increased the size and differentiation of the organoid. |
3. Microfluidics
3.1. Functions and Applications
3.2. Advantages of Organoid Culture
3.3. Applications in Organoid Culture
3.4. Limitations of MFBs with Respect to Organoid Cultures
| Reference | Cell Source | Goal | Bioreactor Setup | Outcomes |
|---|---|---|---|---|
| Zhang et al. [36]. | HepG2, hiPSCs. | Integration of multiple sensors for automated and continuous in situ monitoring. | Layered PDMS microbioreactor integrated with microelectrode array. | Real-time monitoring of microenvironmental parameters, soluble biomarkers, and organoid morphology. |
| Toh et al. [42]. | HepG2, MCF7, BMSCs. | Improvement of cell–cell and cell–matrix interaction via perfusion-seeding and entrapment. | Perfused PDMS substrate equipped with microchannel and micropillar array. | Preservation of 3D cytoarchitecture, differentiation competence, and cell-specific function. Direct microscopy monitoring. |
| Fu et al. [43]. | HepG2 and Balb/c 3T3 fibroblast cells. | Enhanced formation of multicellular spheroids using a microfluidic Bioreactor. | Two-port PDMS perfusion bioreactor with U-shaped hydrogel structures. | Successful trapping of cells and spheroid formation in non-adherent hydrogel structures. |
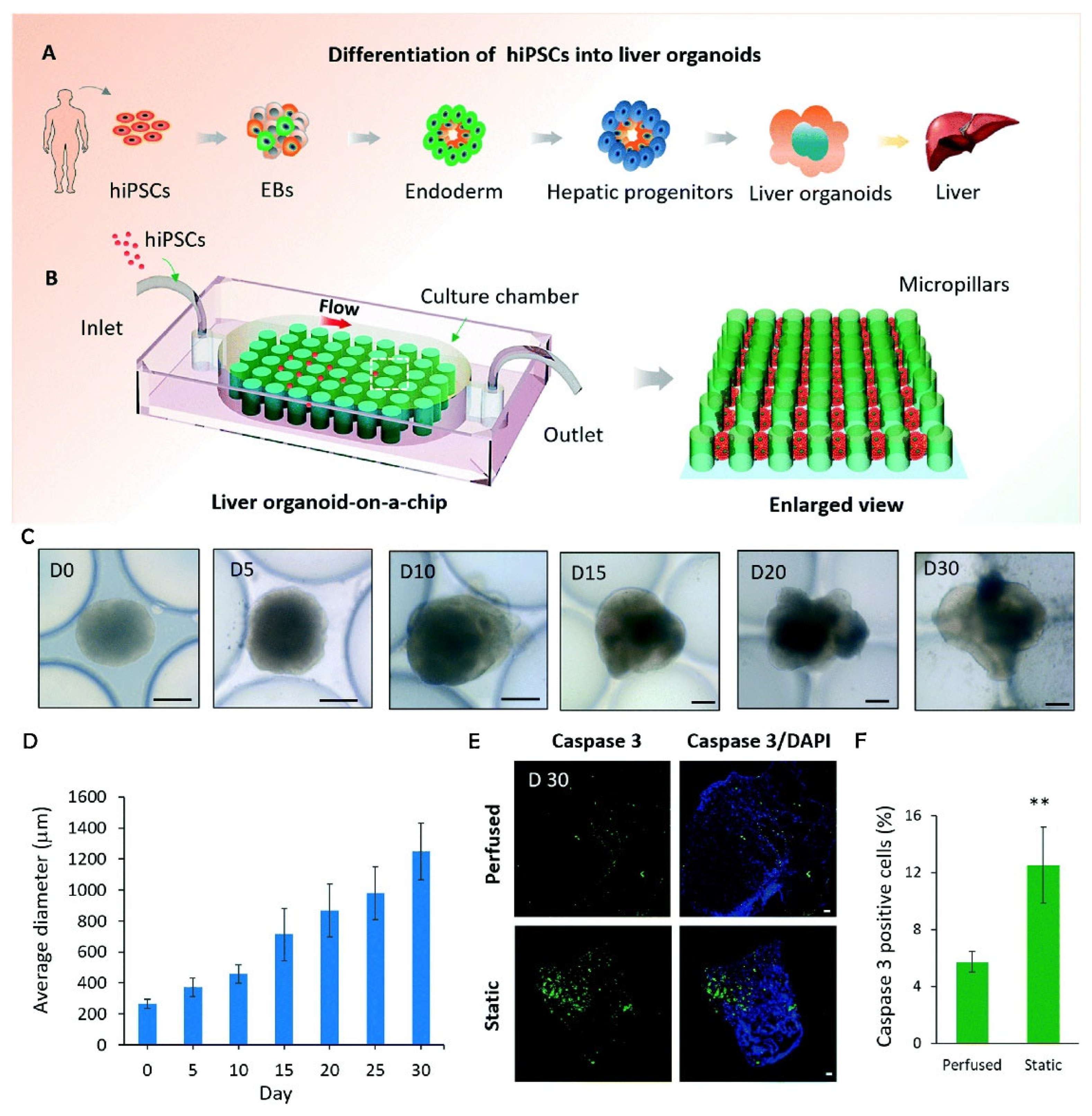
| Wang et al. [44]. | hiPSCs. | Creation of a chip-based liver model. | Perfused PDMS micropillar array. | Better tissue functionality from a perfused chip system than static culture. |
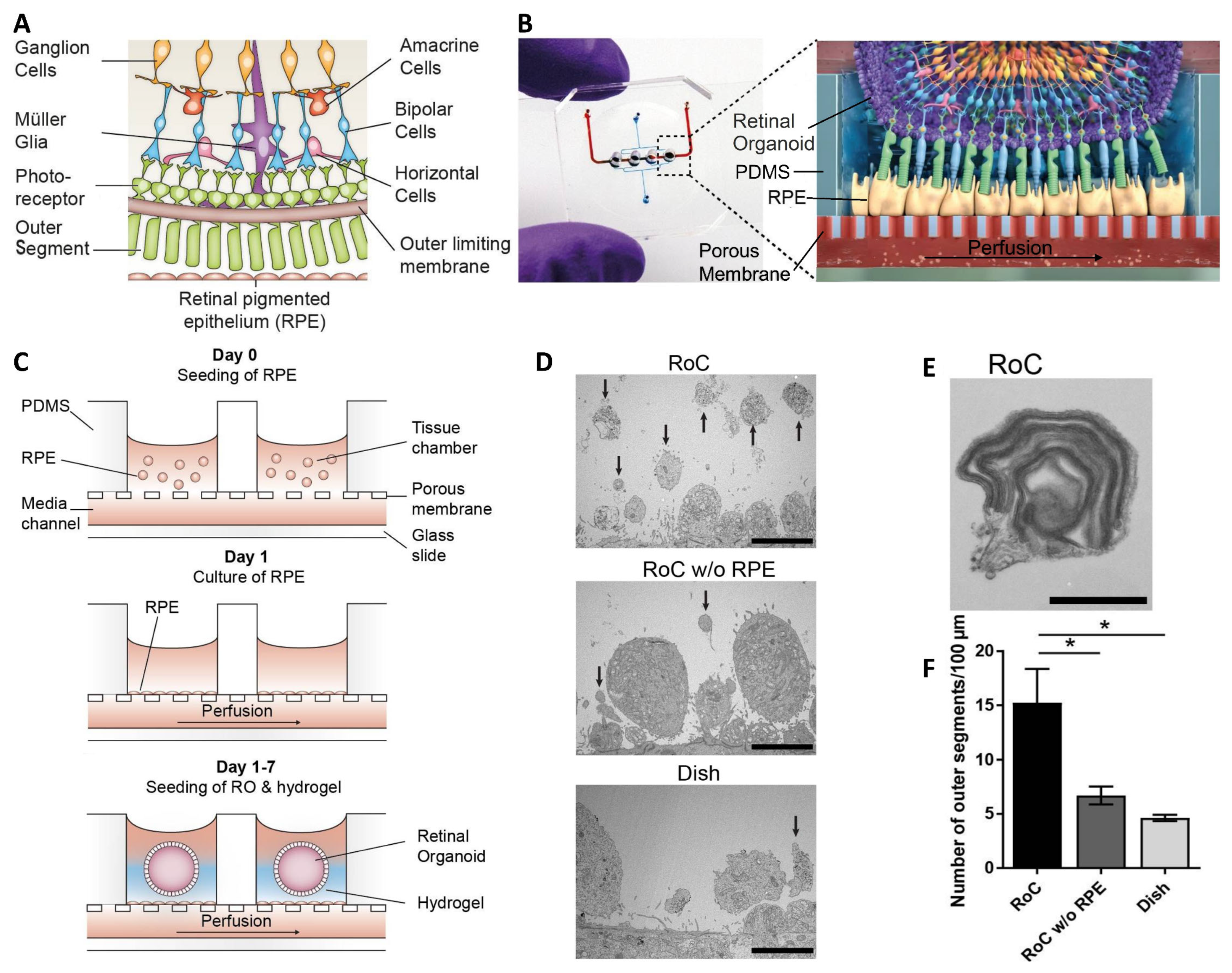
| Achberger et al. [45]. | hiPSCs. | Biomimetic model of subretinal space vascularization and cell interaction. | Hydrogel-laden PDMS chamber with PET membrane on a glass slide. | Vascular-like perfusion improved the interaction of the photoreceptor, and RPE enhanced the outer segment-like structures. |
4. Rotating Wall Vessels
4.1. Uses in Organoid Culture
4.2. Simulation of Microgravity
4.3. Limitations
| Reference | Cell Source | Goal | Bioreactor Setup | Outcomes |
|---|---|---|---|---|
| Rogers et al. [63]. | Human mesenchymal stem cells. | Embryoid Body cell expansion. | 10 mL HARV from Synthecon, GelMA microcarriers. | Cells maintain pluripotency and differentiation capacity as well as or better than monolayer culture. |
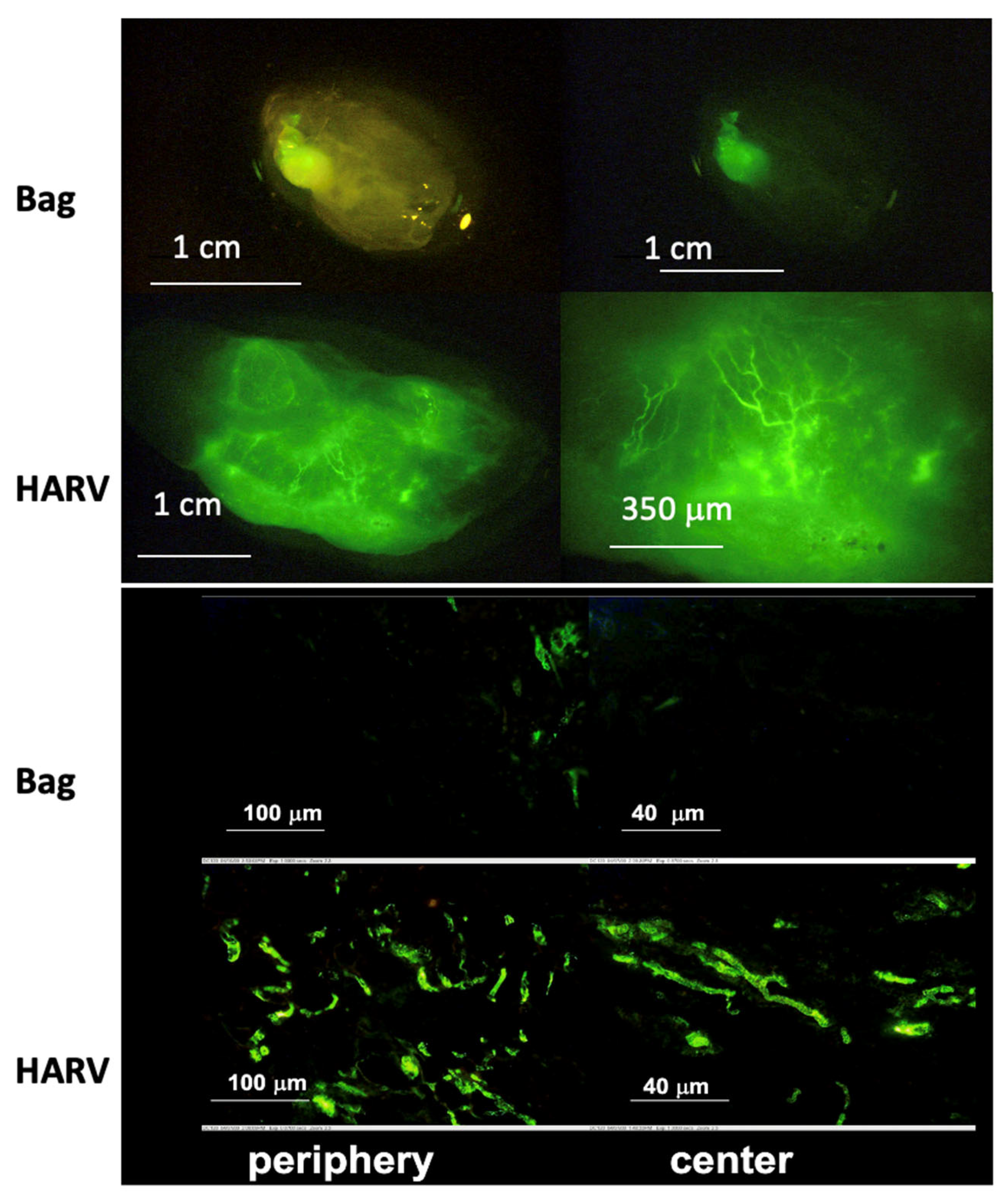
| Botta et al. [61]. | PC12 cells. | Spheroid formation. | 50 mL HARV from Synthecon, microcarrier beads. | Real-time imaging shows an increase in aggregate size over time. |
| Wilkinson et al. [64]. | iPSC-derived lung fibroblasts. | Lung organoid generation. | 4 mL HARV from Synthecon, alginate beads. | Lung organoids generated in a scalable manner appropriate for high-throughput screening. |
| DiStefano et al. [65]. | mESCs. | Improved retinal organoid generation. | 50 mL HARV from Synthecon. | Faster growth and maturation in RWV compared to static, as shown by fluorescence imaging and RNA-seq. |
| Takahashi et al. [74]. | Human iPSCs. | Intestinal organoid. | JTEC CellPet 3-D Ips [75]. | More efficient, robust, and scalable organoid generation. |
| Papadaki et al. [76]. | Rat primary cardiomyocytes. | Cardiac Organoid. | 100 mL HARV from Synthecon. | HARV-cultured organoids have improved contraction and cardiac gene expression. |
| Lelkes et al. [67]. | PC12 cells. | Neuroendocrine organoid culture. | 35 mL HARV from Synthecon. | Increase in neuroendocrine expression over neuronal expression and increased vascularization with RWV culture. |
| Phelan et al. [68]. | A549 human lung adenocarcinoma cells. | Cancer organoid. | Custom 10 mL air bubble-isolating RWV. | RWV without bubbles led to significant increases in organoid size. |
5. Electrical Stimulation Bioreactors
5.1. Organoid Electrical Stimulation
5.2. Commercial vs. Custom-Made Systems
| Reference | Cell Source | Goal | Bioreactor Setup | Outcomes |
|---|---|---|---|---|
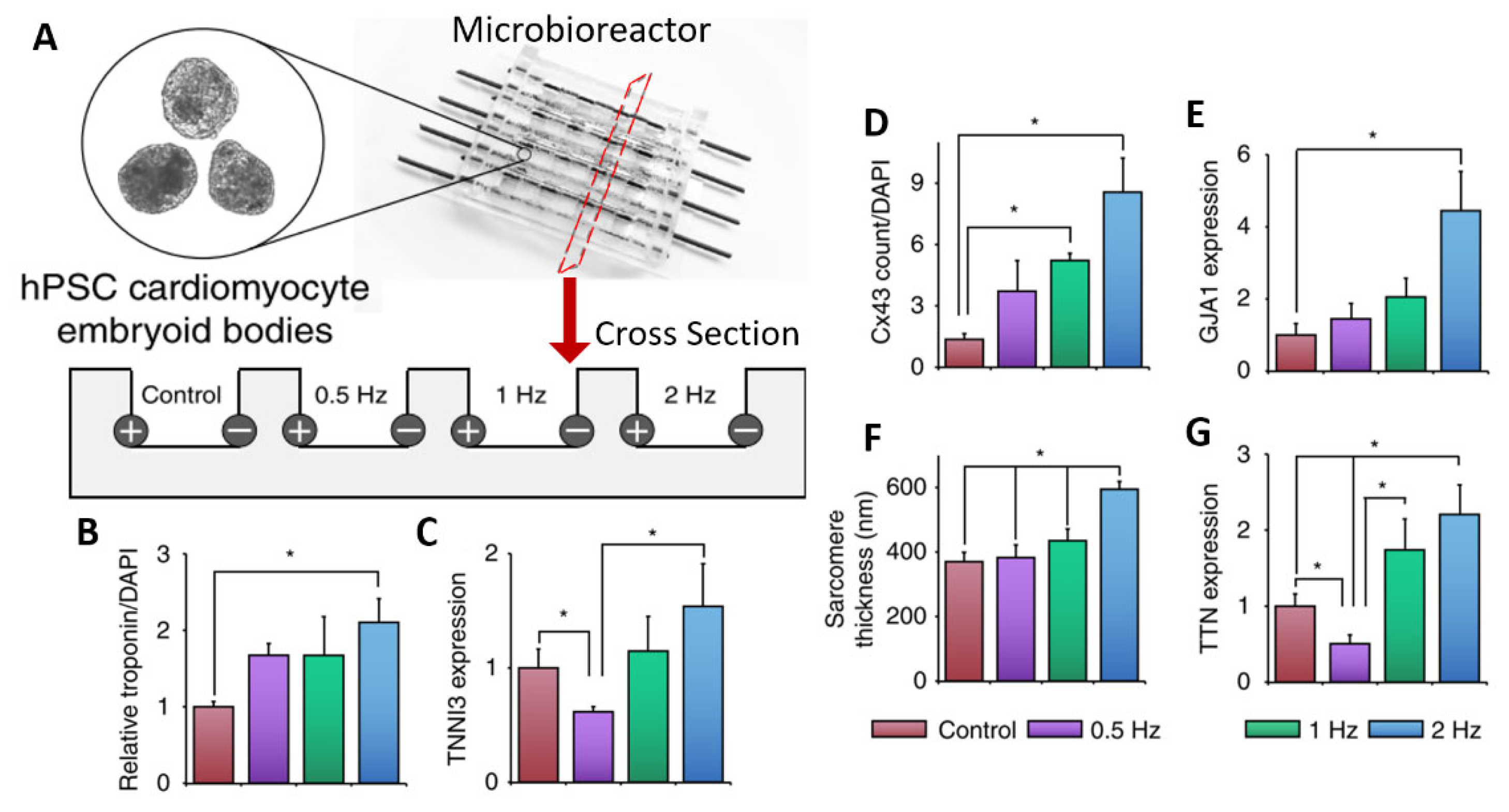
| Eng et al. [85]. | hESCs and hiPSC. | Cardiac organoid differentiation/maturation. | Custom bioreactor with carbon rods in PDMS microchambers. | An increase in frequency leads to increased cardiac gene expression and better beating adaptation. |
| Yoshida et al. [86]. | hiPSC-CMs. | Cardiac organoid maturation. | Organic carbon electrodes in poly(vinyl) alcohol hydrogel chambers. | ES leads to enhanced cardiac troponin expression and sarcomere formation. |
| Ma et al. [88]. | hiPSCs with GCaMP reporter. | Early induction of cardiomyocyte differentiation. | IonOptix C-Pace system. | Organoid contractions were seen after 2 days with ES compared to 7 days without ES. |
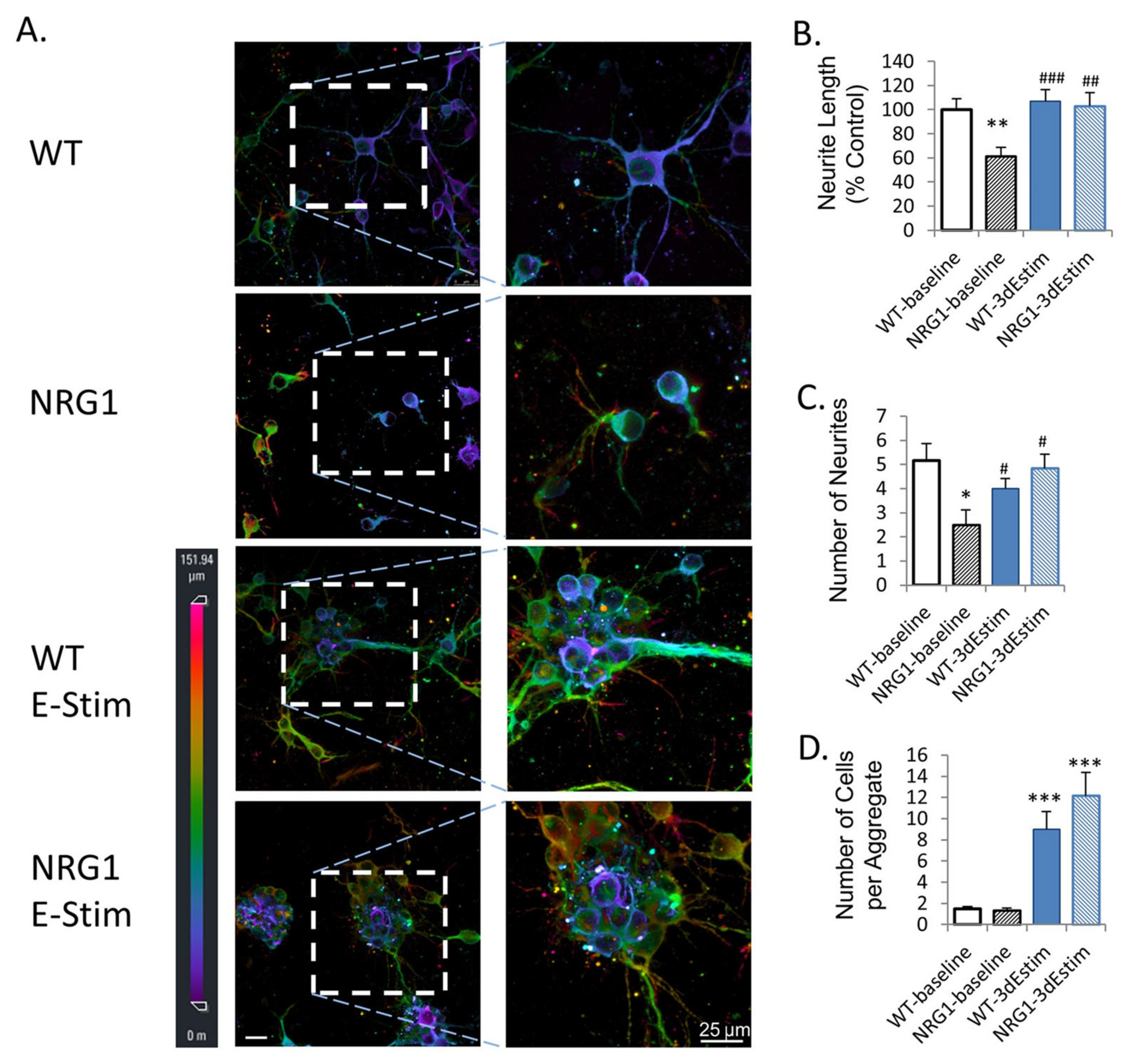
| Zhang et al. [89]. | Primary mouse cortical neurons. | Enhanced Neural Organoid. | 3D interdigitated electrodes coated with polypyrrole. | ES of organoids leads to enhanced neurite outgrowth and synaptogenesis and rescues the phenotype of NRG1-KO neurons. |
| Sefton et al. [90]. | Primary neural precursor cells. | Formation and differentiation of neural organoid. | Agarose salt bridge stimulation. | ES increases survival and spheroid size. It also increases neuronal expression while reducing astrocyte expression. |
| Ahadian et al. [92]. | mESCs. | Cardiac organoid differentiation. | IonOptix C-Pace system. | ES enhanced cardiac gene expression and the beating area of organoids. |
6. Discussion/Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an In Vitro Model of Human Development and Disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef]
- Kim, M.B.; Hwangbo, S.; Jang, S.; Jo, Y.K. Bioengineered Co-Culture of Organoids to Recapitulate Host-Microbe Interactions. Mater. Today BIO 2022, 16, 100345. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Corrò, C.; Novellasdemunt, L.; Li, V.S.W. A Brief History of Organoids. Am. J. Physiol. Cell Physiol. 2020, 319, C151–C165. [Google Scholar] [CrossRef] [PubMed]
- Bacakova, L.; Zarubova, J.; Travnickova, M.; Musilkova, J.; Pajorova, J.; Slepicka, P.; Kasalkova, N.S.; Svorcik, V.; Kolska, Z.; Motarjemi, H.; et al. Stem Cells: Their Source, Potency and Use in Regenerative Therapies with Focus on Adipose-Derived Stem Cells—A Review. Biotechnol. Adv. 2018, 36, 1111–1126. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, G. Embryoid Body Formation from Human Pluripotent Stem Cells in Chemically Defined E8 Media. In StemBook; Harvard Stem Cell Institute: Cambridge, MA, USA, 2008. [Google Scholar] [CrossRef]
- Moradi, S.; Mahdizadeh, H.; Šarić, T.; Kim, J.; Harati, J.; Shahsavarani, H.; Greber, B.; Moore, J.B. Research and Therapy with Induced Pluripotent Stem Cells (IPSCs): Social, Legal, and Ethical Considerations. Stem Cell Res. Ther. 2019, 10, 341. [Google Scholar] [CrossRef]
- Oldak, B.; Wildschutz, E.; Bondarenko, V.; Aguilera-Castrejon, A.; Zhao, C.; Tarazi, S.; Comar, M.-Y.; Ashouokhi, S.; Lokshtanov, D.; Roncato, F.; et al. Transgene-Free Ex Utero Derivation of a Human Post-Implantation Embryo Model Solely from Genetically Unmodified Naïve PSCs. bioRxiv 2023. [Google Scholar] [CrossRef]
- Partridge, E.A.; Davey, M.G.; Hornick, M.A.; McGovern, P.E.; Mejaddam, A.Y.; Vrecenak, J.D.; Mesas-Burgos, C.; Olive, A.; Caskey, R.C.; Weiland, T.R.; et al. An Extra-Uterine System to Physiologically Support the Extreme Premature Lamb. Nat. Commun. 2017, 8, 15112. [Google Scholar] [CrossRef]
- Erickson, L.E. Bioreactors for Commodity Products. In Comprehensive Biotechnology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 683–689. [Google Scholar] [CrossRef]
- Vandermies, M.; Fickers, P. Bioreactor-Scale Strategies for the Production of Recombinant Protein in the Yeast Yarrowia Lipolytica. Microorganisms 2019, 7, 40. [Google Scholar] [CrossRef]
- Wang, D.; Liu, W.; Han, B.; Xu, R. The Bioreactor: A Powerful Tool for Large-Scale Culture of Animal Cells. Curr. Pharm. Biotechnol. 2005, 6, 397–403. [Google Scholar] [CrossRef]
- Grayson, W.; Stephenson, M. Recent Advances in Bioreactors for Cell-Based Therapies. F1000Research 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Borys, B.S.; Dang, T.; So, T.; Rohani, L.; Revay, T.; Walsh, T.; Thompson, M.; Argiropoulos, B.; Rancourt, D.E.; Jung, S.; et al. Overcoming Bioprocess Bottlenecks in the Large-Scale Expansion of High-Quality HiPSC Aggregates in Vertical-Wheel Stirred Suspension Bioreactors. Stem Cell Res. Ther. 2021, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Bellani, C.F.; Ajeian, J.; Duffy, L.; Miotto, M.; Groenewegen, L.; Connon, C.J. Scale-Up Technologies for the Manufacture of Adherent Cells. Front. Nutr. 2020, 7, 178. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning from 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Hansmann, J.; Groeber, F.; Kahlig, A.; Kleinhans, C.; Walles, H. Bioreactors in Tissue Engineering—Principles, Applications and Commercial Constraints. Biotechnol. J. 2013, 8, 298–307. [Google Scholar] [CrossRef]
- Sucosky, P.; Osorio, D.F.; Brown, J.B.; Neitzel, G.P. Fluid Mechanics of a Spinner-Flask Bioreactor. Biotechnol. Bioeng. 2004, 85, 34–46. [Google Scholar] [CrossRef]
- Rouwkema, J.; FJM Koopman, B.; van Blitterswijk, C.A.; Dhert, W.J.; Malda, J. Supply of Nutrients to Cells in Engineered Tissues. Biotechnol. Genet. Eng. Rev. 2009, 26, 163–178. [Google Scholar] [CrossRef]
- Goto-Silva, L.; Ayad, N.M.E.; Herzog, I.L.; Silva, N.P.; Lamien, B.; Orlande, H.R.B.; da Costa Souza, A.; Ribeiro, S.; Martins, M.; Domont, G.B.; et al. Computational Fluid Dynamic Analysis of Physical Forces Playing a Role in Brain Organoid Cultures in Two Different Multiplex Platforms. BMC Dev. Biol. 2019, 19, 3. [Google Scholar] [CrossRef]
- Qian, X.; Jacob, F.; Song, M.M.; Nguyen, H.N.; Song, H.; Ming, G.L. Generation of Human Brain Region–Specific Organoids Using a Miniaturized Spinning Bioreactor. Nat. Protoc. 2018, 13, 565–580. [Google Scholar] [CrossRef]
- Ovando-Roche, P.; West, E.L.; Branch, M.J.; Sampson, R.D.; Fernando, M.; Munro, P.; Georgiadis, A.; Rizzi, M.; Kloc, M.; Naeem, A.; et al. Use of Bioreactors for Culturing Human Retinal Organoids Improves Photoreceptor Yields. Stem Cell Res. Ther. 2018, 9, 156. [Google Scholar] [CrossRef]
- McMurtrey, R.J. Analytic Models of Oxygen and Nutrient Diffusion, Metabolism Dynamics, and Architecture Optimization in Three-Dimensional Tissue Constructs with Applications and Insights in Cerebral Organoids. Tissue Eng. Part. C Methods 2016, 22, 221–249. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, Q.; Cai, H.; Tan, W.S. A Comparative Gene-Expression Analysis of CD34+ Hematopoietic Stem and Progenitor Cells Grown in Static and Stirred Culture Systems. Cell. Mol. Biol. Lett. 2006, 11, 475. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Generation of Cerebral Organoids from Human Pluripotent Stem Cells. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-Bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Fluri, D.A.; Tonge, P.D.; Song, H.; Baptista, R.P.; Shakiba, N.; Shukla, S.; Clarke, G.; Nagy, A.; Zandstra, P.W. Derivation, Expansion and Differentiation of Induced Pluripotent Stem Cells in Continuous Suspension Cultures. Nat. Methods 2012, 9, 509–516. [Google Scholar] [CrossRef]
- Chisti, Y. Bioreactor Design. In Basic Biotechnology, 3rd ed; Cambridge University Press: Cambridge, UK, 2006; pp. 181–200. [Google Scholar] [CrossRef]
- zur Nieden, N.I.; Cormier, J.T.; Rancourt, D.E.; Kallos, M.S. Embryonic Stem Cells Remain Highly Pluripotent Following Long Term Expansion as Aggregates in Suspension Bioreactors. J. Biotechnol. 2007, 129, 421–432. [Google Scholar] [CrossRef]
- Schneeberger, K.; Sánchez-Romero, N.; Ye, S.; van Steenbeek, F.G.; Oosterhoff, L.A.; Pla Palacin, I.; Chen, C.; van Wolferen, M.E.; van Tienderen, G.; Lieshout, R.; et al. Large-Scale Production of LGR5-Positive Bipotential Human Liver Stem Cells. Hepatology 2020, 72, 257. [Google Scholar] [CrossRef]
- Romero-Morales, A.I.; O’Grady, B.J.; Balotin, K.M.; Bellan, L.M.; Lippmann, E.S.; Gama, V. Spin∞: An Updated Miniaturized Spinning Bioreactor Design for the Generation of Human Cerebral Organoids from Pluripotent Stem Cells. HardwareX 2019, 6, e00084. [Google Scholar] [CrossRef]
- van den Berg, A.; Craighead, H.; Yang, P.; Young, E.W.K.; Beebe, D.J. Fundamentals of Microfluidic Cell Culture in Controlled Microenvironments. Chem. Soc. Rev. 2010, 39, 1036–1048. [Google Scholar] [CrossRef]
- Stone, H.A.; Stroock, A.D.; Ajdari, A. Engineering Flows in Small Devices: Microfluidics toward a Lab-on-a-Chip. Annu. Rev. Fluid. Mech. 2004, 36, 381–411. [Google Scholar] [CrossRef]
- Dungchai, W.; Chailapakul, O.; Henry, C.S. Electrochemical Detection for Paper-Based Microfluidics. Anal. Chem. 2009, 81, 5821–5826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Shaegh, S.A.M.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-Integrated Organs-on-Chips Platform for Automated and Continual in Situ Monitoring of Organoid Behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, P.S.; Craighead, H.G. Micro- and Nanomechanical Sensors for Environmental, Chemical, and Biological Detection. Lab Chip 2007, 7, 1238–1255. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic Organs-on-Chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human Gut-on-a-Chip Inhabited by Microbial Flora That Experiences Intestinal Peristalsis-like Motions and Flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef]
- Rodrigues, T.; Kundu, B.; Silva-Correia, J.; Kundu, S.C.; Oliveira, J.M.; Reis, R.L.; Correlo, V.M. Emerging Tumor Spheroids Technologies for 3D in Vitro Cancer Modeling. Pharmacol. Ther. 2018, 184, 201–211. [Google Scholar] [CrossRef]
- An, S.; Han, S.Y.; Cho, S.W. Hydrogel-Integrated Microfluidic Systems for Advanced Stem Cell Engineering. BioChip J. 2019, 13, 306–322. [Google Scholar] [CrossRef]
- Toh, Y.C.; Zhang, C.; Zhang, J.; Khong, Y.M.; Chang, S.; Samper, V.D.; van Noort, D.; Hutmacher, D.W.; Yu, H. A Novel 3D Mammalian Cell Perfusion-Culture System in Microfluidic Channels. Lab Chip 2007, 7, 302–309. [Google Scholar] [CrossRef]
- Fu, C.Y.; Tseng, S.Y.; Yang, S.M.; Hsu, L.; Liu, C.H.; Chang, H.Y. A Microfluidic Chip with a U-Shaped Microstructure Array for Multicellular Spheroid Formation, Culturing and Analysis. Biofabrication 2014, 6, 015009. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Deng, P.; Chen, W.; Guo, Y.; Tao, T.; Qin, J. In Situ Differentiation and Generation of Functional Liver Organoids from Human IPSCs in a 3D Perfusable Chip System. Lab Chip 2018, 18, 3606–3616. [Google Scholar] [CrossRef] [PubMed]
- Achberger, K.; Probst, C.; Haderspeck, J.C.; Bolz, S.; Rogal, J.; Chuchuy, J.; Nikolova, M.; Cora, V.; Antkowiak, L.; Haq, W.; et al. Merging Organoid and Organ-on-a-Chip Technology to Generate Complex Multi-Layer Tissue Models in a Human Retina-on-a-Chip Platform. eLife 2019, 8, e46188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Radisic, M. Organ-on-a-Chip Devices Advance to Market. Lab Chip 2017, 17, 2395–2420. [Google Scholar] [CrossRef]
- Xia, Y.; Whitesides, G.M. Soft Lithography. Annu. Rev. Mater. Sci. 1998, 28, 153–184. [Google Scholar] [CrossRef]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M.T. Advantages and Challenges of Microfluidic Cell Culture in Polydimethylsiloxane Devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Valadez, A.V.; Zuo, P.; Nie, Z. Microfluidic 3D Cell Culture: Potential Application for Tissue-Based Bioassays. Bioanalysis 2012, 4, 1509–1525. [Google Scholar] [CrossRef] [PubMed]
- Toepke, M.W.; Beebe, D.J. PDMS Absorption of Small Molecules and Consequences in Microfluidic Applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef]
- Whitesides, G.M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Kiss, J.Z.; Wolverton, C.; Wyatt, S.E.; Hasenstein, K.H.; van Loon, J.J.W.A. Comparison of Microgravity Analogs to Spaceflight in Studies of Plant Growth and Development. Front. Plant Sci. 2019, 10, 1577. [Google Scholar] [CrossRef]
- Schwarz, R.P.; Wolf, D.A. Rotating Bio-Reactor Cell Culture Apparatus. US4988623A, 29 January 1991. Google Patents. Available online: https://patents.google.com/patent/US4988623A/en (accessed on 10 January 2023).
- Hammond, T.G.; Hammond, J.M. Optimized Suspension Culture: The Rotating-Wall Vessel. Am. J. Physiol. Ren. Physiol. 2001, 281, F12–F25. [Google Scholar] [CrossRef]
- Radtke, A.L.; Herbst-Kralovetz, M.M. Culturing and Applications of Rotating Wall Vessel Bioreactor Derived 3D Epithelial Cell Models. J. Vis. Exp. 2012, 3, 3868. [Google Scholar] [CrossRef]
- Goodwin, T.J.; Prewett, T.L.; Wolf, D.A.; Spaulding, G.F. Reduced Shear Stress: A Major Component in the Ability of Mammalian Tissues to Form Three-Dimensional Assemblies in Simulated Microgravity. J. Cell Biochem. 1993, 51, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Navran, S. The Application of Low Shear Modeled Microgravity to 3-D Cell Biology and Tissue Engineering. Biotechnol. Annu. Rev. 2008, 14, 275–296. [Google Scholar] [CrossRef] [PubMed]
- Navran, S. Rotary Bioreactor for Recombinant Protein Production. In Cell Technology for Cell Products; Springer: Berlin/Heidelberg, Germany, 2007; Volume 567–569. [Google Scholar] [CrossRef]
- Cinbiz, M.N.; Tiǧli, R.S.; Beşkardeş, I.G.; Gümüşderelioglu, M.; Çolak, Ü. Computational Fluid Dynamics Modeling of Momentum Transport in Rotating Wall Perfused Bioreactor for Cartilage Tissue Engineering. J. Biotechnol. 2010, 150, 389–395. [Google Scholar] [CrossRef]
- Yin, X.; Mead, B.E.; Safaee, H.; Langer, R.; Karp, J.M.; Levy, O. Stem Cell Organoid Engineering. Cell Stem Cell 2016, 18, 25. [Google Scholar] [CrossRef]
- Botta, G.P.; Manley, P.; Miller, S.; Lelkes, P.I. Real-Time Assessment of Three-Dimensional Cell Aggregation in Rotating Wall Vessel Bioreactors In Vitro. Nat. Protoc. 2006, 1, 2116–2128. [Google Scholar] [CrossRef]
- Harrison, R.P.; Medcalf, N.; Rafiq, Q.A. Cell Therapy-Processing Economics: Small-Scale Microfactories as a Stepping Stone toward Large-Scale Macrofactories. Regen. Med. 2018, 13, 159–173. [Google Scholar] [CrossRef]
- Rogers, R.E.; Haskell, A.; White, B.P.; Dalal, S.; Lopez, M.; Tahan, D.; Pan, S.; Kaur, G.; Kim, H.; Barreda, H.; et al. A Scalable System for Generation of Mesenchymal Stem Cells Derived from Induced Pluripotent Cells Employing Bioreactors and Degradable Microcarriers. Stem Cells Transl. Med. 2021, 10, 1650–1665. [Google Scholar] [CrossRef]
- Wilkinson, D.C.; Alva-Ornelas, J.A.; Sucre, J.M.S.; Vijayaraj, P.; Durra, A.; Richardson, W.; Jonas, S.J.; Paul, M.K.; Karumbayaram, S.; Dunn, B.; et al. Development of a Three-Dimensional Bioengineering Technology to Generate Lung Tissue for Personalized Disease Modeling. Stem Cells Transl. Med. 2017, 6, 622. [Google Scholar] [CrossRef]
- DiStefano, T.; Chen, H.Y.; Panebianco, C.; Kaya, K.D.; Brooks, M.J.; Gieser, L.; Morgan, N.Y.; Pohida, T.; Swaroop, A. Accelerated and Improved Differentiation of Retinal Organoids from Pluripotent Stem Cells in Rotating-Wall Vessel Bioreactors. Stem Cell Rep. 2018, 10, 300–313. [Google Scholar] [CrossRef]
- Lelkes, P.I.; Galvan, D.L.; Thomas Hayman, G.; Goodwin, T.J.; Chatman, D.Y.; Cherian, S.; Garcia, R.M.G.; Unsworth, B.R. Simulated Microgravity Conditions Enhance Differentiation of Cultured PC12 Cells towards the Neuroendocrine Phenotype. Cell Dev. Biol. Anim. 1998, 34, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Lelkes, P.I.; Akhtar, N.; Lelkes, E.; Maltz, L.; Arthur, R.; Wiederholt, J.; Unsworth, B.R. Neuroendocrine Tissue Engineering in Rotating Wall Vessel Bioreactors under Simulated Microgravity Conditions. In Proceedings of the 2001 Conference Proceedings of the 23rd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Istanbul, Turkey, 25–28 October 2001; Volume 3, pp. 2987–2990. [CrossRef]
- Phelan, M.A.; Gianforcaro, A.L.; Gerstenhaber, J.A.; Lelkes, P.I. An Air Bubble-Isolating Rotating Wall Vessel Bioreactor for Improved Spheroid/Organoid Formation. Tissue Eng. Part. C Methods 2019, 25, 479. [Google Scholar] [CrossRef]
- Phelan, M.; Lelkes, P.; Swaroop, A. Customized Rotating Wall Vessel Bioreactors Produce Improved Retinal Organoids with Reduced Operational Costs and Less Frequent Experimental Failure. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3316. [Google Scholar]
- Wnorowski, A.; Sharma, A.; Chen, H.; Wu, H.; Shao, N.Y.; Sayed, N.; Liu, C.; Countryman, S.; Stodieck, L.S.; Rubins, K.H.; et al. Effects of Spaceflight on Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Structure and Function. Stem Cell Rep. 2019, 13, 960. [Google Scholar] [CrossRef] [PubMed]
- Mayer-Wagner, S.; Hammerschmid, F.; Redeker, J.I.; Schmitt, B.; Holzapfel, B.M.; Jansson, V.; Betz, O.B.; Müller, P.E. Simulated Microgravity Affects Chondrogenesis and Hypertrophy of Human Mesenchymal Stem Cells. Int. Orthop. 2014, 38, 2615–2621. [Google Scholar] [CrossRef]
- Gershovich, P.M.; Gershovich, J.G.; Zhambalova, A.P.; Romanov, Y.A.; Buravkova, L.B. Cytoskeletal Proteins and Stem Cell Markers Gene Expression in Human Bone Marrow Mesenchymal Stromal Cells after Different Periods of Simulated Microgravity. Acta Astronaut. 2012, 70, 36–42. [Google Scholar] [CrossRef]
- Juhl, O.J.; Buettmann, E.G.; Friedman, M.A.; DeNapoli, R.C.; Hoppock, G.A.; Donahue, H.J. Update on the Effects of Microgravity on the Musculoskeletal System. npj Microgravity 2021, 7, 28. [Google Scholar] [CrossRef]
- Takahashi, J.; Mizutani, T.; Sugihara, H.Y.; Nagata, S.; Kato, S.; Hiraguri, Y.; Takeoka, S.; Tsuchiya, M.; Kuno, R.; Kakinuma, S.; et al. Suspension Culture in a Rotating Bioreactor for Efficient Generation of Human Intestinal Organoids. Cell Rep. Methods 2022, 2, 100337. [Google Scholar] [CrossRef]
- “CELLFLOAT” Products—J-Tec Corporation. Available online: https://www.j-tec.co.jp/english/life-science/cellfloat/ (accessed on 19 March 2023).
- Papadaki, M.; Bursac, N.; Langer, R.; Merok, J.; Vunjak-Novakovic, G.; Freed, L.E. Tissue Engineering of Functional Cardiac Muscle: Molecular, Structural, and Electrophysiological Studies. Am. J. Physiol. Heart Circ. Physiol. 2001, 280, H168–H178. [Google Scholar] [CrossRef]
- Chen, C.; Bai, X.; Ding, Y.; Lee, I.S. Electrical Stimulation as a Novel Tool for Regulating Cell Behavior in Tissue Engineering. Biomater. Res. 2019, 23, 25. [Google Scholar] [CrossRef]
- Basser, P.J.; Roth, B.J. New Currents in Electrical Stimulation of Excitable Tissues1. Annu. Rev. Biomed. Eng. 2003, 2, 377–397. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Pezzulo, G.; Finkelstein, J.M. Endogenous Bioelectric Signaling Networks: Exploiting Voltage Gradients for Control of Growth and Form. Annu. Rev. Biomed. Eng. 2017, 19, 353–387. [Google Scholar] [CrossRef] [PubMed]
- Casella, A.; Panitch, A.; Leach, J.K. Endogenous Electric Signaling as a Blueprint for Conductive Materials in Tissue Engineering. Bioelectricity 2020, 3, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Pascoal-Faria, P.; Castelo Ferreira, P.; Datta, A.; Amado, S.; Moura, C.; Alves, N. Electrical Stimulation Optimization in Bioreactors for Tissue Engineering Applications. Appl. Mech. Mater. 2019, 890, 314–323. [Google Scholar] [CrossRef]
- Sekirnjak, C.; Hottowy, P.; Sher, A.; Dabrowski, W.; Litke, A.M.; Chichilnisky, E.J. Electrical Stimulation of Mammalian Retinal Ganglion Cells with Multielectrode Arrays. J. Neurophysiol. 2006, 95, 3311–3327. [Google Scholar] [CrossRef]
- Huang, Q.; Tang, B.; Romero, J.C.; Yang, Y.; Elsayed, S.K.; Pahapale, G.; Lee, T.J.; Pantoja, I.E.M.; Han, F.; Berlinicke, C.; et al. Shell Microelectrode Arrays (MEAs) for Brain Organoids. Sci. Adv. 2022, 8, 5031. [Google Scholar] [CrossRef]
- Karbassi, E.; Fenix, A.; Marchiano, S.; Muraoka, N.; Nakamura, K.; Yang, X.; Murry, C.E. Cardiomyocyte Maturation: Advances in Knowledge and Implications for Regenerative Medicine. Nat. Rev. Cardiol. 2020, 17, 341–359. [Google Scholar] [CrossRef]
- Eng, G.; Lee, B.W.; Protas, L.; Gagliardi, M.; Brown, K.; Kass, R.S.; Keller, G.; Robinson, R.B.; Vunjak-Novakovic, G. Autonomous Beating Rate Adaptation in Human Stem Cell-Derived Cardiomyocytes. Nat. Commun. 2016, 7, 10312. [Google Scholar] [CrossRef]
- Yoshida, S.; Sumomozawa, K.; Nagamine, K.; Nishizawa, M. Hydrogel Microchambers Integrated with Organic Electrodes for Efficient Electrical Stimulation of Human IPSC-Derived Cardiomyocytes. Macromol. Biosci. 2019, 19, 1900060. [Google Scholar] [CrossRef]
- Culture Pacing System—IonOptix. Available online: https://www.ionoptix.com/products/systems/cell-culture-pacing/ (accessed on 31 January 2023).
- Ma, R.; Liang, J.; Huang, W.; Guo, L.; Cai, W.; Wang, L.; Paul, C.; Yang, H.T.; Kim, H.W.; Wang, Y. Electrical Stimulation Enhances Cardiac Differentiation of Human Induced Pluripotent Stem Cells for Myocardial Infarction Therapy. Antioxid. Redox Signal 2018, 28, 371. [Google Scholar] [CrossRef]
- Zhang, Q.; Beirne, S.; Shu, K.; Esrafilzadeh, D.; Huang, X.F.; Wallace, G.G. Electrical Stimulation with a Conductive Polymer Promotes Neurite Outgrowth and Synaptogenesis in Primary Cortical Neurons in 3D. Sci. Rep. 2018, 8, 9855. [Google Scholar] [CrossRef] [PubMed]
- Sefton, E.; Iwasa, S.N.; Morrison, T.; Naguib, H.E.; Popovic, M.R.; Morshead, C.M. Electric Field Application In Vivo Regulates Neural Precursor Cell Behavior in the Adult Mammalian Forebrain. eNeuro 2020, 7, 1–12. [Google Scholar] [CrossRef]
- LaBarge, W.; Mattappally, S.; Kannappan, R.; Fast, V.G.; Pretorius, D.; Berry, J.L.; Zhang, J. Maturation of Three-Dimensional, HiPSC-Derived Cardiomyocyte Spheroids Utilizing Cyclic, Uniaxial Stretch and Electrical Stimulation. PLoS ONE 2019, 14, e0219442. [Google Scholar] [CrossRef]
- Ahadian, S.; Yamada, S.; Estili, M.; Liang, X.; Banan Sadeghian, R.; Nakajima, K.; Shiku, H.; Matsue, T.; Khademhosseini, A. Carbon Nanotubes Embedded in Embryoid Bodies Direct Cardiac Differentiation. Biomed. Microdevices 2017, 19, 57. [Google Scholar] [CrossRef] [PubMed]
- Ahadian, S.; Zhou, Y.; Yamada, S.; Estili, M.; Liang, X.; Nakajima, K.; Shiku, H.; Matsue, T. Graphene Induces Spontaneous Cardiac Differentiation in Embryoid Bodies. Nanoscale 2016, 8, 7075–7084. [Google Scholar] [CrossRef] [PubMed]
- Tandon, N.; Marsano, A.; Maidhof, R.; Wan, L.; Park, H.; Vunjak-Novakovic, G. Optimization of Electrical Stimulation Parameters for Cardiac Tissue Engineering. J. Tissue Eng. Regen. Med. 2011, 5, e115–e125. [Google Scholar] [CrossRef]
- Barash, Y.; Dvir, T.; Tandeitnik, P.; Ruvinov, E.; Guterman, H.; Cohen, S. Electric Field Stimulation Integrated into Perfusion Bioreactor for Cardiac Tissue Engineering. Tissue Eng. Part. C Methods 2010, 16, 1417–1426. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.J.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced Maturation of Human Cardiac Tissue Grown from Pluripotent Stem Cells. Nature 2018, 556, 239. [Google Scholar] [CrossRef]
- Visone, R.; Talò, G.; Lopa, S.; Rasponi, M.; Moretti, M. Enhancing All-in-One Bioreactors by Combining Interstitial Perfusion, Electrical Stimulation, on-Line Monitoring and Testing within a Single Chamber for Cardiac Constructs. Sci. Rep. 2018, 8, 16944. [Google Scholar] [CrossRef]
- Ruan, J.L.; Tulloch, N.L.; Razumova, M.v.; Saiget, M.; Muskheli, V.; Pabon, L.; Reinecke, H.; Regnier, M.; Murry, C.E. Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue. Circulation 2016, 134, 1557–1567. [Google Scholar] [CrossRef]
- Carlos-Oliveira, M.; Lozano-Juan, F.; Occhetta, P.; Visone, R.; Rasponi, M. Current Strategies of Mechanical Stimulation for Maturation of Cardiac Microtissues. Biophys. Rev. 2021, 13, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.; Renteria, E.S.; Sime, D.S.; Ju, Y.M.; Kim, J.H.; Criswell, T.; Shupe, T.D.; Atala, A.; Marini, F.C.; Gurcan, M.N.; et al. Bioreactor Design and Validation for Manufacturing Strategies in Tissue Engineering. Bio-Des. Manuf. 2022, 5, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Sood, P.; Sood, N.; Gokhale, T. Microwaves: An Alternative Bacterial Sterilization Technique? GSTF J. BioSciences 2015, 3, 3. [Google Scholar] [CrossRef]
- Lelkes, P.; Unsworth, B.R. Neuroendocrine Tissue Engineering in RWV Bioreactors. In Methods of Tissue Engineering; Atala, A., Ed.; Academic Press: San Diego, CA, USA, 2002; pp. 371–382. [Google Scholar]
- Yin, Y.; Liu, P.Y.; Shi, Y.; Li, P. Single-Cell Sequencing and Organoids: A Powerful Combination for Modelling Organ Development and Diseases. In Reviews of Physiology, Biochemistry and Pharmacology; Springer: Berlin/Heidelberg, Germany, 2021; Volume 179, pp. 189–210. [Google Scholar]
- Tang, X.Y.; Wu, S.; Wang, D.; Chu, C.; Hong, Y.; Tao, M.; Hu, H.; Xu, M.; Guo, X.; Liu, Y. Human Organoids in Basic Research and Clinical Applications. Signal Transduct. Target. Ther. 2022, 7, 168. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-Derived Organoids Model Treatment Response of Metastatic Gastrointestinal Cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Hernández, D.; Rooney, L.A.; Daniszewski, M.; Gulluyan, L.; Liang, H.H.; Cook, A.L.; Hewitt, A.W.; Pébay, A. Culture Variabilities of Human IPSC-Derived Cerebral Organoids Are a Major Issue for the Modelling of Phenotypes Observed in Alzheimer’s Disease. Stem Cell Rev. Rep. 2022, 18, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Doi, D.; Morizane, A.; Kikuchi, T.; Onoe, H.; Hayashi, T.; Kawasaki, T.; Motono, M.; Sasai, Y.; Saiki, H.; Gomi, M.; et al. Prolonged Maturation Culture Favors a Reduction in the Tumorigenicity and the Dopaminergic Function of Human ESC-Derived Neural Cells in a Primate Model of Parkinson’s Disease. Stem Cells 2012, 30, 935–945. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Licata, J.P.; Schwab, K.H.; Har-el, Y.-e.; Gerstenhaber, J.A.; Lelkes, P.I. Bioreactor Technologies for Enhanced Organoid Culture. Int. J. Mol. Sci. 2023, 24, 11427. https://doi.org/10.3390/ijms241411427
Licata JP, Schwab KH, Har-el Y-e, Gerstenhaber JA, Lelkes PI. Bioreactor Technologies for Enhanced Organoid Culture. International Journal of Molecular Sciences. 2023; 24(14):11427. https://doi.org/10.3390/ijms241411427
Chicago/Turabian StyleLicata, Joseph P., Kyle H. Schwab, Yah-el Har-el, Jonathan A. Gerstenhaber, and Peter I. Lelkes. 2023. "Bioreactor Technologies for Enhanced Organoid Culture" International Journal of Molecular Sciences 24, no. 14: 11427. https://doi.org/10.3390/ijms241411427
APA StyleLicata, J. P., Schwab, K. H., Har-el, Y.-e., Gerstenhaber, J. A., & Lelkes, P. I. (2023). Bioreactor Technologies for Enhanced Organoid Culture. International Journal of Molecular Sciences, 24(14), 11427. https://doi.org/10.3390/ijms241411427





