Enzymatic Characterization of the Isocitrate Dehydrogenase with Dual Coenzyme Specificity from the Marine Bacterium Umbonibacter marinipuiceus
Abstract
1. Introduction
2. Results
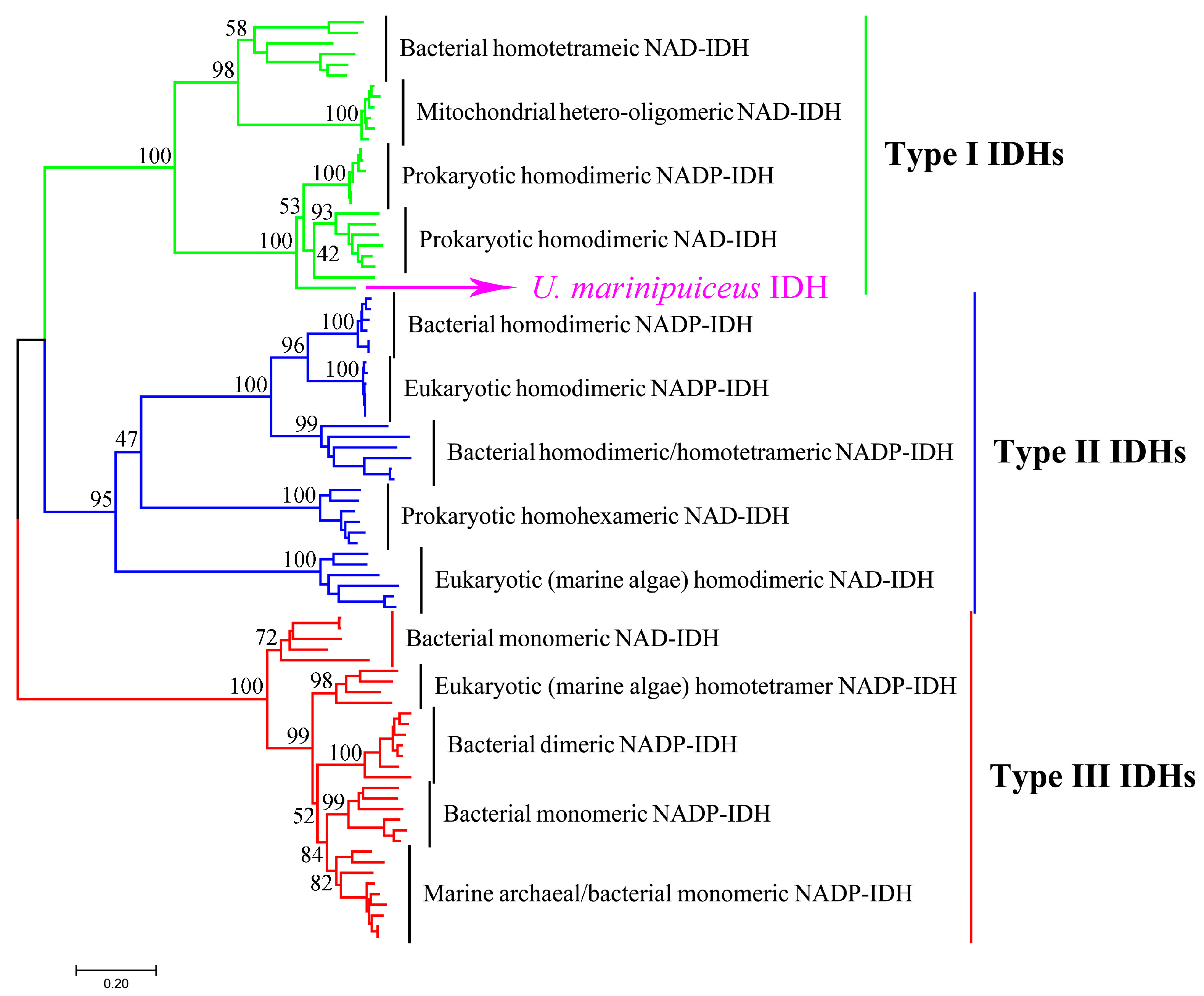
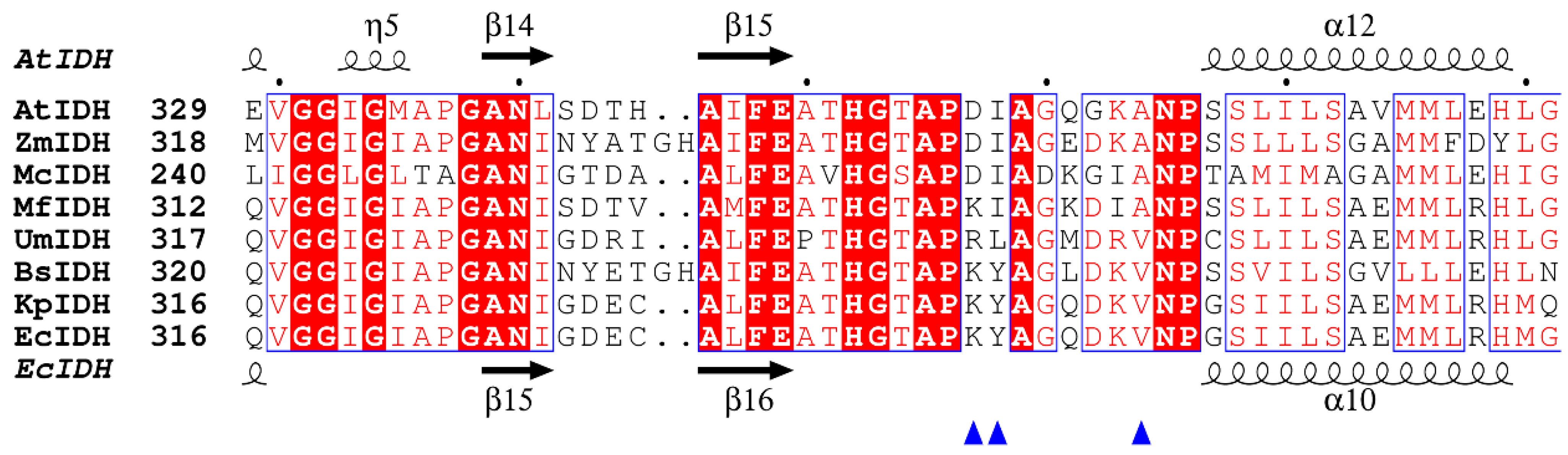
2.1. Bioinformatics Analysis
2.2. Expression and Purification
2.3. The Effects of pH, Temperature, and Metal Ions
2.4. Kinetics Analysis
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids and Reagents
4.2. Site-Directed Mutagenesis
4.3. Recombinant Protein Expression and Purification
4.4. SDS-PAGE and Gel Filtration Chromatography
4.5. Circular Dichroism Spectroscopy
4.6. Enzyme Characterization
4.7. Enzyme Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, R.; Jeong, S.S. Functional prediction: Identification of protein orthologs and paralogs. Protein Sci. 2000, 9, 2344–2353. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Liu, Y.; Ma, T.; Ding, J. Structure and allosteric regulation of human NAD+-dependent isocitrate dehydrogenase. Cell Discov. 2020, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- Spaans, S.K.; Weusthuis, R.A.; van der Oost, J.; Kengen, S.W. NADPH-generating systems in bacteria and archaea. Front. Microbiol. 2015, 6, 742. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Dang, L.; Yen, K.; Attar, E.C. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann. Oncol. 2016, 27, 599–608. [Google Scholar] [CrossRef]
- Eniafe, J.; Jiang, S. The functional roles of TCA cycle metabolites in cancer. Oncogene 2021, 40, 3351–3363. [Google Scholar] [CrossRef]
- Wang, P.; Lv, C.; Zhu, G. Novel type II and monomeric NAD+-specific isocitrate dehydrogenases: Phylogenetic affinity, enzymatic characterization, and evolutionary implication. Sci. Rep. 2015, 5, 9150. [Google Scholar] [CrossRef]
- Wang, P.; Wu, Y.; Liu, J.; Song, P.; Li, S.; Zhou, X.; Zhu, G. Crystal structure of the isocitrate dehydrogenase 2 from Acinetobacter baumannii (AbIDH2) reveals a novel dimeric structure with two monomeric IDH-like subunits. Int. J. Mol. Sci. 2018, 19, 1131. [Google Scholar] [CrossRef]
- Huang, S.; Zhao, J.; Li, W.; Wang, P.; Xue, Z.; Zhu, G. Biochemical and Phylogenetic Characterization of a Novel NADP+-Specific Isocitrate Dehydrogenase from the Marine Microalga Iphaeodactylum trcornutum. Front. Mol. Biosci. 2021, 8, 702083. [Google Scholar] [CrossRef]
- Zhu, G.; Golding, G.B.; Dean, A.M. The selective cause of an ancient adaptation. Science 2005, 307, 1279–1282. [Google Scholar] [CrossRef]
- Wang, P.; Jin, M.; Su, R.; Song, P.; Wang, M.; Zhu, G. Enzymatic characterization of isocitrate dehydrogenase from an emerging zoonotic pathogen Streptococcus suis. Biochimie 2011, 93, 1470–1475. [Google Scholar] [CrossRef]
- Wang, P.; Jin, M.; Zhu, G. Biochemical and molecular characterization of NAD+-dependent isocitrate dehydrogenase from the ethanologenic bacterium Zymomonas mobilis. FEMS Microbiol. Lett. 2012, 327, 134–141. [Google Scholar] [CrossRef]
- Dean, A.M.; Golding, G.B. Protein engineering reveals ancient adaptive replacements in isocitrate dehydrogenase. Proc. Natl. Acad. Sci. USA 1997, 94, 3104–3109. [Google Scholar] [CrossRef] [PubMed]
- Leyland, M.L.; Kelly, D.J. Purification and characterization of a monomeric isocitrate dehydrogenase with dual coenzyme specificity from the photosynthetic bacterium Rhodomicrobium vannielii. Eur. J. Biochem. 1991, 202, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Potter, S. Evidence for a dual-specificity isocitrate dehydrogenase in the euryarchaeotan Thermoplasma acidophilum. Can. J. Microbiol. 1993, 39, 262–264. [Google Scholar] [CrossRef]
- Kim, H.; Mozaffar, Z.; Weete, J.D. A dual cofactor-specific isocitrate dehydrogenase from Pythium ultimum. Can J. Microbiol. 1996, 42, 1241–1247. [Google Scholar] [CrossRef]
- Romkina, A.Y.; Kiriukhin, M.Y. Biochemical and molecular characterization of the isocitrate dehydrogenase with dual coenzyme specificity from the obligate methylotroph Methylobacillus Flagellatus. PLoS ONE 2017, 12, e0176056. [Google Scholar] [CrossRef]
- Gao, H.; Li, J.; Sivakumar, D.; Kim, T.S.; Patel, S.K.S.; Kalia, V.C.; Kim, I.W.; Zhang, Y.W.; Lee, J.K. NADH oxidase from Lactobacillus reuteri: A versatile enzyme for oxidized cofactor regeneration. Int. J. Biol. Macromol. 2019, 123, 629–636. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Tanaka, N.; Frolova, G.M. Umboniibacter marinipuniceus gen. nov., sp. nov., a marine gammaproteobacterium isolated from the mollusc Umbonium costatum from the Sea of Japan. Int. J. Syst. Evol. Microbiol. 2010, 60, 603–609. [Google Scholar] [CrossRef]
- Aoshima, M.; Ishii, M.; Igarashi, Y. A novel biotin protein required for reductive carboxylation of 2-oxoglutarate by isocitrate dehydrogenase in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 2004, 51, 791–798. [Google Scholar] [CrossRef]
- Wu, M.C.; Tian, C.Q.; Cheng, H.M.; Xu, L.; Wang, P.; Zhu, G.P. A novel type II NAD+-specific isocitrate dehydrogenase from the marine bacterium Congregibacter litoralis KT71. PLoS ONE 2015, 10, e0125229. [Google Scholar] [CrossRef]
- Hirota, R.; Tsubouchi, K.; Takada, Y. NADP+-dependent isocitrate dehydrogenase from a psychrophilic bacterium, Psychromonas marina. Extremophiles 2017, 21, 711–721. [Google Scholar] [CrossRef]
- Yasuda, W.; Kobayashi, M.; Takada, Y. Analysis of amino acid residues involved in cold activity of monomeric isocitrate dehydrogenase from psychrophilic bacteria, Colwellia maris and Colwellia psychrerythraea. J. Biosci. Bioeng. 2013, 116, 567–572. [Google Scholar] [CrossRef]
- Inoue, H.; Tamura, T.; Ehara, N.; Nishito, A.; Nakayama, Y.; Maekawa, M.; Imada, K.; Tanaka, H.; Inagaki, K. Biochemical and molecular characterization of the NAD(+)-dependent isocitrate dehydrogenase from the chemolithotroph Acidithiobacillus thiooxidans. FEMS Microbiol. Lett. 2002, 214, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Song, P.; Jin, M.; Zhu, G. Isocitrate dehydrogenase from Streptococcus mutans: Biochemical properties and evaluation of a putative phosphorylation site at Ser102. PLoS ONE 2013, 8, e58918. [Google Scholar] [CrossRef] [PubMed]
- Steen, I.H.; Madern, D.; Karlstrom, M.; Lien, T.; Ladenstein, R.; Birkeland, N.K. Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state, and phylogenetic affiliation. J. Biol. Chem. 2001, 276, 43924–43931. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Greer, A.; Dean, A.M. A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity. Proc Natl Acad Sci. USA 1995, 92, 11666–11670. [Google Scholar] [CrossRef]
- Jin, M.M.; Wang, P.; Li, X.; Zhao, X.Y.; Xu, L.; Song, P.; Zhu, G.P. Biochemical characterization of NADP(+)-dependent isocitrate dehydrogenase from Microcystis aeruginosa PCC7806. Mol. Biol. Rep. 2013, 40, 2995–3002. [Google Scholar] [CrossRef]
- Huang, S.P.; Zhou, L.C.; Wen, B.; Wang, P.; Zhu, G.P. Biochemical Characterization and Crystal Structure of a Novel NAD+-Dependent Isocitrate Dehydrogenase from Phaeodactylum tricornutum. Int. J. Mol. Sci. 2020, 21, 5915. [Google Scholar] [CrossRef]
- Ellington, A.D.; Bull, J.J. Evolution. Changing the cofactor diet of an enzyme. Science 2005, 310, 454–455. [Google Scholar] [CrossRef]
- Golding, G.B.; Dean, A.M. The structural basis of molecular adaptation. Mol. Biol. Evol. 1998, 15, 355–369. [Google Scholar] [CrossRef]
- Tang, W.G.; Song, P.; Cao, Z.Y.; Wang, P.; Zhu, G.P. A unique homodimeric NAD⁺-linked isocitrate dehydrogenase from the smallest autotrophic eukaryote Ostreococcus tauri. FASEB J. 2015, 29, 2462–2472. [Google Scholar] [CrossRef]
- Lv, C.; Wang, P.; Wang, W.; Su, R.; Ge, Y.; Zhu, Y.; Zhu, G. Two isocitrate dehydrogenases from a plant pathogen Xanthomonas campestris pv. campestris 8004. Bioinformatic analysis, enzymatic characterization, and implication in virulence. J. Basic Microbiol. 2016, 56, 975–985. [Google Scholar] [CrossRef]
- Huang, S.P.; Cheng, H.M.; Wang, P.; Zhu, G.P. Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from Bifidobacterium longum. Int. J. Mol. Sci. 2016, 17, 296. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, Y.; Guo, X.; Huang, S.; Zhu, G. Biochemical and phylogenetic characterization of a monomeric isocitrate dehydrogenase from a marine methanogenic archaeon Methanococcoides methylutens. Extremophiles 2020, 24, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Edlin, J.D.; Sundaram, T.K. Regulation of isocitrate dehydrogenase by phosphorylation in Escherichia coli K-12 and a simple method for determining the amount of inactive phosphoenzyme. J Bacteriol. 1989, 171, 2634–2638. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Thorsness, P.E.; Ramalingam, V.; Helmers, N.H.; Koshland, D.E.; Stroud, R.M. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc. Natl. Acad. Sci. USA 1989, 86, 8635–8639. [Google Scholar] [CrossRef]
- Cortay, J.C.; Bleicher, F.; Rieul, C.; Reeves, H.C.; Cozzone, A.J. Nucleotide sequence and expression of the aceK gene coding for isocitrate dehydrogenase kinase/phosphatase in Escherichia coli. J. Bacteriol. 1988, 170, 89–97. [Google Scholar] [CrossRef]
- Cozzone, A.J.; El-Mansi, M. Control of isocitrate dehydrogenase catalytic activity by protein phosphorylation in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2005, 9, 132–146. [Google Scholar] [CrossRef]
- Yu, B.J.; Kim, J.A.; Moon, J.H.; Ryu, S.E.; Pan, J.G. The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 2008, 18, 1529–1536. [Google Scholar]
- Zhang, Z.; Tan, M.; Xie, Z.; Dai, L.; Chen, Y.; Zhao, Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011, 7, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Patel, S.K.S.; Tiwari, M.K.; Lai, C.; Kumar, A.; Kim, Y.S.; Kalia, V.C.; Lee, J.K. Phe-140 Determines the catalytic efficiency of arylacetonitrilase from Alcaligenes faecalis. Int. J. Mol. Sci. 2020, 21, 7859. [Google Scholar] [CrossRef] [PubMed]
- Raussens, V.; Ruysschaert, J.M.; Goormaghtigh, E. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: A new scaling method. Anal. Biochem. 2003, 319, 114–121. [Google Scholar] [CrossRef]


| Metal Ions | Relative Activity (%) | Metal Ions | Relative Activity (%) | Metal Ions | Relative Activity (%) |
|---|---|---|---|---|---|
| None | 0 | ||||
| Mn2+ | 100 ± 0 | Mn2+ | 100 ± 0 | ||
| Mg2+ | 19.51 ± 4.66 | Mn2+ + Mg2+ | 114.97 ± 5.13 | Mg2+ | 100 ± 0 |
| Ca2+ | 15.90 ± 1.32 | Mn2+ + Ca2+ | 67.10 ± 4.27 | Mg2+ + Ca2+ | 20.75 ± 1.34 |
| Co2+ | 0.66 ± 1.29 | Mn2+ + Co2+ | 20.04 ± 2.80 | Mg2+ + Co2+ | 7.68 ± 0.74 |
| Cu2+ | 0.31 ± 0.27 | Mn2+ + Cu2+ | 4.04 ± 0.33 | Mg2+ + Cu2+ | 6.97 ± 0.61 |
| Ni2+ | 2.29 ± 0.64 | Mn2+ + Ni2+ | 103.51 ± 3.42 | Mg2+ + Ni2+ | 102.07 ± 0.59 |
| K+ | 8.30 ± 0.43 | Mn2+ + K+ | 106.13 ± 4.37 | Mg2+ + K+ | 110.91 ± 1.10 |
| Na+ | 2.31 ± 0.75 | Mn2+ + Na+ | 104.88 ± 3.92 | Mg2+ + Na+ | 101.03 ± 2.11 |
| Li+ | 7.69 ± 1.07 | Mn2+ + Li+ | 103.53 ± 0.98 | Mg2+ + Li+ | 107.04 ± 1.20 |
| Enzyme | NAD+ | NADP+ | ||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) | |
| UmIDH | 1800.0 ± 64.4 | 7.5 ± 0.3 | 0.0042 | 1167.7 ± 113.0 | 11.1 ± 0.4 | 0.0095 |
| R345K/L346Y | - | - | - | 605.9 ± 38.2 | 15.3 ± 0.2 | 0.025 |
| R345D/L346I/V352A | 369.7 ± 11.84 | 17.7 ± 0.9 | 0.048 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, M.; Zhao, J.; Xu, W.; Han, X.; Chen, X.; Wang, P.; Zhu, G. Enzymatic Characterization of the Isocitrate Dehydrogenase with Dual Coenzyme Specificity from the Marine Bacterium Umbonibacter marinipuiceus. Int. J. Mol. Sci. 2023, 24, 11428. https://doi.org/10.3390/ijms241411428
Bian M, Zhao J, Xu W, Han X, Chen X, Wang P, Zhu G. Enzymatic Characterization of the Isocitrate Dehydrogenase with Dual Coenzyme Specificity from the Marine Bacterium Umbonibacter marinipuiceus. International Journal of Molecular Sciences. 2023; 24(14):11428. https://doi.org/10.3390/ijms241411428
Chicago/Turabian StyleBian, Mingjie, Jiaxin Zhao, Wenqiang Xu, Xueyang Han, Xuefei Chen, Peng Wang, and Guoping Zhu. 2023. "Enzymatic Characterization of the Isocitrate Dehydrogenase with Dual Coenzyme Specificity from the Marine Bacterium Umbonibacter marinipuiceus" International Journal of Molecular Sciences 24, no. 14: 11428. https://doi.org/10.3390/ijms241411428
APA StyleBian, M., Zhao, J., Xu, W., Han, X., Chen, X., Wang, P., & Zhu, G. (2023). Enzymatic Characterization of the Isocitrate Dehydrogenase with Dual Coenzyme Specificity from the Marine Bacterium Umbonibacter marinipuiceus. International Journal of Molecular Sciences, 24(14), 11428. https://doi.org/10.3390/ijms241411428






