A Random Forest Model for Peptide Classification Based on Virtual Docking Data
Abstract
1. Introduction
2. Results
2.1. Dataset Characterization
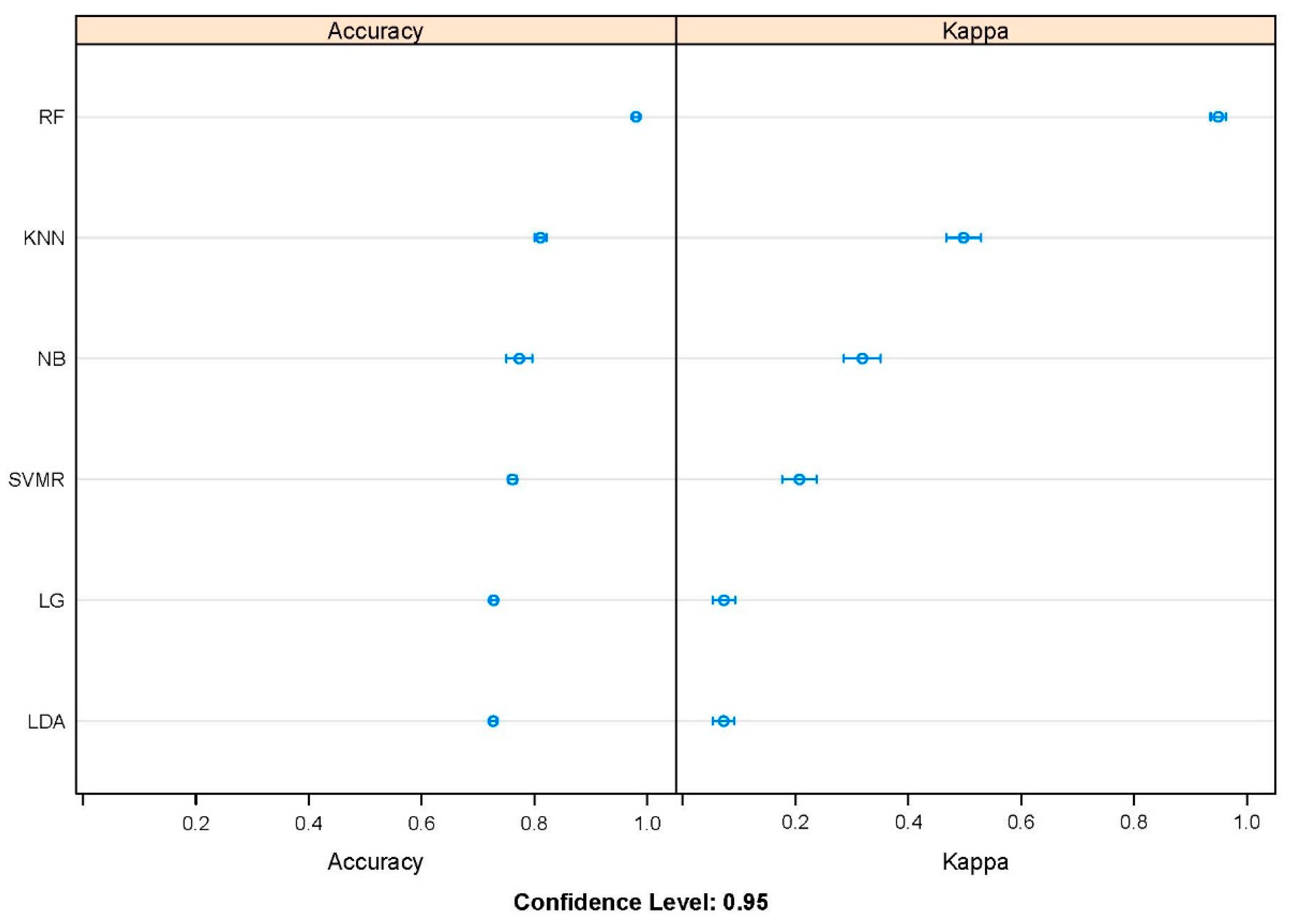
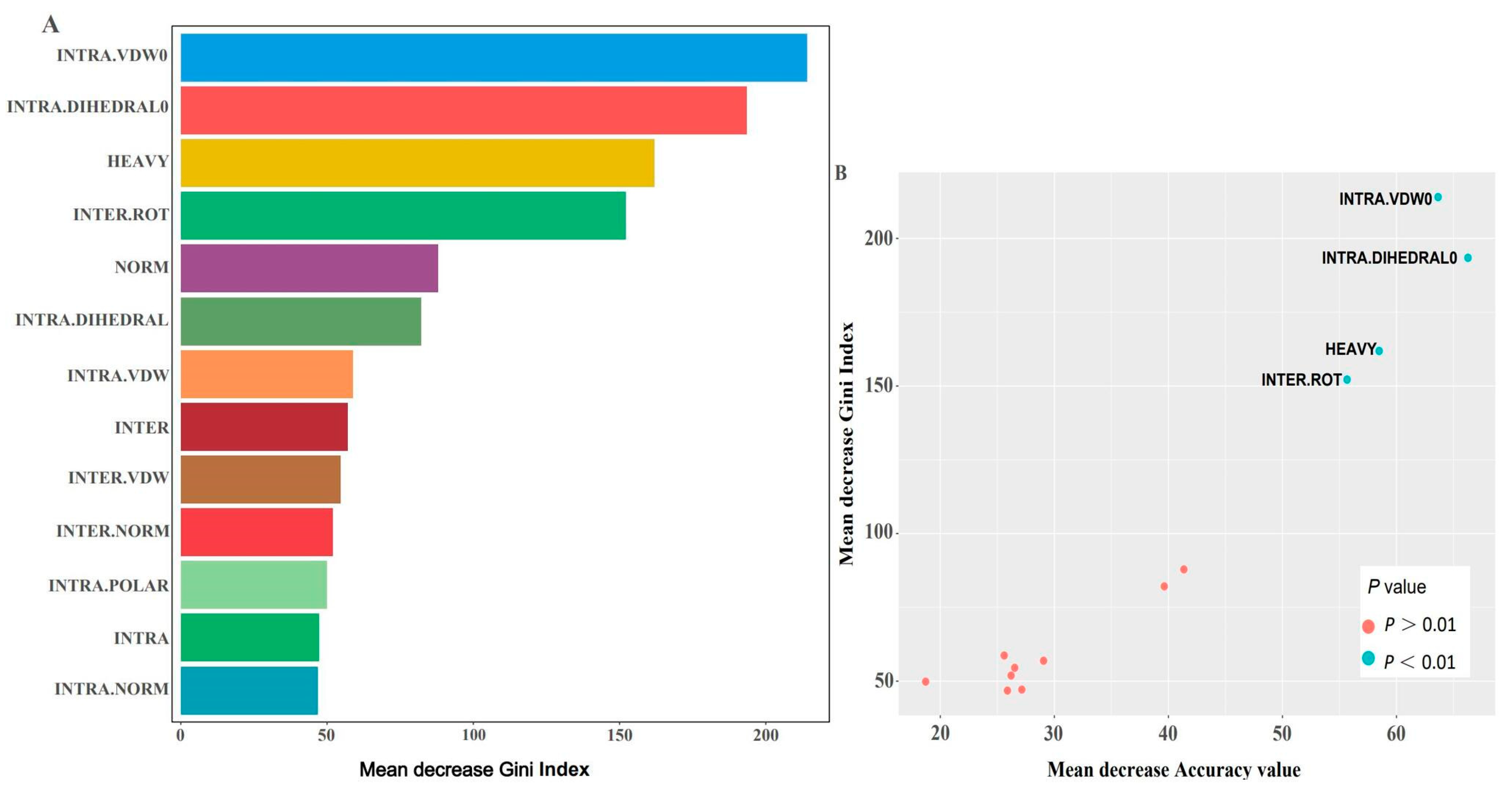
2.2. Algorithm Selection and Feature Importance
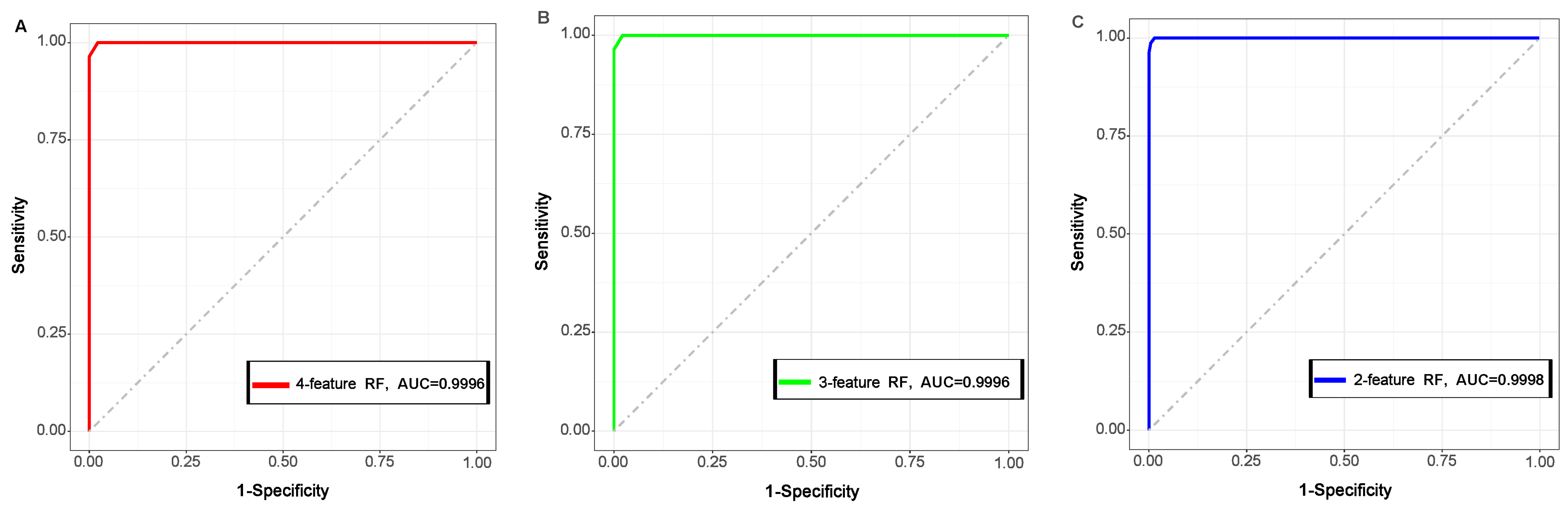
2.3. Construction of RF Model
2.4. Performance of RF Model on Independent Data
3. Discussion
4. Materials and Methods
4.1. Dataset Collection
4.1.1. Affinity Assay between Peptides and Proteins by SPR
4.1.2. Structure Preparation
4.1.3. Molecular Docking
4.2. Pre-Selection of Different ML Algorithms
4.3. Selection of Important Features
4.4. RF Model Reconstruction Using the Important Features
4.5. Performance Evaluation of the Constructed Model
4.6. Performance of the RF Models on an Unknown Peptide Dataset
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lei, Y.; Li, S.; Liu, Z.; Wan, F.; Tian, T.; Li, S.; Zhao, D.; Zeng, J. A deep-learning framework for multi-level peptide–protein interaction prediction. Nat. Commun. 2021, 12, 5465. [Google Scholar] [CrossRef] [PubMed]
- Johansson-Åkhe, I.; Mirabello, C.; Wallner, B. Predicting protein-peptide interaction sites using distant protein complexes as structural templates. Sci. Rep. 2019, 9, 4267. [Google Scholar] [CrossRef] [PubMed]
- Johansson-Åkhe, I.; Mirabello, C.; Wallner, B. InterPep2: Global peptide–protein docking using interaction surface templates. Bioinformatics 2020, 36, 2458–2465. [Google Scholar] [CrossRef] [PubMed]
- Caporale, A.; Adorinni, S.; Lamba, D.; Saviano, M. Peptide-Protein Interactions: From Drug Design to Supramolecular Biomaterials. Molecules 2021, 26, 1219. [Google Scholar] [CrossRef]
- Lee, A.C.; Harris, J.L.; Khanna, K.K.; Hong, J.H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef]
- Tripathi, N.M.; Bandyopadhyay, A. High throughput virtual screening (HTVS) of peptide library: Technological advancement in ligand discovery. Eur. J. Med. Chem. 2022, 243, 114766. [Google Scholar] [CrossRef]
- London, N.; Raveh, B.; Cohen, E.; Fathi, G.; Schueler-Furman, O. Rosetta FlexPepDock web server—High resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011, 39, W249–W253. [Google Scholar] [CrossRef]
- Bielza, C.; Larrañaga, P. Discrete Bayesian Network Classifiers: A Survey. ACM Comput. Surv. 2014, 47, 1–43. [Google Scholar] [CrossRef]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.-Y. HPEPDOCK: A web server for blind peptide–protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Lee, H.; Heo, L.; Lee, M.S.; Seok, C. GalaxyPepDock: A protein–peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 2015, 43, W431–W435. [Google Scholar] [CrossRef]
- Carmona, S.R.; Alvarez-Garcia, D.; Foloppe, N.; Garmendia-Doval, A.B.; Juhos, S.; Schmidtke, P.; Barril, X.; Hubbard, R.E.; Morley, S.D. rDock: A Fast, Versatile and Open Source Program for Docking Ligands to Proteins and Nucleic Acids. PLoS Comput. Biol. 2014, 10, e1003571. [Google Scholar]
- Patel, L.; Shukla, T.; Huang, X.; Ussery, D.W.; Wang, S. Machine Learning Methods in Drug Discovery. Molecules 2020, 25, 5277. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef]
- Gupta, P.; Mohanty, D. SMMPPI: A machine learning-based approach for prediction of modulators of protein-protein interactions and its application for identification of novel inhibitors for RBD:hACE2 interactions in SARS-CoV-2. Brief. Bioinform. 2021, 22, bbab111. [Google Scholar] [CrossRef] [PubMed]
- Bukhari SN, H.; Jain, A.; Haq, E.; Mehbodniya, A.; Webber, J. Machine Learning Techniques for the Prediction of B-Cell and T-Cell Epitopes as Potential Vaccine Targets with a Specific Focus on SARS-CoV-2 Pathogen: A Review. Pathogens 2022, 11, 146. [Google Scholar] [CrossRef]
- Kumari, M.; Subbarao, N. Deep learning model for virtual screening of novel 3C-like protease enzyme inhibitors against SARS coronavirus diseases. Comput. Biol. Med. 2021, 132, 104317. [Google Scholar] [CrossRef]
- Jabeen, A.; de March, C.A.; Matsunami, H.; Ranganathan, S. Machine Learning Assisted Approach for Finding Novel High Activity Agonists of Human Ectopic Olfactory Receptors. Int. J. Mol. Sci. 2021, 22, 11546. [Google Scholar] [CrossRef]
- Danishuddin; Kumar, V.; Parate, S.; Bahuguna, A.; Lee, G.; Kim, M.O.; Lee, K.W. Development of Machine Learning Models for Accurately Predicting and Ranking the Activity of Lead Molecules to Inhibit PRC2 Dependent Cancer. Pharmaceuticals 2021, 14, 699. [Google Scholar] [CrossRef]
- Jana, T.; Ghosh, A.; Das Mandal, S.; Banerjee, R.; Saha, S. PPIMpred: A web server for high-throughput screening of small molecules targeting protein–protein interaction. R. Soc. Open Sci. 2017, 4, 160501. [Google Scholar] [CrossRef]
- Abella, J.R.; Antunes, D.A.; Clementi, C.; Kavraki, L.E. Large-Scale Structure-Based Prediction of Stable Peptide Binding to Class I HLAs Using Random Forests. Front. Immunol. 2020, 11, 1583. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y. Improving scoring-docking-screening powers of protein-ligand scoring functions using random forest. J. Comput. Chem. 2017, 38, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Alnammi, M.; Ericksen, S.S.; Voter, A.F.; Ananiev, G.E.; Keck, J.L.; Hoffmann, F.M.; Wildman, S.A.; Gitter, A. Practical Model Selection for Prospective Virtual Screening. J. Chem. Inf. Model. 2019, 59, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Machado, G.; Vilalta, C.; Recamonde-Mendoza, M.; Corzo, C.; Torremorell, M.; Perez, A.; VanderWaal, K. Identifying outbreaks of Porcine Epidemic Diarrhea virus through animal movements and spatial neighborhoods. Sci. Rep. 2019, 9, 457. [Google Scholar] [CrossRef]
- Wei, Y.; Li, J.; Qing, J.; Huang, M.; Wu, M.; Gao, F.; Li, D.; Hong, Z.; Kong, L.; Huang, W.; et al. Discovery of Novel Hepatitis C Virus NS5B Polymerase Inhibitors by Combining Random Forest, Multiple e-Pharmacophore Modeling and Docking. PLoS ONE 2016, 11, e0148181. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Poongavanam, V.; Kongsted, J. Virtual Screening Models for Prediction of HIV-1 RT Associated RNase H Inhibition. PLoS ONE 2013, 8, e73478. [Google Scholar] [CrossRef]
- Cao, S. Research Onthe Design and Function of Peptide Targeting Aβ1-42 Protein. Master’s Thesis, Henan Agricultural University, Zhengzhou, China, 2021. (In Chinese). [Google Scholar]
- Hao, J. Rarional Design, Identification and Application of Affinity Peptide Ligands of Porcine Circovirus Type 2 Cap Protein. PhD’s Thesis, Sichuan Agricultural University, Chengdu, China, 2020. (In Chinese). [Google Scholar]
- Hu, M. Antigen-Display Nanoparticles Mediated by Affinity Peptides Targeting Classical Swine Fever Virus E2 Protein and Porcine Circovirus 2 Capsid Protein. PhD’s Thesis, Jilin University, Changchun, China, 2020. (In Chinese). [Google Scholar]
- Wang, F.; Li, N.; Wang, C.; Xing, G.; Cao, S.; Xu, Q.; Zhang, Y.; Hu, M.; Zhang, G. DPL: A comprehensive database on sequences, structures, sources and functions of peptide ligands. Database 2020, 2020, baaa089. [Google Scholar]
- Hu, M.; Wang, F.; Li, N.; Xing, G.; Sun, X.; Zhang, Y.; Cao, S.; Cui, N.; Zhang, G. An antigen display system of GEM nanoparticles based on affinity peptide ligands. Int. J. Biol. Macromol. 2021, 193 Pt A, 574–584. [Google Scholar] [CrossRef]
- Duffy, F.J.; Verniere, M.; Devocelle, M.; Bernard, E.; Shields, D.C.; Chubb, A.J. CycloPs: Generating virtual libraries of cyclized and constrained peptides including nonnatural amino acids. J. Chem. Inf. Model. 2011, 51, 829–836. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Svetnik, V.; Liaw, A.; Tong, C.; Christopher, C.J.; Sheridan, R.P.; Feuston, B.P. Random Forest: A Classification and Regression Tool for Compound Classification and QSAR Modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Eric, A. EricArcher/rfPermute, Version 2.5 (v2.5); Zenodo: Geneva, Switzerland, 2021. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 13 May 2023).

| Selected Variables | mtry | Accuracy | Kappa | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|---|
| INTRA.VDW0 INTRA.DIHEDRAL0 HEAVY INTER.ROT | 4 | 0.9915 | 0.9778 | 0.9932 | 0.9858 | 0.9997 |
| INTRA.VDW0 INTRA.DIHEDRAL0 HEAVY | 3 | 0.9902 | 0.9753 | 0.9934 | 0.9817 | 0.9995 |
| INTRA.VDW0 INTRA.DIHEDRAL0 | 2 | 0.9912 | 0.9781 | 0.9978 | 0.9743 | 0.9997 |
| Selected Variables | Accuracy | Kappa | Sensitivity | Specificity | F1 | MCC |
|---|---|---|---|---|---|---|
| INTRA.VDW0 INTRA.DIHEDRAL0 HEAVY INTER.ROT | 0.9880 | 0.9707 | 0.9928 | 0.9762 | 0.9916 | 0.9707 |
| INTRA.VDW0 INTRA.DIHEDRAL0 HEAVY | 0.9880 | 0.9707 | 0.9928 | 0.9762 | 0.9916 | 0.9707 |
| INTRA.VDW0 INTRA.DIHEDRAL0 | 0.9897 | 0.9737 | 1 | 0.9623 | 0.9930 | 0.9741 |
| Models | Class | Prediction Affinity (n) | Accuracy | ||
|---|---|---|---|---|---|
| A | UA | ||||
| 4-feature model | Actual affinity (n) | A | 760 | 180 | 0.714 |
| UA | 140 | 40 | |||
| 3-feature model | A | 680 | 260 | 0.661 | |
| UA | 120 | 60 | |||
| 2-feature model | A | 620 | 320 | 0.607 | |
| UA | 120 | 60 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, H.; Wang, F.; Li, N.; Xu, Q.; Zheng, G.; Sun, X.; Hu, M.; Xing, G.; Zhang, G. A Random Forest Model for Peptide Classification Based on Virtual Docking Data. Int. J. Mol. Sci. 2023, 24, 11409. https://doi.org/10.3390/ijms241411409
Feng H, Wang F, Li N, Xu Q, Zheng G, Sun X, Hu M, Xing G, Zhang G. A Random Forest Model for Peptide Classification Based on Virtual Docking Data. International Journal of Molecular Sciences. 2023; 24(14):11409. https://doi.org/10.3390/ijms241411409
Chicago/Turabian StyleFeng, Hua, Fangyu Wang, Ning Li, Qian Xu, Guanming Zheng, Xuefeng Sun, Man Hu, Guangxu Xing, and Gaiping Zhang. 2023. "A Random Forest Model for Peptide Classification Based on Virtual Docking Data" International Journal of Molecular Sciences 24, no. 14: 11409. https://doi.org/10.3390/ijms241411409
APA StyleFeng, H., Wang, F., Li, N., Xu, Q., Zheng, G., Sun, X., Hu, M., Xing, G., & Zhang, G. (2023). A Random Forest Model for Peptide Classification Based on Virtual Docking Data. International Journal of Molecular Sciences, 24(14), 11409. https://doi.org/10.3390/ijms241411409







