1. Introduction
The growing utilization of various chemicals, including medicines, pesticides, and chemical fertilizers, has sparked increasing interest due to their considerable toxicity [
1]. Apart from criticism for animal testing, assessing the toxicity and risk of chemicals is a complex task that often requires significant time and effort [
2]. Therefore, the research for chemical toxicity assessment is crucial in the advancement of alternative methods to animal testing and will support the development of predictive modeling [
3].
Many drugs also fail clinical trials due to their unmanageable toxicity, which is a significant cause of the high failure rate in clinical drug development. To alleviate this problem in an already long and costly process, it would be ideal to gauge toxicity directly from the molecular structure [
4]. However, this is a challenging task for humans because chemical systems are complex and such properties are not always immediately evident from molecular substructures. In fact, determining effective methods to represent molecules for toxicity prediction remains challenging, though many exist, including theoretical, empirical, and semi-empirical representations. Descriptors representing a molecular structure may be divided into graph-based and geometry-based types. Graph-based representations primarily describe the connectivity between atoms, while geometry-based representations include information about the molecule’s physical geometry, including bond lengths and angles [
5].
Traditionally, many quantitative structure–toxicity relationship (QSTR) models have been introduced as a means to evaluate and predict chemical risk, relying solely on molecular structure information [
6,
7,
8]. The molecular structure is featurized by physical and chemical properties such as size, covalent bonds, and polarity, and it can be effectively characterized through quantitative molecular descriptors. Among the most common descriptors for toxicity prediction are Molecular Access System keys (MACCS Keys), PubChem Substructure Fingerprints, Klekotha–Roth, and Extended-Connectivity Fingerprints (ECFPs) [
9]. However, these models require predefined features and molecular descriptors, and a lack of exhaustiveness would lead to poor performance. With more than 5000 molecular descriptors available for characterizing molecules, it is not always clear how to select the most appropriate representation while avoiding selection bias. In place of such general descriptors, there has been growing interest in alternative data-driven representations via deep learning [
5,
10,
11].
With the development of molecular graph representations and deep learning, graph neural networks (GNNs) are one of the leading research areas for molecular property prediction. In a molecular graph, atoms are treated as vertices and bonds are treated as edges. By functioning directly on molecular graphs, GNNs provide a more accurate description of the chemical structure with connectivity information and can allow for more relevant representations of molecules without having to consider many-body interactions or complex electronic structures [
5,
12,
13]. Starting from message-passing neural networks (MPNNs) [
14], many variations have been developed with higher complexity or additional attention mechanisms, such as the graph convolution network (GCN), graph attention network (GAT), path-augmented graph transformer network (PAGTN) [
15], and Attentive FP model [
5]. These architectures employ various techniques to overcome the complexity and irregularity of molecular graphs.
In recent years, deep learning has grown popular for toxicity prediction due to its successful performance and because it avoids a challenging feature selection process. For example, Mayr et al.’s DeepTox pipeline proved effective on the Tox21 data challenge and outperformed shallower machine learning methods [
16]. The choice of molecular representation has also shown to be important in this domain, and graphs have been shown to be useful in encoding a molecular structure, as vertices can represent atoms and edges can represent connections between atoms [
17]. In 2017, Xu et al., introduced molecular graph-encoding convolutional neural networks for acute oral toxicity (AOT) prediction via their deepAOT model [
10]. This model uses graph convolutions to represent molecules, removing the need to calculate fingerprints and allowing the algorithm to learn more directly from the data. Wu and Wei presented another topology-based molecular representation in 2018 that paired element-specific persistent homology with multitask deep learning and proved successful on four benchmark toxicity datasets [
18].
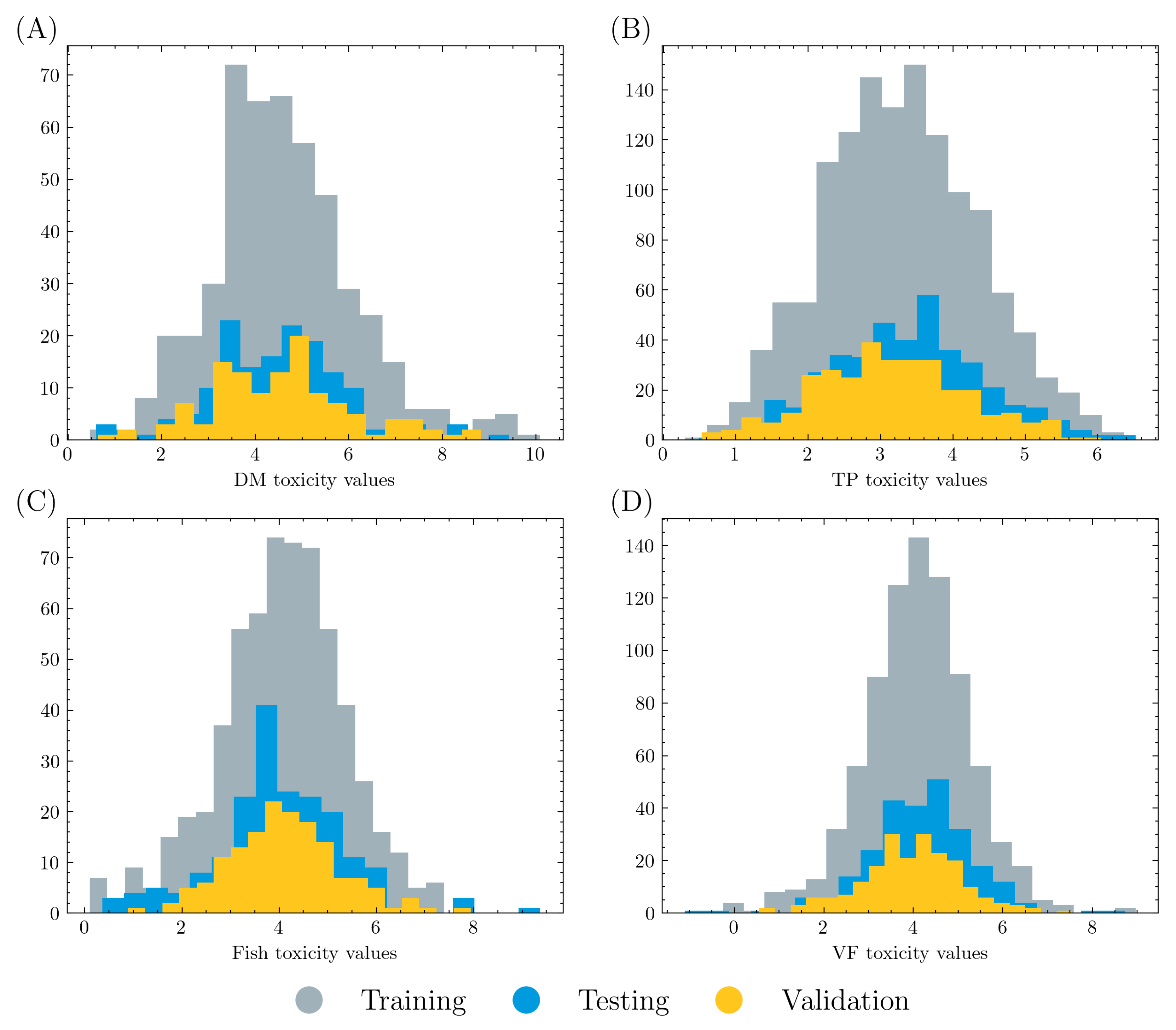
In this study, the performance of five graph models (MPNN, GCN, GAT, PAGTN, and Attentive FP) was evaluated on four acute toxicity tasks (fish,
Daphnia magna,
Tetrahymena pyriformis, and
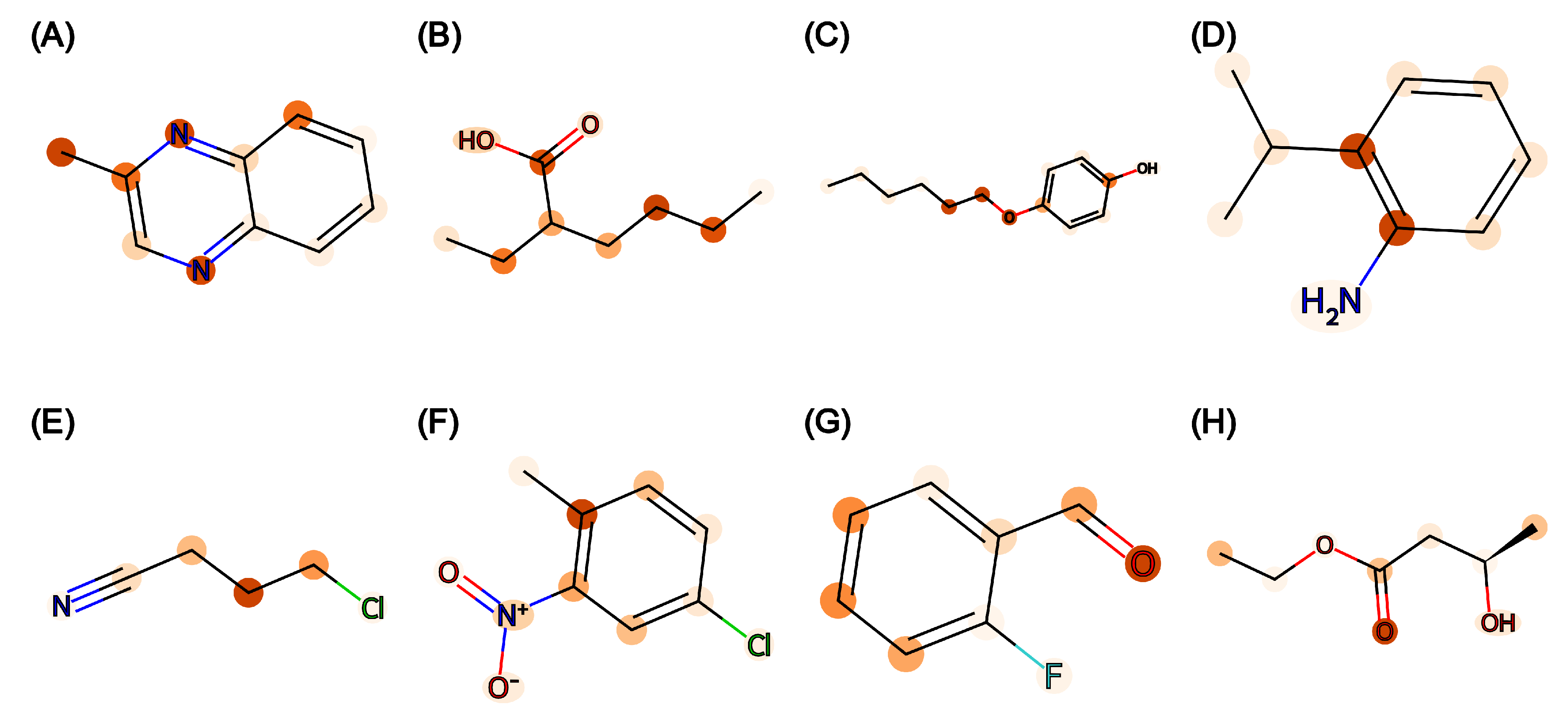
Vibrio fischeri). With rigorous training and testing, Attentive FP was identified as the best-performing model with low prediction error. Furthermore, the attention weights from the trained Attentive FP model can help visualize atom importance with a molecular heatmap. This interpretability is especially important in molecular property prediction for understanding the model’s internal workings and gaining new scientific insights [
19].
3. Discussion
Appropriate parameters are needed to reach the best performance. As shown in
Figure 3, model performance varies with different parameter settings. For example, in the Attentive FP model on the DM task, the model performed poorly with a large batch size, large number of layers, and low dropout rate. These indicate that increased model complexity might not always lead to better performance, and each model must be tuned appropriately. For the GCN model on the TP task, the model performed better with a smaller batch size, large channel width, and suitable dropout rate. The V shape in the dropout (
Figure 3F) implies that a reasonable dropout rate is required to overcome overfitting, while a large dropout rate might lead to insufficient training. In this work, a grid search cross-validation method was used to explore the hyperparameter space. However, it is not possible to explore all possible configurations, and the grid search method does not necessarily lead to the optimum set. A Bayesian optimization approach, as implemented in the hyperopt package [
20], may prove useful for future work.
Sample size is an important factor for machine learning models. As reported in this study, graph models reached the best and worst performance in the TP and DM tasks, respectively. This aligns with the comparison of the training size, where the TP task includes 2035 molecules while the DM task has only 753 molecules. The VF and fish tasks have similar sample sizes of 1271 and 956, and thus have comparable performance. Therefore, it is reasonable to conclude that a larger dataset would lead to better performance. In addition, the training datasets in this work lacked a significant number of nontoxic molecules. Real-world applications may not reflect this data distribution, so these models may be limited in such environments and should be used with caution where a large number of molecules have low or zero toxicity. Future research may investigate graph model performance on a dataset curated with regard to real-life scenarios.
In the current study, each graph model was trained independently on four tasks, which does not take advantage of the fact that some tasks share common molecules. Transfer learning should be considered in future applications where a low-level embedding layer can be trained to learn common molecular structures with high-level task-specific layers.
It should be noted that the primary purpose of this study is to evaluate and compare the five selected graph models quantitatively instead of using model interpretation and molecular structural explanation. Attentive FP is highlighted to be unique in producing atomic weights from which a heatmap may be generated for further analysis, but the study is limited in analyzing the structural basis of toxicity.
5. Conclusions
The development of graph models provides an alternative method to QSTR models for acute toxicity prediction. In this study, a pipeline was provided to compare different graph models for this purpose. Performance metrics were evaluated and key differences were reported among five popular graph models, namely, MPNNs, GCNs, GATs, PAGTNs, and Attentive FP. As reported, the Attentive FP model performed the best with a low prediction error, a high coefficient of determination R2, and good interpretability. The importance of parameter fine-tuning and sample size was discussed, along with the potential application of transfer learning.