Smad3 Mediates Diabetic Dyslipidemia and Fatty Liver in db/db Mice by Targeting PPARδ
Abstract
1. Introduction
2. Results
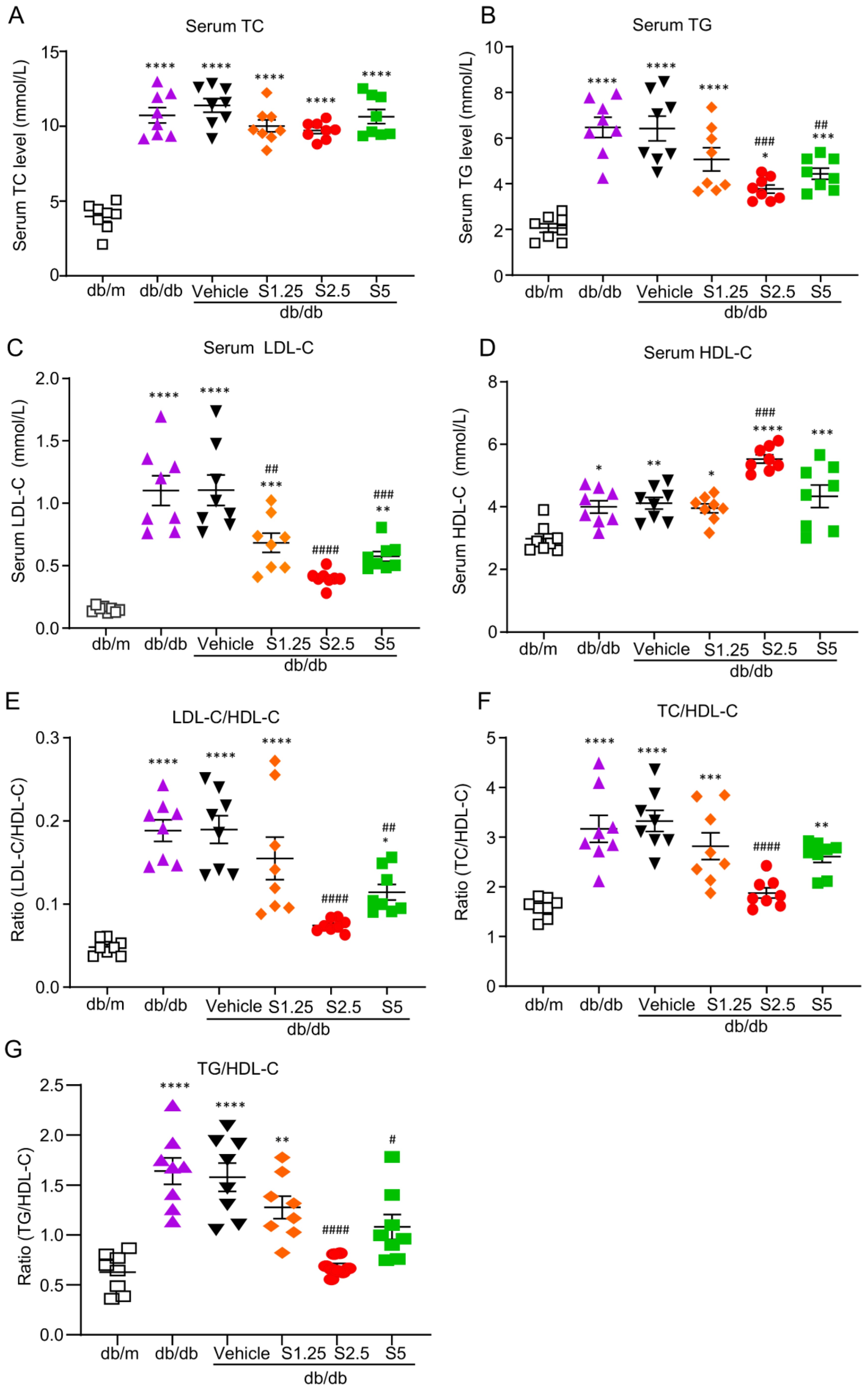
2.1. Smad3 Deficiency Inhibits Dyslipidemia and Fatty Liver in db/db Mice
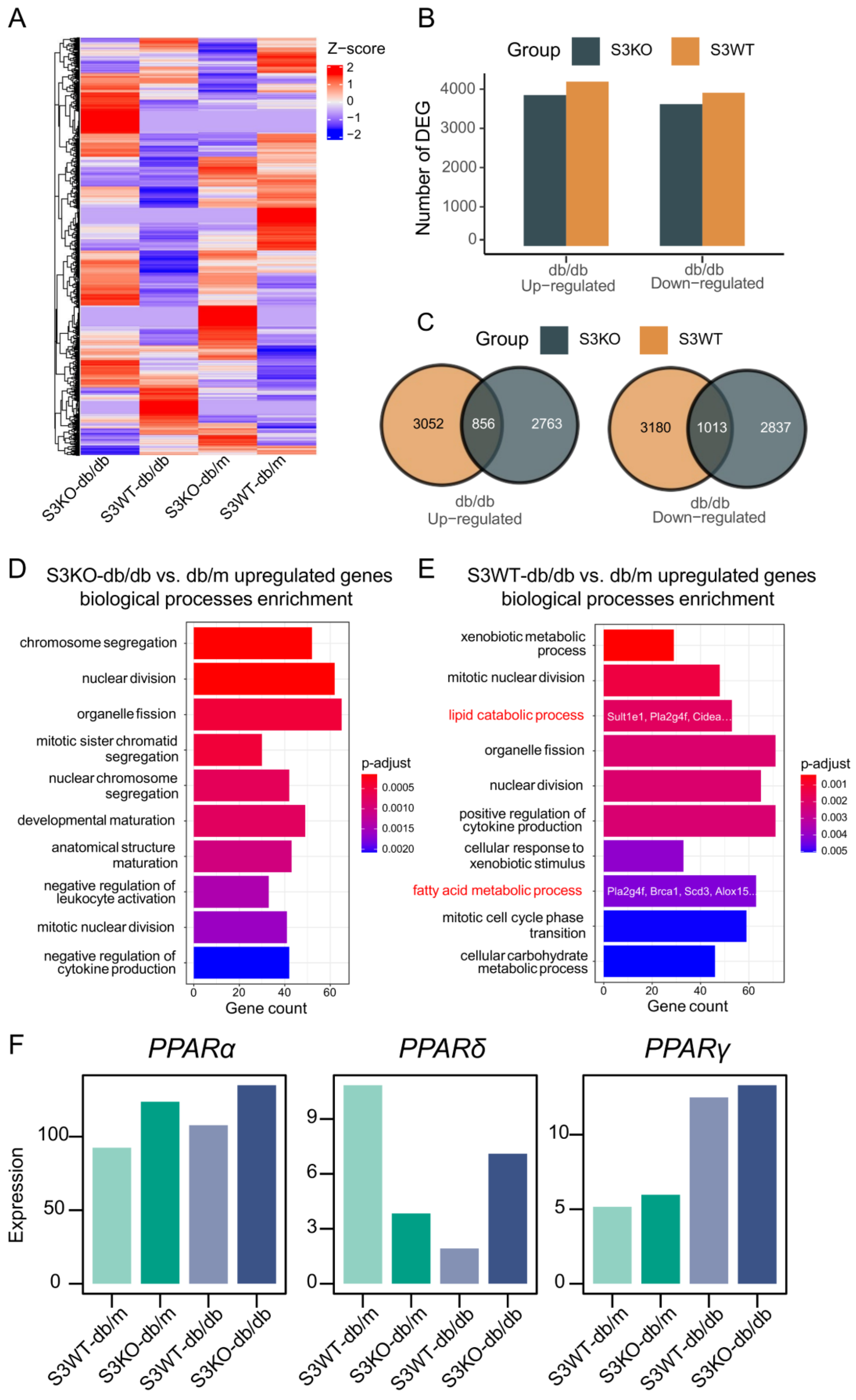
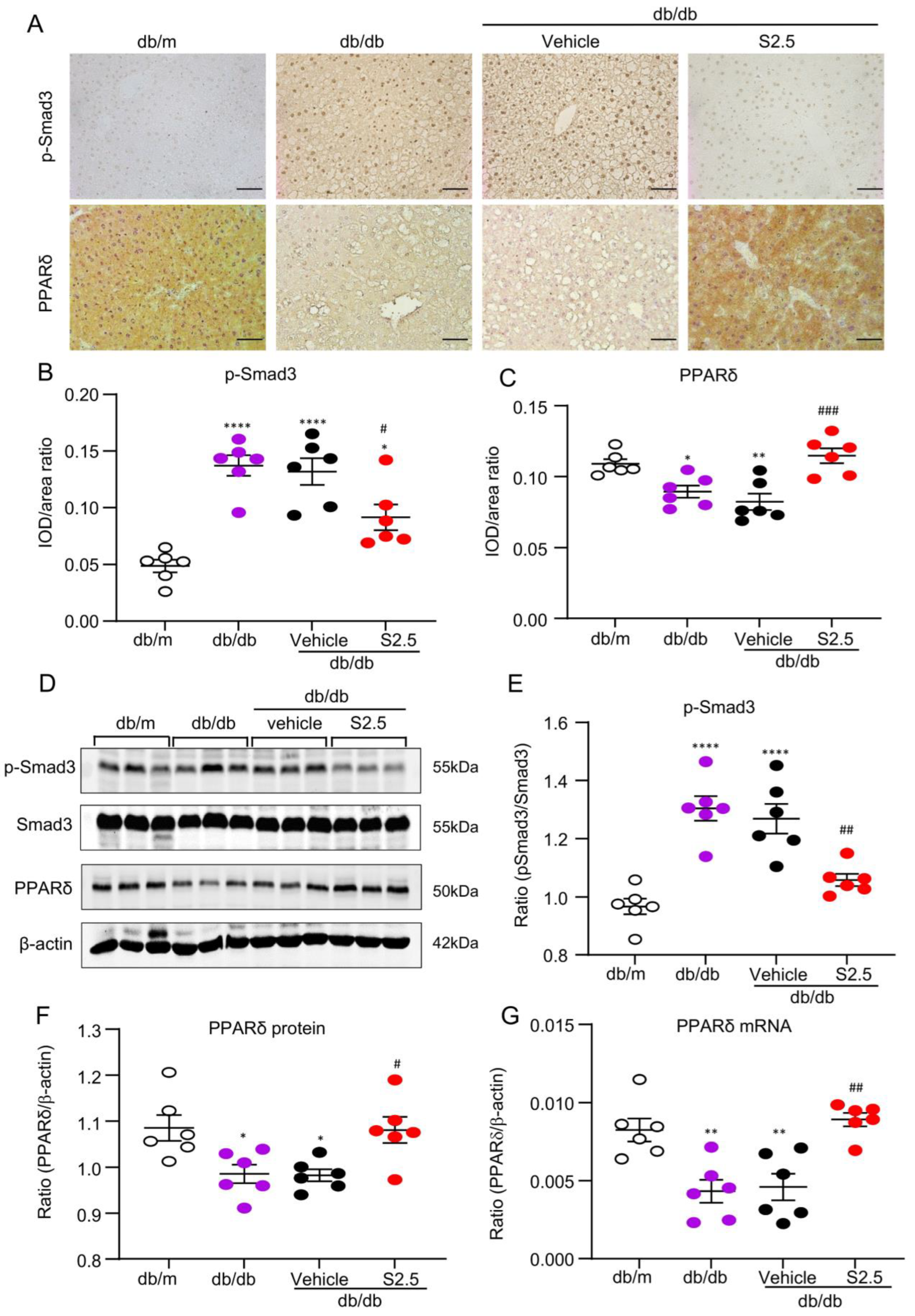
2.2. Deletion of Smad3 from db/db Mice Prevents the Development of Fatty Liver by Restoring PPARδ
2.3. Treatment with SIS3 Significantly Improves Diabetic Dyslipidemia in db/db Mice
2.4. Treatment with SIS3 Significantly Attenuates Fatty Liver in T2D Mice
2.5. Treatment with SIS3 Increases PPARδ Expression in the Liver Tissues of db/db Mice
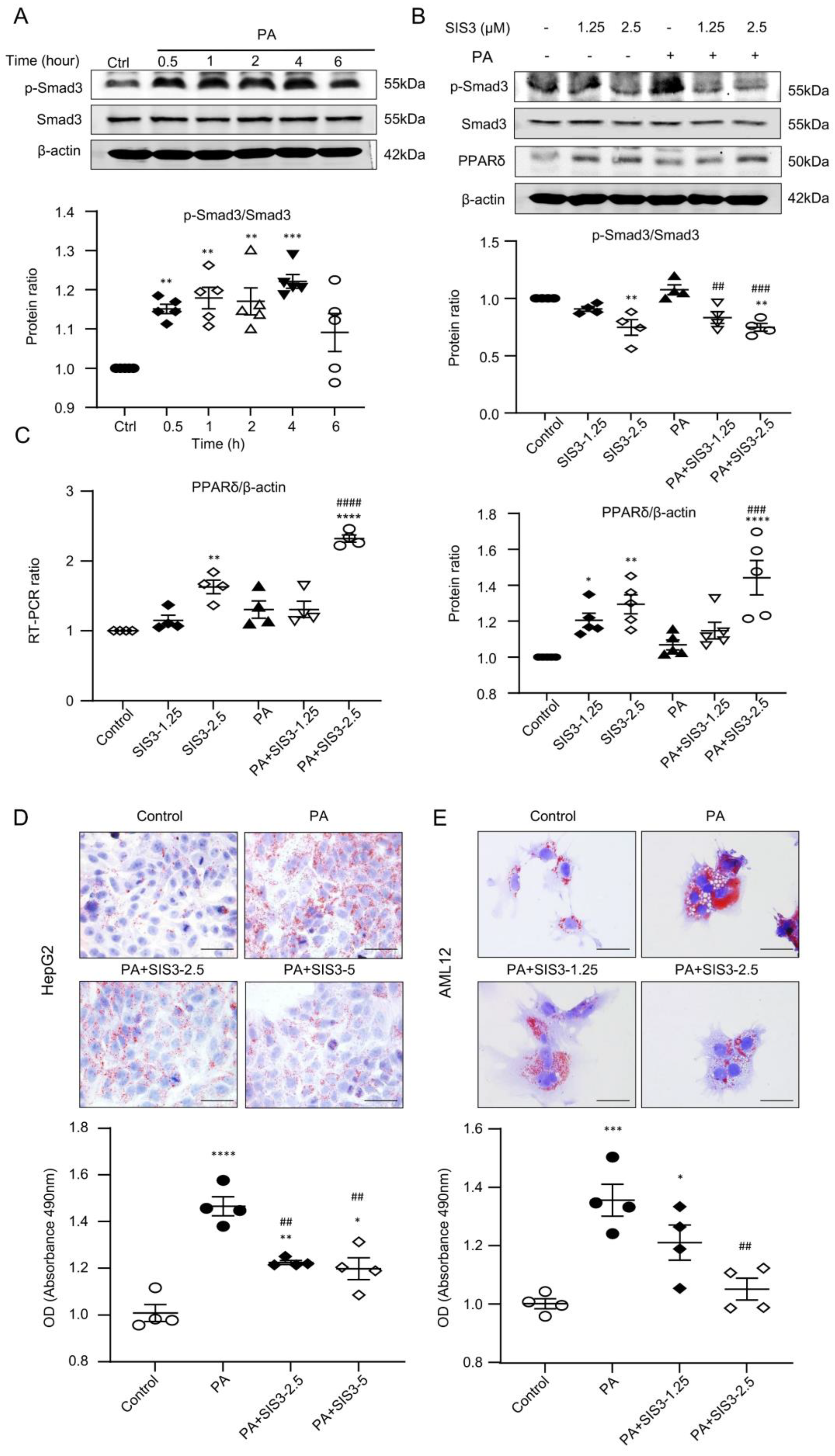
2.6. SIS3 Treatment Inhibits Fatty Liver and Reduces Lipid Accumulation by Blocking Smad3-Mediated Inhibition of PPARδ Expression In Vivo and In Vitro
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. SIS3 Treatment
4.3. Fasting Blood Glucose, Body Weight, and Weight of Liver Tissues
4.4. Glucose and Insulin Tolerance Tests
4.5. Hematoxylin and Eosin Staining and Immunohistochemistry Staining
4.6. Oil Red O Staining
4.7. BODIPY (493/503) Staining
4.8. Filipin III Staining
4.9. Measurement of Blood Lipids and Liver Functions
4.10. Western Blotting Analysis
4.11. RNA Extraction and Quantitative Real-Time PCR
4.12. Cell Culture
4.13. RNA Sequencing and Functional Enrichment Analysis
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Goldberg, R.B. Dyslipidemia in Diabetes: When and How to Treat? Endocrinol. Metab. Clin. N. Am. 2022, 51, 603–624. [Google Scholar] [CrossRef] [PubMed]
- Chehade, J.M.; Gladysz, M.; Mooradian, A.D. Dyslipidemia in type 2 diabetes: Prevalence, pathophysiology, and management. Drugs 2013, 73, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Kaze, A.D.; Santhanam, P.; Musani, S.K.; Ahima, R.; Echouffo-Tcheugui, J.B. Metabolic Dyslipidemia and Cardiovascular Outcomes in Type 2 Diabetes Mellitus: Findings from the Look AHEAD Study. J. Am. Heart Assoc. 2021, 10, e016947. [Google Scholar] [CrossRef] [PubMed]
- Hodis, H.N.; Mack, W.J.; Azen, S.P.; Alaupovic, P.; Pogoda, J.M.; LaBree, L.; Hemphill, L.C.; Kramsch, D.M.; Blankenhorn, D.H. Triglyceride- and cholesterol-rich lipoproteins have a differential effect on mild/moderate and severe lesion progression as assessed by quantitative coronary angiography in a controlled trial of lovastatin. Circulation 1994, 90, 42–49. [Google Scholar] [CrossRef]
- Mosenzon, O.; Alguwaihes, A.; Leon, J.L.A.; Bayram, F.; Darmon, P.; Davis, T.M.E.; Dieuzeide, G.; Eriksen, K.T.; Hong, T.; Kaltoft, M.S.; et al. CAPTURE: A multinational, cross-sectional study of cardiovascular disease prevalence in adults with type 2 diabetes across 13 countries. Cardiovasc. Diabetol. 2021, 20, 154. [Google Scholar] [CrossRef]
- Su, W.; Cao, R.; He, Y.C.; Guan, Y.F.; Ruan, X.Z. Crosstalk of Hyperglycemia and Dyslipidemia in Diabetic Kidney Disease. Kidney Dis. 2017, 3, 171–180. [Google Scholar] [CrossRef]
- Lemieux, I.; Lamarche, B.; Couillard, C.; Pascot, A.; Cantin, B.; Bergeron, J.; Dagenais, G.R.; Despres, J.P. Total cholesterol/HDL cholesterol ratio vs LDL cholesterol/HDL cholesterol ratio as indices of ischemic heart disease risk in men: The Quebec Cardiovascular Study. Arch. Intern. Med. 2001, 161, 2685–2692. [Google Scholar] [CrossRef]
- Zhou, M.; Zhu, L.; Cui, X.; Feng, L.; Zhao, X.; He, S.; Ping, F.; Li, W.; Li, Y. The triglyceride to high-density lipoprotein cholesterol (TG/HDL-C) ratio as a predictor of insulin resistance but not of beta cell function in a Chinese population with different glucose tolerance status. Lipids Health Dis. 2016, 15, 104. [Google Scholar] [CrossRef]
- Williamson, R.M.; Price, J.F.; Glancy, S.; Perry, E.; Nee, L.D.; Hayes, P.C.; Frier, B.M.; Van Look, L.A.; Johnston, G.I.; Reynolds, R.M.; et al. Prevalence of and risk factors for hepatic steatosis and nonalcoholic Fatty liver disease in people with type 2 diabetes: The Edinburgh Type 2 Diabetes Study. Diabetes Care 2011, 34, 1139–1144. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Targher, G.; Day, C.P. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 330–344. [Google Scholar] [CrossRef]
- Bril, F.; Cusi, K. Management of Nonalcoholic Fatty Liver Disease in Patients with Type 2 Diabetes: A Call to Action. Diabetes Care 2017, 40, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, H.L.; Liu, T.T.; Lan, H.Y. TGF-Beta as a Master Regulator of Diabetic Nephropathy. Int. J. Mol. Sci. 2021, 22, 7881. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Wang, L.; Tang, P.M.; Wang, H.L.; Li, J.C.; Xu, B.H.; Xue, V.W.; Tan, R.Z.; Jin, N.; Chan, T.F.; et al. Smad3 deficiency promotes beta cell proliferation and function in db/db mice via restoring Pax6 expression. Theranostics 2021, 11, 2845–2859. [Google Scholar] [CrossRef]
- Xu, B.H.; Sheng, J.; You, Y.K.; Huang, X.R.; Ma, R.C.W.; Wang, Q.; Lan, H.Y. Deletion of Smad3 prevents renal fibrosis and inflammation in type 2 diabetic nephropathy. Metabolism 2020, 103, 154013. [Google Scholar] [CrossRef]
- Dong, L.; Li, J.C.; Hu, Z.J.; Huang, X.R.; Wang, L.; Wang, H.L.; Ma, R.C.W.; Lan, H.Y.; Yang, S.J. Deletion of Smad3 protects against diabetic myocardiopathy in db/db mice. J. Cell Mol. Med. 2021, 25, 4860–4869. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Wei, B.; He, H.J.; Huang, X.R.; Sheng, J.Y.; Chen, X.C.; Wang, L.; Tan, R.Z.; Li, J.C.; Liu, J.; et al. Smad3 deficiency improves islet-based therapy for diabetes and diabetic kidney injury by promoting beta cell proliferation via the E2F3-dependent mechanism. Theranostics 2022, 12, 379–395. [Google Scholar] [CrossRef]
- Chen, H.Y.; Huang, X.R.; Wang, W.; Li, J.H.; Heuchel, R.L.; Chung, A.C.; Lan, H.Y. The protective role of Smad7 in diabetic kidney disease: Mechanism and therapeutic potential. Diabetes 2011, 60, 590–601. [Google Scholar] [CrossRef]
- Chung, J.Y.; Tang, P.M.; Chan, M.K.; Wang, L.; Huang, X.R.; To, K.F.; Ma, R.C.; Lan, H.Y. AANG Prevents Smad3-dependent Diabetic Nephropathy by Restoring Pancreatic beta-Cell Development in db/db Mice. Int. J. Biol. Sci. 2022, 18, 5489–5502. [Google Scholar] [CrossRef]
- Li, J.; Qu, X.; Yao, J.; Caruana, G.; Ricardo, S.D.; Yamamoto, Y.; Yamamoto, H.; Bertram, J.F. Blockade of endothelial-mesenchymal transition by a Smad3 inhibitor delays the early development of streptozotocin-induced diabetic nephropathy. Diabetes 2010, 59, 2612–2624. [Google Scholar] [CrossRef]
- Gross, B.; Pawlak, M.; Lefebvre, P.; Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 2017, 13, 36–49. [Google Scholar] [CrossRef]
- Ruan, X.; Zheng, F.; Guan, Y. PPARs and the kidney in metabolic syndrome. Am. J. Physiol. Renal Physiol. 2008, 294, F1032-47. [Google Scholar] [CrossRef] [PubMed]
- Arguello, G.; Balboa, E.; Arrese, M.; Zanlungo, S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim. Biophys. Acta 2015, 1852, 1765–1778. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Montagner, A.; Tan, N.S.; Wahli, W. Insights into the Role of PPARbeta/delta in NAFLD. Int. J. Mol. Sci. 2018, 19, 1893. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.; Aguilar-Recarte, D.; Palomer, X.; Vazquez-Carrera, M. Revealing the role of peroxisome proliferator-activated receptor beta/delta in nonalcoholic fatty liver disease. Metabolism 2021, 114, 154342. [Google Scholar] [CrossRef]
- Liu, S.; Hatano, B.; Zhao, M.; Yen, C.C.; Kang, K.; Reilly, S.M.; Gangl, M.R.; Gorgun, C.; Balschi, J.A.; Ntambi, J.M.; et al. Role of peroxisome proliferator-activated receptor {delta}/{beta} in hepatic metabolic regulation. J. Biol. Chem. 2011, 286, 1237–1247. [Google Scholar] [CrossRef]
- Tong, L.; Wang, L.; Yao, S.; Jin, L.; Yang, J.; Zhang, Y.; Ning, G.; Zhang, Z. PPARdelta attenuates hepatic steatosis through autophagy-mediated fatty acid oxidation. Cell Death Dis. 2019, 10, 197. [Google Scholar] [CrossRef]
- Bojic, L.A.; Telford, D.E.; Fullerton, M.D.; Ford, R.J.; Sutherland, B.G.; Edwards, J.Y.; Sawyez, C.G.; Gros, R.; Kemp, B.E.; Steinberg, G.R.; et al. PPARdelta activation attenuates hepatic steatosis in Ldlr-/- mice by enhanced fat oxidation, reduced lipogenesis, and improved insulin sensitivity. J. Lipid Res. 2014, 55, 1254–1266. [Google Scholar] [CrossRef]
- Wang, Y.X.; Lee, C.H.; Tiep, S.; Yu, R.T.; Ham, J.; Kang, H.; Evans, R.M. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell 2003, 113, 159–170. [Google Scholar] [CrossRef]
- Wagner, N.; Wagner, K.D. The Role of PPARs in Disease. Cells 2020, 9, 2367. [Google Scholar] [CrossRef]
- Palomer, X.; Barroso, E.; Pizarro-Delgado, J.; Pena, L.; Botteri, G.; Zarei, M.; Aguilar, D.; Montori-Grau, M.; Vazquez-Carrera, M. PPARbeta/delta: A Key Therapeutic Target in Metabolic Disorders. Int. J. Mol. Sci. 2018, 19, 913. [Google Scholar] [CrossRef]
- Lange, N.F.; Graf, V.; Caussy, C.; Dufour, J.F. PPAR-Targeted Therapies in the Treatment of Non-Alcoholic Fatty Liver Disease in Diabetic Patients. Int. J. Mol. Sci. 2022, 23, 4305. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Ding, G.; Qin, Q.; Huang, Y.; Lewis, W.; He, N.; Evans, R.M.; Schneider, M.D.; Brako, F.A.; Xiao, Y.; et al. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat. Med. 2004, 10, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Choy, L.; Derynck, R. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J. Biol. Chem. 2003, 278, 9609–9619. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.S.; Michalik, L.; Di-Poi, N.; Ng, C.Y.; Mermod, N.; Roberts, A.B.; Desvergne, B.; Wahli, W. Essential role of Smad3 in the inhibition of inflammation-induced PPARbeta/delta expression. EMBO J. 2004, 23, 4211–4221. [Google Scholar] [CrossRef]
- Tan, C.K.; Leuenberger, N.; Tan, M.J.; Yan, Y.W.; Chen, Y.; Kambadur, R.; Wahli, W.; Tan, N.S. Smad3 deficiency in mice protects against insulin resistance and obesity induced by a high-fat diet. Diabetes 2011, 60, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.; Quijano, C.; Kamaraju, A.K.; Gavrilova, O.; Malek, R.; Chen, W.; Zerfas, P.; Zhigang, D.; Wright, E.C.; Stuelten, C.; et al. Protection from obesity and diabetes by blockade of TGF-beta/Smad3 signaling. Cell Metab. 2011, 14, 67–79. [Google Scholar] [CrossRef]
- Grzych, G.; Pekar, J.D.; Chevalier-Curt, M.J.; Decoin, R.; Vergriete, P.; Henry, H.; Odou, P.; Maboudou, P.; Brousseau, T.; Vamecq, J. Antioxidants other than vitamin C may be detected by glucose meters: Immediate relevance for patients with disorders targeted by antioxidant therapies. Clin. Biochem. 2021, 92, 71–76. [Google Scholar] [CrossRef]
- Weidinger, A.; Kozlov, A.V. Biological Activities of Reactive Oxygen and Nitrogen Species: Oxidative Stress versus Signal Transduction. Biomolecules 2015, 5, 472–484. [Google Scholar] [CrossRef]
- Liu, R.M.; Desai, L.P. Reciprocal regulation of TGF-beta and reactive oxygen species: A perverse cycle for fibrosis. Redox Biol. 2015, 6, 565–577. [Google Scholar] [CrossRef]
- Lin, C.W.; Zhang, H.; Li, M.; Xiong, X.; Chen, X.; Chen, X.; Dong, X.C.; Yin, X.M. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J. Hepatol. 2013, 58, 993–999. [Google Scholar] [CrossRef]
- Ko, B.; Van Raamsdonk, J.M. RNA Sequencing of Pooled Samples Effectively Identifies Differentially Expressed Genes. Biology 2023, 12, 812. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]



| Group (n = 6) | AUC-FBG (mmol/L × min) | AUC-IPGTT (mmol/L × min) | AUC-IPITT (mmol/L × min) |
|---|---|---|---|
| db/m | 61.01 ± 8.39 | 45.14 ± 6.49 | 11.44 ± 1.73 |
| db/db | 128.33 ± 19.37 p < 0.0001 **** | 164.70 ± 7.17 p < 0.0001 **** | 102.25 ± 14.48 p < 0.0001 **** |
| db/db + vehicle | 128.82 ± 13.09 p < 0.0001 **** | 165.20 ± 8.14 p < 0.0001 **** | 109.62 ± 10.77 p < 0.0001 **** |
| db/db + S2.5 | 83.08 ± 2.58 p < 0.05 * p < 0.0001 #### | 118.47 ± 6.25 p < 0.0001 **** p < 0.0001 #### | 80.01 ± 11.57 p < 0.0001 **** p < 0.0001 #### |
| Group (n = 8) | LDH (U/L) | Body Weight (g) |
|---|---|---|
| db/m | 774.27 ± 88.11 | 25.13 ± 1.64 |
| db/db | 801.11 ± 61.97 p < 0.0001 **** | 48.75 ± 4.17 p < 0.0001 **** |
| db/db + vehicle | 780.17 ± 68.26 p < 0.0001 **** | 45.88 ± 3.27 p < 0.0001 **** |
| db/db + S2.5 | 757.06 ± 103.62 p < 0.0001 **** | 46.75 ± 1.83 p < 0.0001 **** |
| Group (n = 6) | ALT (IU/L) | AST (IU/L) |
|---|---|---|
| db/m | 10.82 ± 2.29 | 17.36 ± 4.65 |
| db/db | 93.17 ± 24.04 p < 0.0001 **** | 170.18 ± 48.17 p < 0.0001 **** |
| db/db + vehicle | 88.06 ± 21.29 p < 0.0001 **** | 169.81 ± 39.77 p < 0.0001 **** |
| db/db + S2.5 | 41.19 ± 11.49 p < 0.01 ** p < 0.001 ### | 59.17 ± 15.76 p < 0.05 * p < 0.0001 #### |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, H.; Zhong, Y.; Wang, H.; Tang, P.M.-K.; Xue, V.W.; Chen, X.; Chen, J.; Huang, X.; Wang, C.; Lan, H. Smad3 Mediates Diabetic Dyslipidemia and Fatty Liver in db/db Mice by Targeting PPARδ. Int. J. Mol. Sci. 2023, 24, 11396. https://doi.org/10.3390/ijms241411396
He H, Zhong Y, Wang H, Tang PM-K, Xue VW, Chen X, Chen J, Huang X, Wang C, Lan H. Smad3 Mediates Diabetic Dyslipidemia and Fatty Liver in db/db Mice by Targeting PPARδ. International Journal of Molecular Sciences. 2023; 24(14):11396. https://doi.org/10.3390/ijms241411396
Chicago/Turabian StyleHe, Huijun, Yu Zhong, Honglian Wang, Patrick Ming-Kuen Tang, Vivian Weiwen Xue, Xiaocui Chen, Jiaoyi Chen, Xiaoru Huang, Cheng Wang, and Huiyao Lan. 2023. "Smad3 Mediates Diabetic Dyslipidemia and Fatty Liver in db/db Mice by Targeting PPARδ" International Journal of Molecular Sciences 24, no. 14: 11396. https://doi.org/10.3390/ijms241411396
APA StyleHe, H., Zhong, Y., Wang, H., Tang, P. M.-K., Xue, V. W., Chen, X., Chen, J., Huang, X., Wang, C., & Lan, H. (2023). Smad3 Mediates Diabetic Dyslipidemia and Fatty Liver in db/db Mice by Targeting PPARδ. International Journal of Molecular Sciences, 24(14), 11396. https://doi.org/10.3390/ijms241411396








