RNAseq Analysis of Novel 1,3,4-Oxadiazole Chalcogen Analogues Reveals Anti-Tubulin Properties on Cancer Cell Lines
Abstract
1. Introduction
2. Results
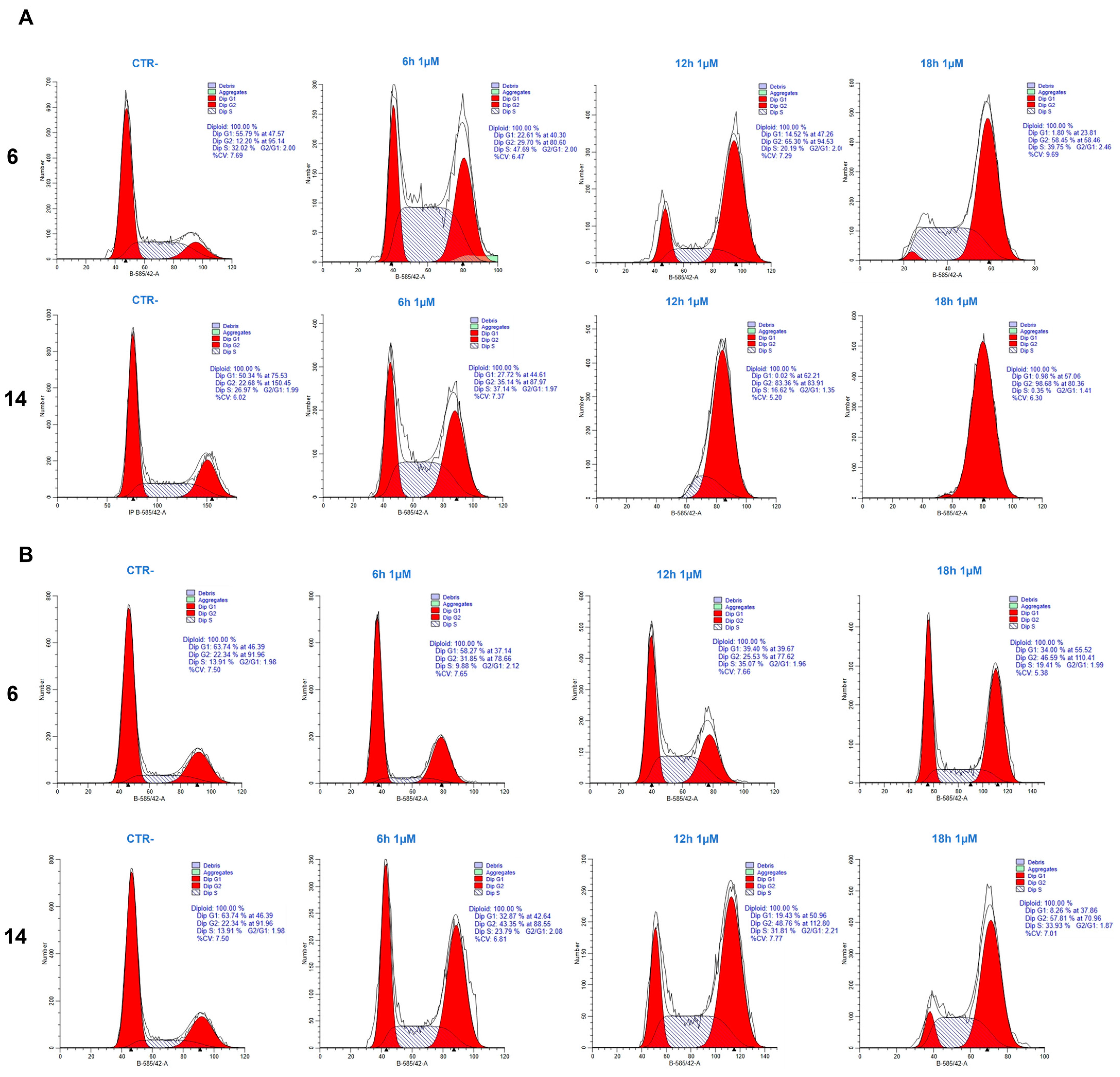
2.1. Cell Cycle Analysis
2.2. Analysis of Gene Expression after Treatment of HeLa and PC-3 Cells with Compounds 6 and 14
2.3. GO Enrichment and KEGG Pathway Analysis of DEGs
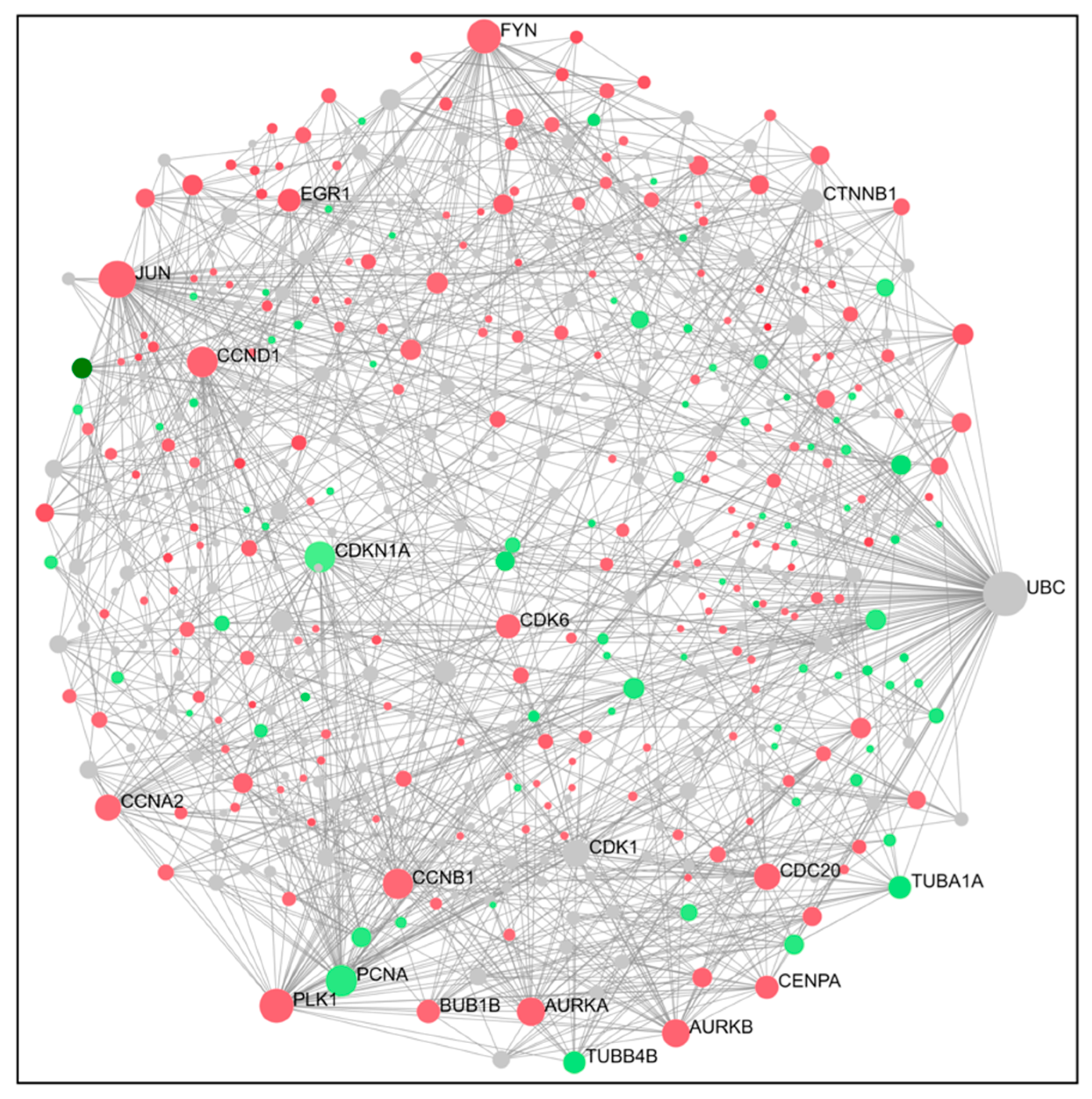
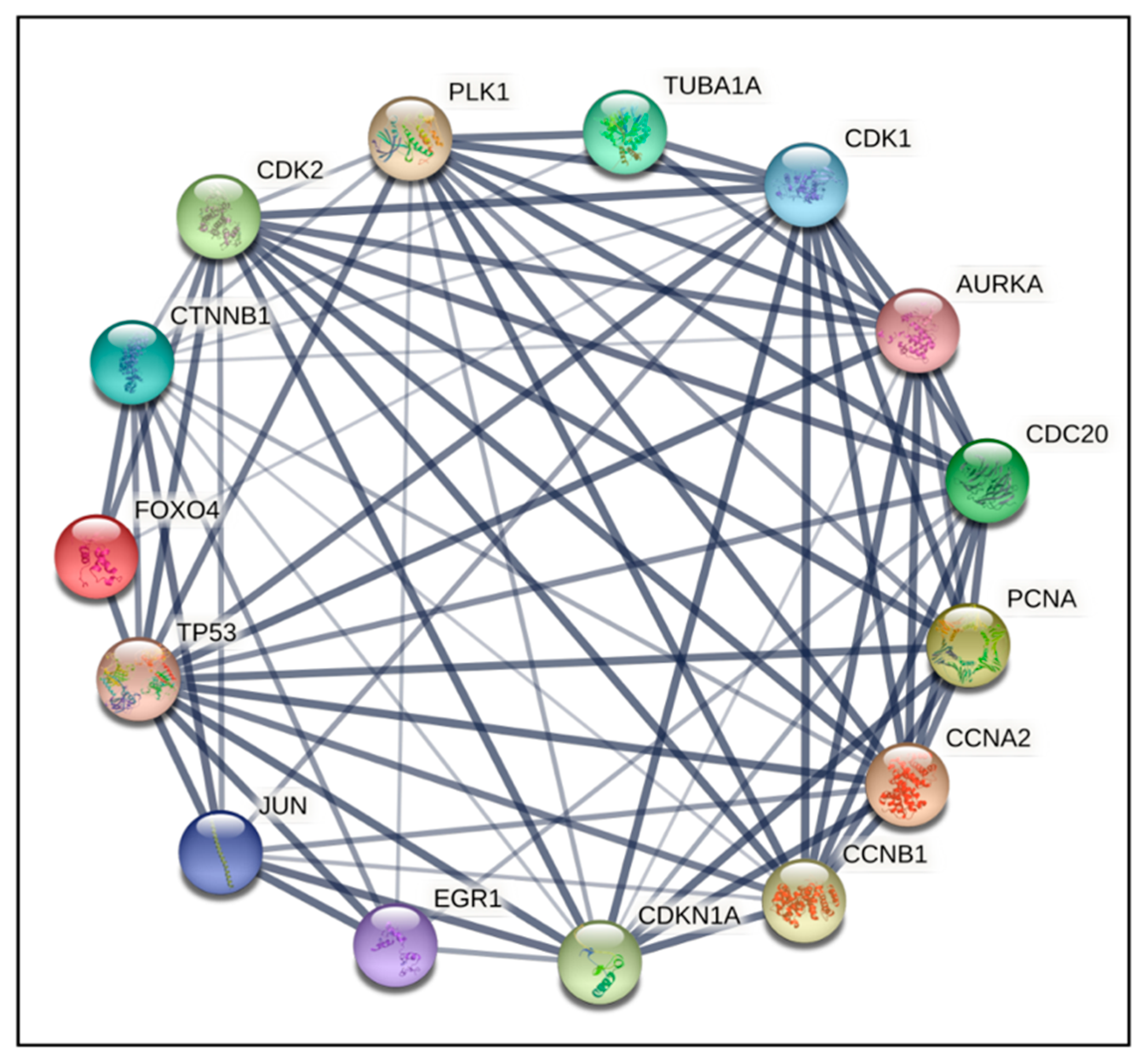
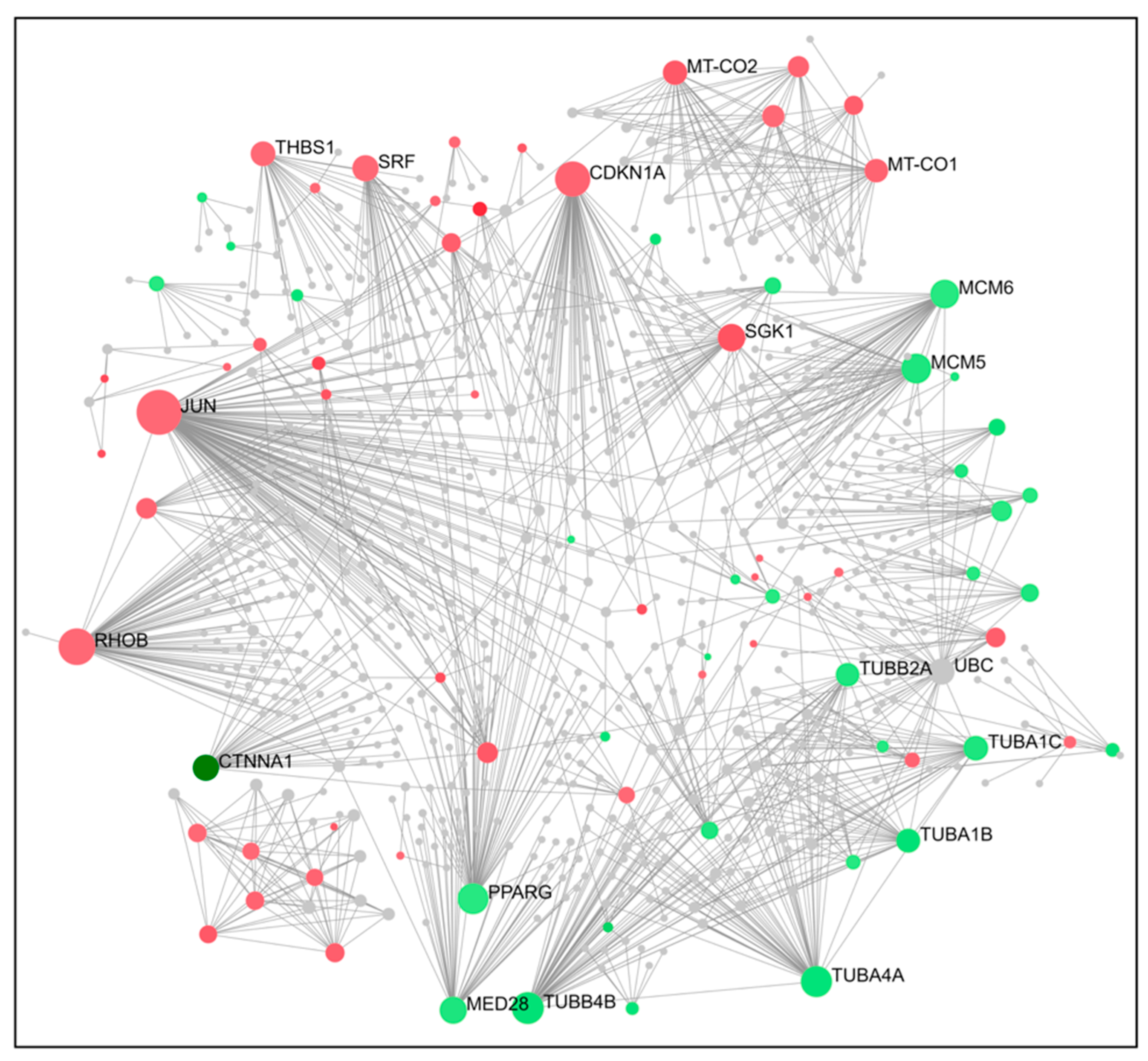
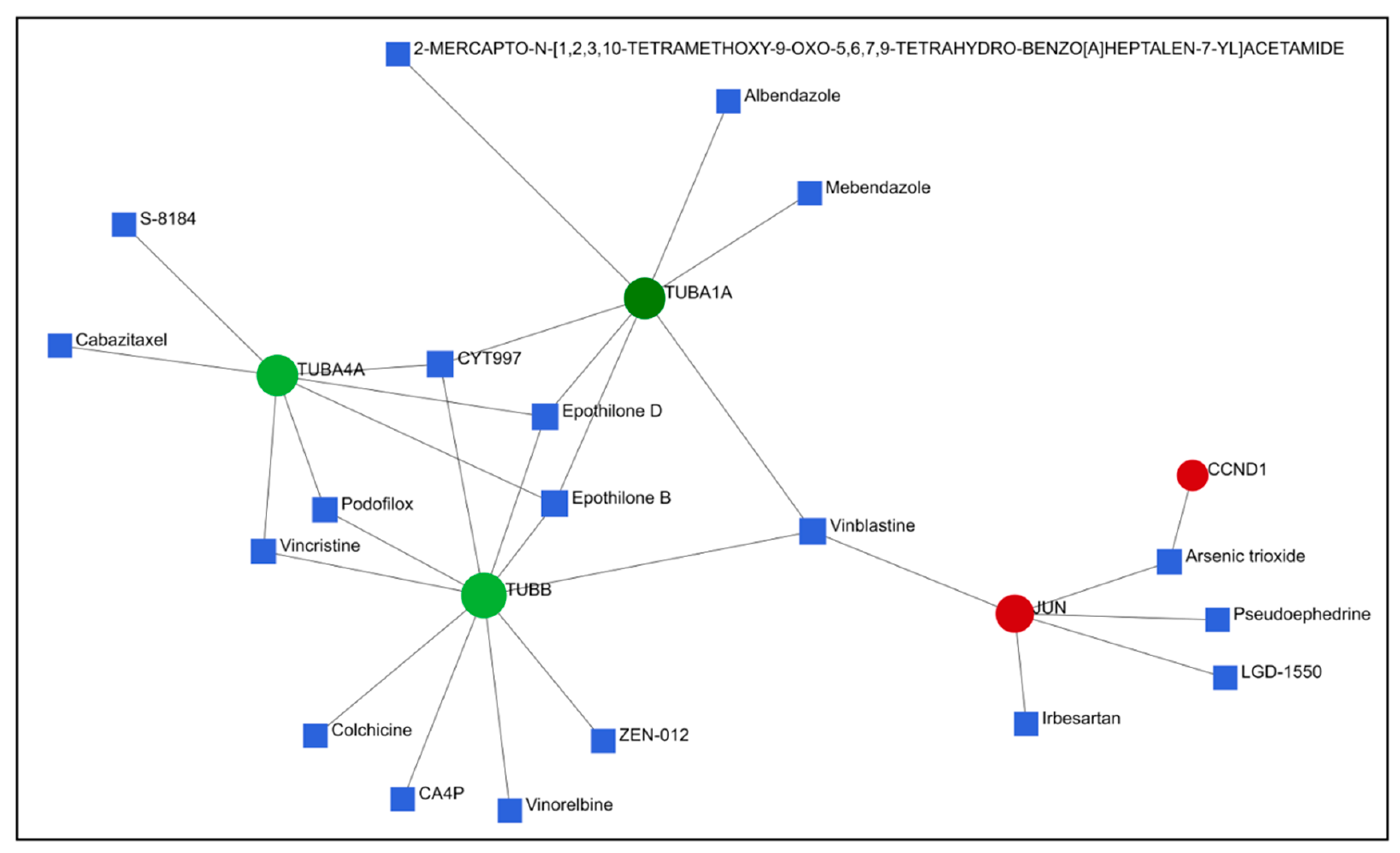
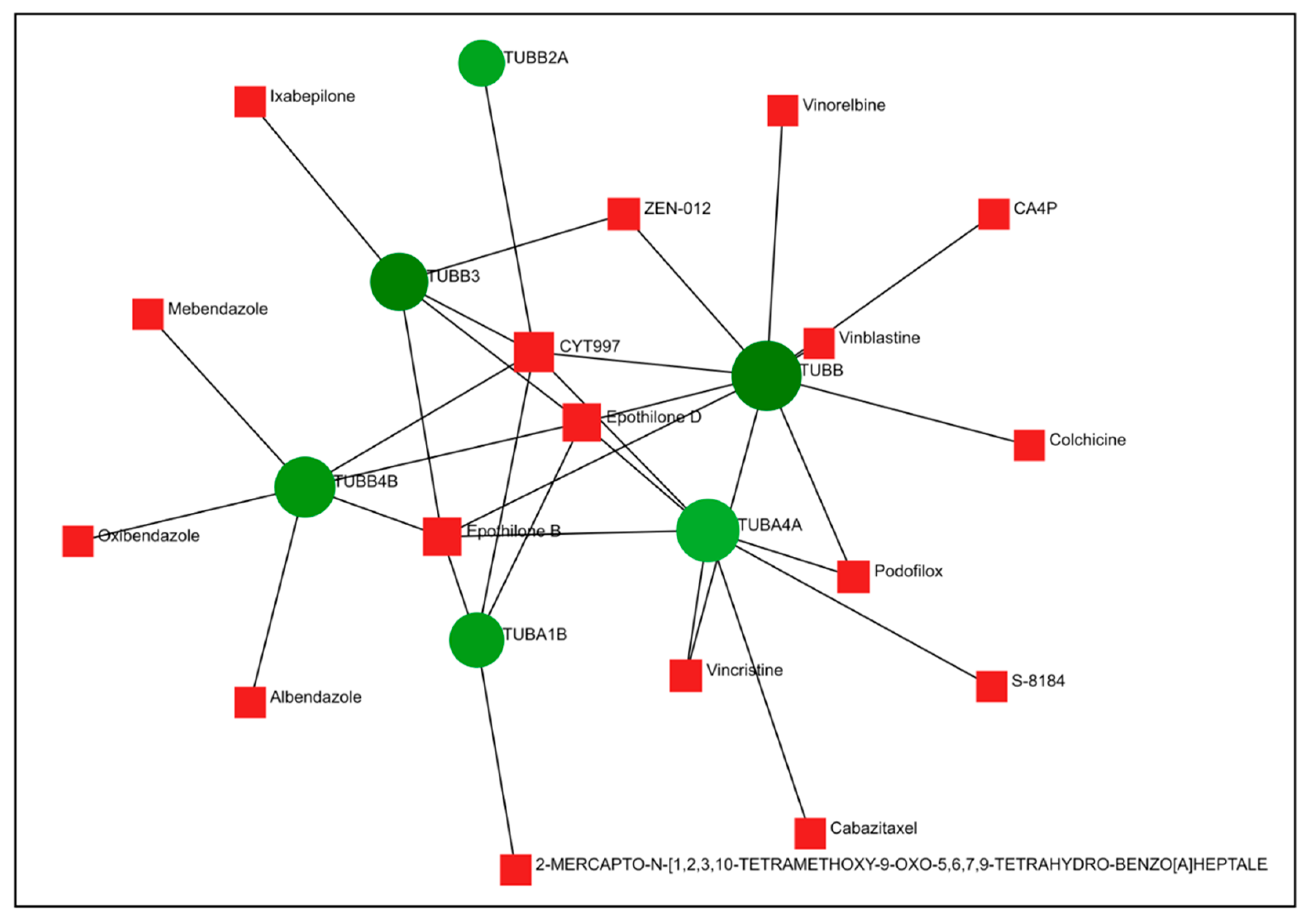
2.4. PPI Network Construction and Hub Genes Screening for Compounds 14 and 6
2.5. Comparison of Gene Expression Changes Caused by Compounds 6 and 14 with Gene Expression Profiles Induced by Treatment of Cancer Cells with Known Drugs
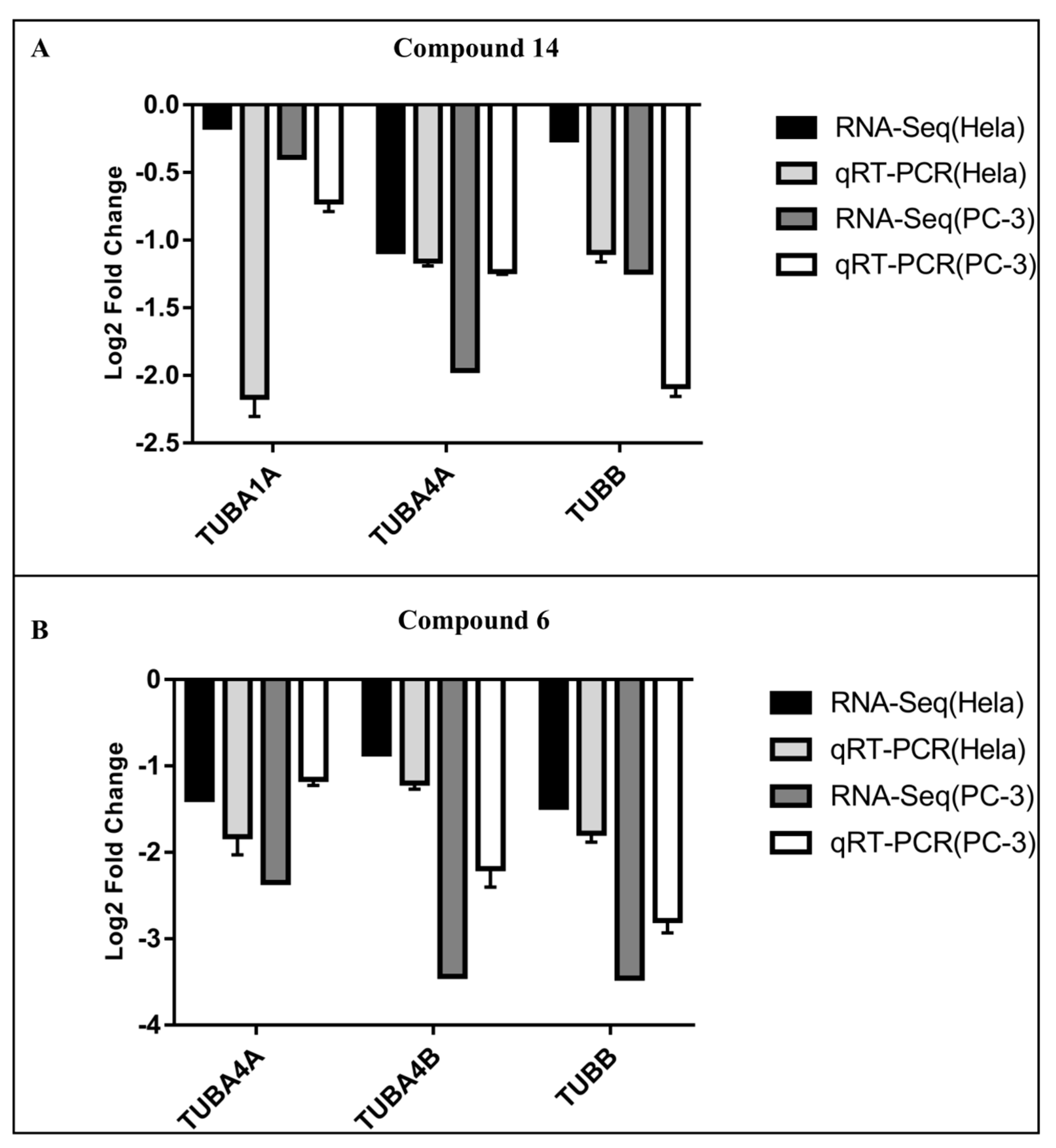
2.6. qRT-PCR Validation of Hub Genes
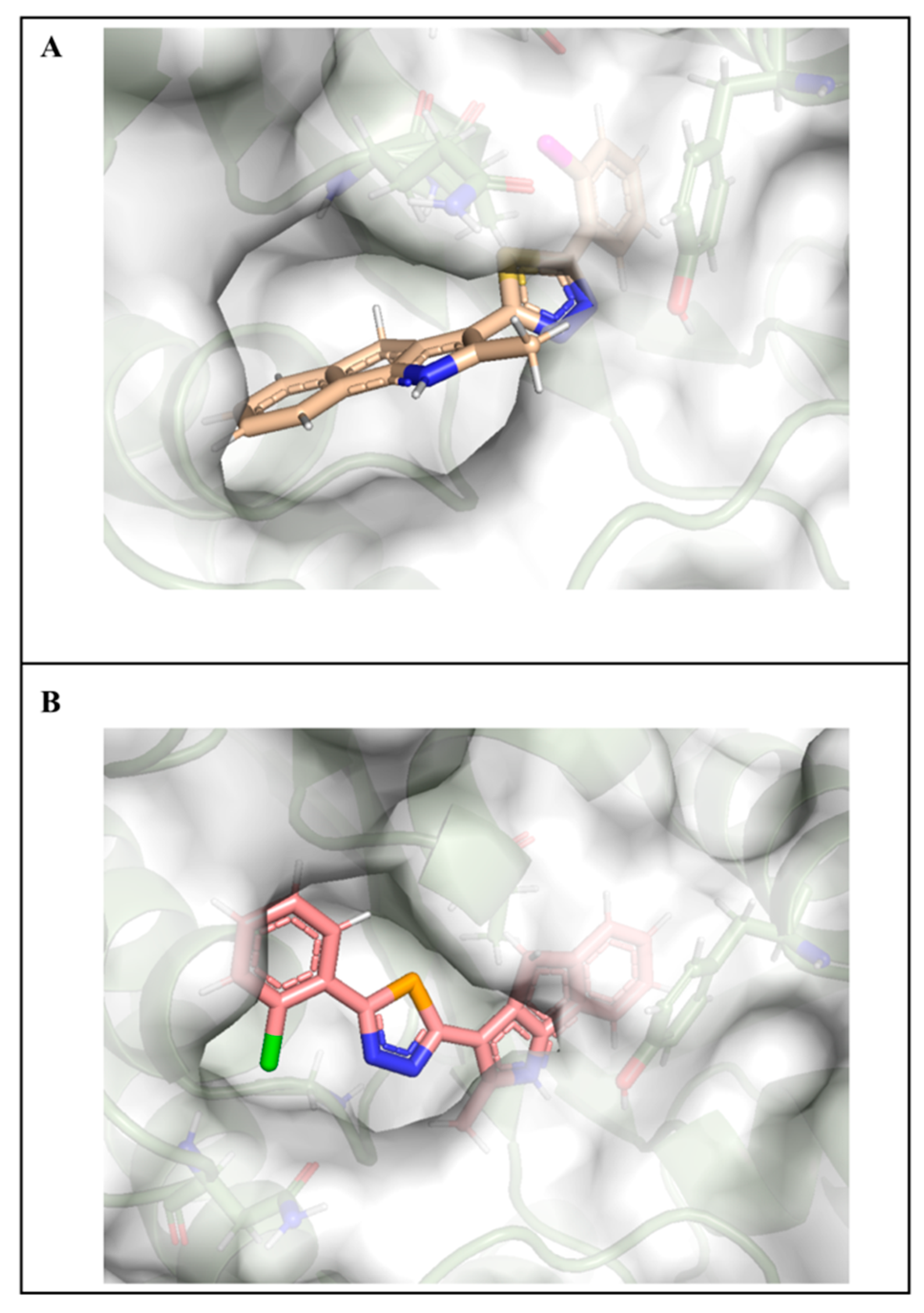
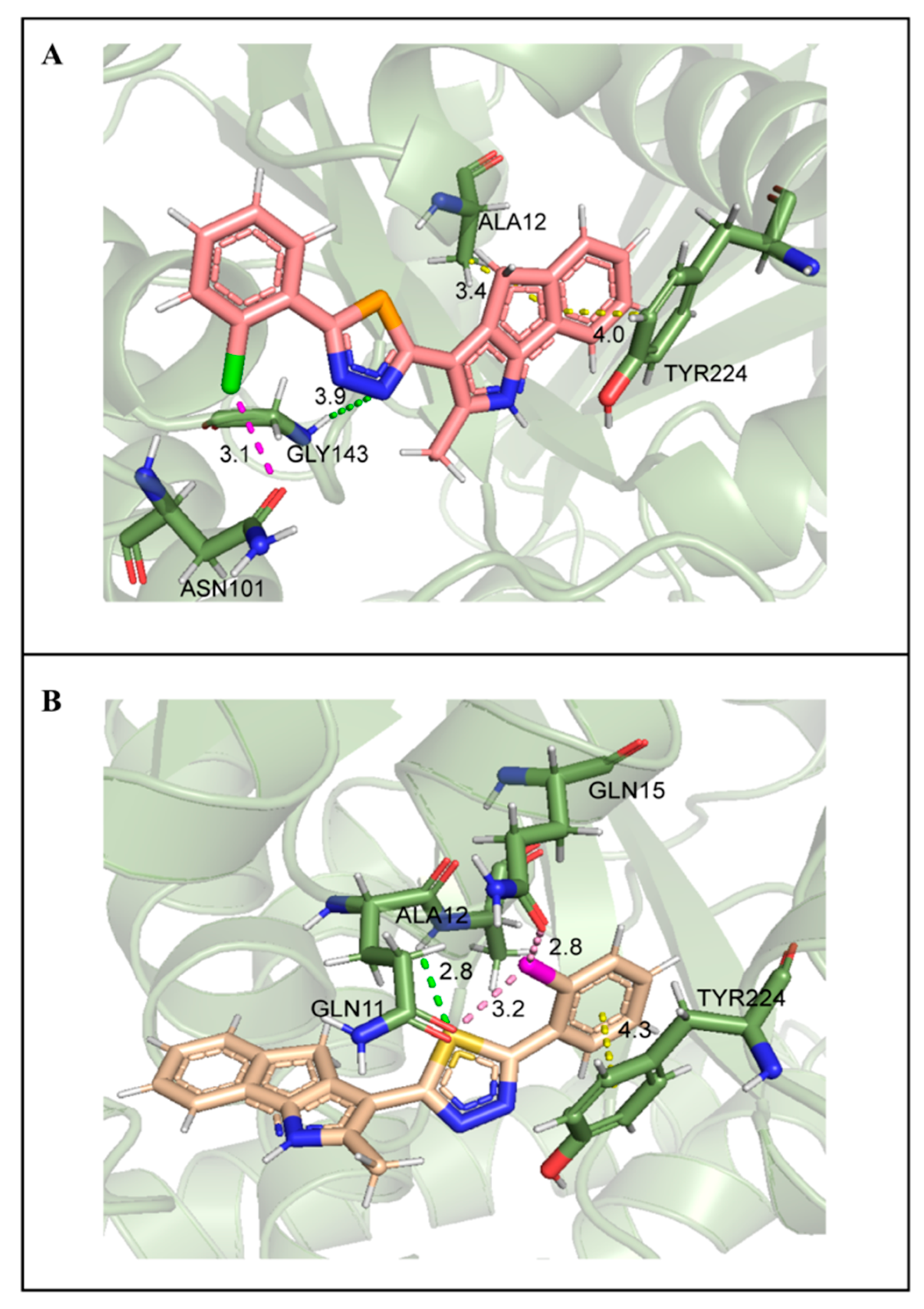
2.7. Molecular Docking Studies
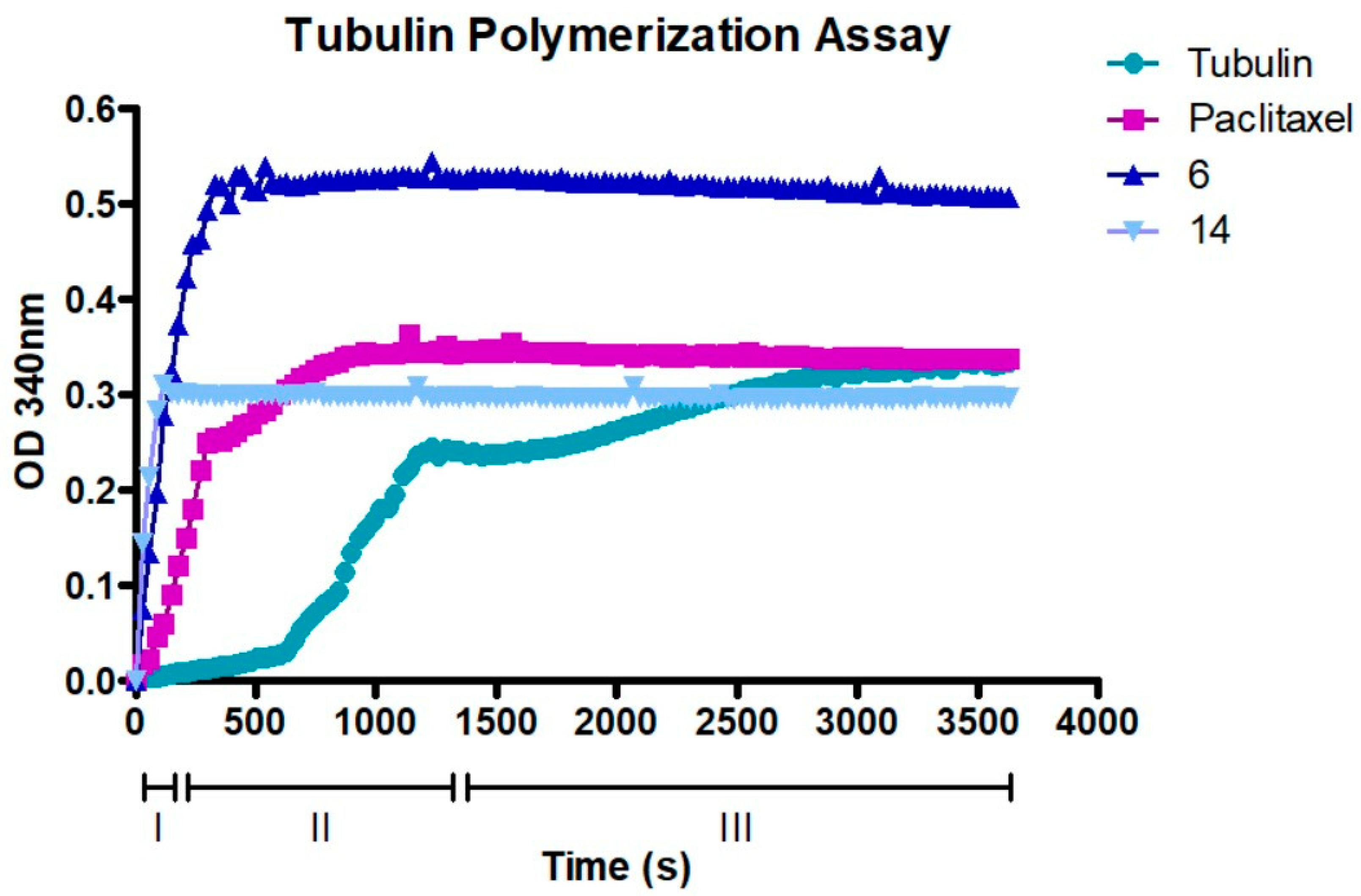
2.8. Compounds 6 and 14 Interact with the Tubulin Polymerization Process
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Cycle Analysis
4.3. RNA-Seq Library Construction and Sequencing
4.4. Identification of Differentially Expressed Genes
4.5. GO Functional Annotation Classification and KEGG Enrichment Analysis
4.6. Protein–Protein Interaction (PPI) Networks and Hub Gene Analysis
4.7. Connectivity Map and Drugbank Analysis of DEGs
4.8. qRT-PCR Validation of DEGs
4.9. Molecular Docking Verification
4.10. Tubulin Polymerization Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zoroddu, S.; Corona, P.; Sanna, L.; Borghi, F.; Bordoni, V.; Asproni, B.; Pinna, G.A.; Bagella, L.; Murineddu, G. Novel 1,3,4-oxadiazole chalcogen analogues: Synthesis and cytotoxic activity. Eur. J. Med. Chem. 2022, 238, 114440. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Goldman, R.D. Overview of the Cytoskeleton from an Evolutionary Perspective. Cold Spring Harb. Perspect. Biol. 2018, 10, a030288. [Google Scholar] [CrossRef]
- Goodson, H.V.; Jonasson, E.M. Microtubules and Microtubule-Associated Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10, a022608. [Google Scholar] [CrossRef]
- Kavallaris, M. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer 2010, 10, 194–204. [Google Scholar] [CrossRef]
- Barlan, K.; Gelfand, V.I. Microtubule-Based Transport and the Distribution, Tethering, and Organization of Organelles. Cold Spring Harb. Perspect. Biol. 2017, 9, a025817. [Google Scholar] [CrossRef]
- Kaul, R.; Risinger, A.L.; Mooberry, S.L. Microtubule-Targeting Drugs: More than Antimitotics. J. Nat. Prod. 2019, 82, 680–685. [Google Scholar] [CrossRef]
- Nieddu, V.; Pinna, G.; Marchesi, I.; Sanna, L.; Asproni, B.; Pinna, G.A.; Bagella, L.; Murineddu, G. Synthesis and Antineoplastic Evaluation of Novel Unsymmetrical 1,3,4-Oxadiazoles. J. Med. Chem. 2016, 59, 10451–10469. [Google Scholar] [CrossRef]
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Rohena, C.C.; Mooberry, S.L. Recent progress with microtubule stabilizers: New compounds, binding modes and cellular activities. Nat. Prod. Rep. 2014, 31, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Sanna, L.; Piredda, R.; Marchesi, I.; Bordoni, V.; Forcales, S.V.; Calvisi, D.F.; Bagella, L. “Verteporfin exhibits anti-proliferative activity in embryonal and alveolar rhabdomyosarcoma cell lines”. Chem. Biol. Interact. 2019, 312, 108813. [Google Scholar] [CrossRef] [PubMed]
- Dumontet, C.; Jordan, M.A. Microtubule-binding agents: A dynamic field of cancer therapeutics. Nat. Rev. Drug Discov. 2010, 9, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Tangutur, A.D.; Kumar, D.; Krishna, K.V.; Kantevari, S. Microtubule Targeting Agents as Cancer Chemotherapeutics: An Overview of Molecular Hybrids as Stabilizing and Destabilizing Agents. Curr. Top. Med. Chem. 2017, 17, 2523–2537. [Google Scholar] [CrossRef] [PubMed]
- Haanstra, J.R.; Bakker, B.M. Drug target identification through systems biology. Drug Discov. Today Technol. 2015, 15, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Weidong, L.; Sanna, L.; Bordoni, V.; Tiansheng, Z.; Chengxun, L.; Murineddu, G.; Pinna, G.A.; Kelvin, D.J.; Bagella, L. Target identification of a novel unsymmetrical 1,3,4-oxadiazole derivative with antiproliferative properties. J. Cell Physiol. 2021, 236, 3789–3799. [Google Scholar] [CrossRef] [PubMed]
- Arnst, K.E.; Wang, Y.; Hwang, D.-J.; Xue, Y.; Costello, T.; Hamilton, D.; Chen, Q.; Yang, J.; Park, F.; Dalton, J.T.; et al. Potent, Metabolically Stable Tubulin Inhibitor Targets the Colchicine Binding Site and Overcomes Taxane Resistance. Cancer Res. 2018, 78, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Urvoas, A.; Cantos-Fernandes, S.; Aumont-Nicaise, M.; Arteni, A.-A.; Velours, C.; Valerio-Lepiniec, M.; Dreier, B.; Plückthun, A.; Pilon, A.; et al. Insight into microtubule nucleation from tubulin-capping proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 9859–9864. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M. Anti-Cancer Agents in Medicinal Chemistry; Bentham Science: Abu Dhabi, United Arab Emirates, 2018; Volume 10, pp. 1–15. [Google Scholar]
- Conde, C.; Cáceres, A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 2009, 10, 319–332. [Google Scholar] [CrossRef]
- Sun, X.; Li, F.; Dong, B.; Suo, S.; Liu, M.; Li, D.; Zhou, J. Regulation of tumor angiogenesis by the microtubule-binding protein CLIP-170. Protein Cell 2013, 4, 266–276. [Google Scholar] [CrossRef]
- Glomb, T.; Karolina, S.; Piotr, Ś. Anti-Cancer Activity of Derivatives of 1,3,4-Oxadiazole. Molecules 2018, 23, 3361. [Google Scholar] [CrossRef]
- Bakr, R.B. Design and synthesis of new egfr-tyrosine kinase inhibitors containing pyrazolo[3,4-d]pyrimidine cores as anticancer agents. Bull. Pharm. Sci. Assiut 2012, 35, 27–42. [Google Scholar] [CrossRef]
- Bhanushali, U.; Kalekar-Joshi, S.; Kulkarni-Munshi, R.; Yellanki, S.; Medishetty, R.; Kulkarni, P.; Chelakara, R.S. Design, Synthesis and Evaluation of 5-pyridin-4-yl-2-thioxo-[1,3,4]oxadiazol-3-yl Derivatives as Anti-angiogenic Agents Targeting VEGFR-2. Anticancer. Agents Med. Chem. 2017, 17, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhu, H.; Yang, Z.-M.; Zhu, H.-L. Synthesis, molecular modeling and biological evaluation of 2-aminomethyl-5-(quinolin-2-yl)-1,3,4-oxadiazole-2(3H)-thione quinolone derivatives as novel anticancer agent. Eur. J. Med. Chem. 2013, 60, 23–28. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Xiao, M.; Li, W.; Miller, D.D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm. Res. 2012, 29, 2943–2971. [Google Scholar] [CrossRef] [PubMed]
- Bondallaz, P.; Barbier, A.; Soehrman, S.; Grenningloh, G.; Riederer, B.M. The control of microtubule stability in vitro and in transfected cells by MAP1B and SCG10. Cell Motil. Cytoskelet. 2006, 63, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Buscaglia, G.; Northington, K.R.; Moore, J.K.; Bates, E.A. Reduced TUBA1A Tubulin Causes Defects in Trafficking and Impaired Adult Motor Behavior. eNeuro 2020, 7. [Google Scholar] [CrossRef]
- Sobierajska, K.; Ciszewski, W.M.; Wawro, M.E.; Wieczorek-Szukała, K.; Boncela, J.; Papiewska-Pajak, I.; Niewiarowska, J.; Kowalska, M.A. TUBB4B Downregulation Is Critical for Increasing Migration of Metastatic Colon Cancer Cells. Cells 2019, 8, 810. [Google Scholar] [CrossRef]
- Zhang, W.; Dong, Y.; Sartor, O.; Zhang, K. Deciphering the Increased Prevalence of TP53 Mutations in Metastatic Prostate Cancer. Cancer Inform. 2022, 21, 11769351221087046. [Google Scholar] [CrossRef]
- Kuno, I.; Takayanagi, D.; Asami, Y.; Murakami, N.; Matsuda, M.; Shimada, Y.; Kato, T. TP53 mutants and non-HPV16/18 genotypes are poor prognostic factors for concurrent chemoradiotherapy in locally advanced cervical cancer. Sci. Rep. 2021, 11, 19261. [Google Scholar] [CrossRef]
- Izadi, S.; Nikkhoo, A.; Hojjat-Farsangi, M.; Namdar, A.; Azizi, G.; Mohammadi, H.; Yousefi, M.; Jadidi-Niaragh, F. CDK1 in Breast Cancer: Implications for Theranostic Potential. Anticancer. Agents Med. Chem. 2020, 20, 758–767. [Google Scholar] [CrossRef]
- Nyberg, T.; Tischkowitz, M.; Antoniou, A.C. BRCA1 and BRCA2 pathogenic variants and prostate cancer risk: Systematic review and meta-analysis. Br. J. Cancer 2022, 126, 1067–1081. [Google Scholar] [CrossRef]
- Hatziieremia, S.; Mohammed, Z.; McCall, P.; Willder, J.M.; Roseweir, A.K.; Underwood, M.A.; Edwards, J. Loss of signal transducer and activator of transcription 1 is associated with prostate cancer recurrence. Mol. Carcinog. 2016, 55, 1667–1677. [Google Scholar] [CrossRef]
- Dogra, N.; Kumar, A.; Mukhopadhyay, T. Fenbendazole acts as a moderate microtubule destabilizing agent and causes cancer cell death by modulating multiple cellular pathways. Sci. Rep. 2018, 8, 11926. [Google Scholar] [CrossRef] [PubMed]
- Rushworth, L.K.; Hewit, K.; Munnings-Tomes, S.; Somani, S.; James, D.; Shanks, E.; Dufès, C.; Straube, A.; Patel, R.; Leung, H.Y. Repurposing screen identifies mebendazole as a clinical candidate to synergise with docetaxel for prostate cancer treatment. Br. J. Cancer 2020, 122, 517–527. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, E.C.; O’Boyle, N.M. Colchicine-Binding Site Inhibitors from Chemistry to Clinic: A Review. Pharmaceuticals 2020, 13, 8, Erratum in Pharmaceuticals 2020, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Lafanechère, L. The microtubule cytoskeleton: An old validated target for novel therapeutic drugs. Front. Pharmacol. 2022, 13, 969183. [Google Scholar] [CrossRef]


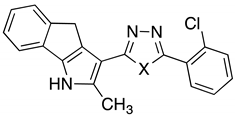
| Compound | X | Compound | X |
|---|---|---|---|
| 6 | S | 14 | Se |
| Compound | X | IC50 HeLa (µM) | IC50 PC-3 (µM) |
|---|---|---|---|
| 6 | S | 0.145 ± 0.06 | 0.176 ± 0.08 |
| 14 | Se | 0.005 ± 0.03 | 0.067 ± 0.06 |
| Name | Belongs to | Score | |
|---|---|---|---|
| 14 | fenbendazole | Tubulin inhibitor | 99.81 |
| oxibendazole | Tubulin inhibitor | 99.81 | |
| flubendazole | Tubulin inhibitor | 99.8 | |
| mebendazole | Tubulin inhibitor | 99.78 | |
| vincristine | Tubulin inhibitor | 99.68 | |
| 6 | mebendazole | Tubulin inhibitor | 99.87 |
| vindesine | Tubulin inhibitor | 99.87 | |
| fenbendazole | Tubulin inhibitor | 99.82 | |
| oxibendazole | Tubulin inhibitor | 99.81 | |
| flubendazole | Tubulin inhibitor | 99.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zoroddu, S.; Sanna, L.; Bordoni, V.; Lyu, W.; Murineddu, G.; Pinna, G.A.; Forcales, S.V.; Sala, A.; Kelvin, D.J.; Bagella, L. RNAseq Analysis of Novel 1,3,4-Oxadiazole Chalcogen Analogues Reveals Anti-Tubulin Properties on Cancer Cell Lines. Int. J. Mol. Sci. 2023, 24, 11263. https://doi.org/10.3390/ijms241411263
Zoroddu S, Sanna L, Bordoni V, Lyu W, Murineddu G, Pinna GA, Forcales SV, Sala A, Kelvin DJ, Bagella L. RNAseq Analysis of Novel 1,3,4-Oxadiazole Chalcogen Analogues Reveals Anti-Tubulin Properties on Cancer Cell Lines. International Journal of Molecular Sciences. 2023; 24(14):11263. https://doi.org/10.3390/ijms241411263
Chicago/Turabian StyleZoroddu, Stefano, Luca Sanna, Valentina Bordoni, Weidong Lyu, Gabriele Murineddu, Gerard A. Pinna, Sonia Vanina Forcales, Arturo Sala, David J. Kelvin, and Luigi Bagella. 2023. "RNAseq Analysis of Novel 1,3,4-Oxadiazole Chalcogen Analogues Reveals Anti-Tubulin Properties on Cancer Cell Lines" International Journal of Molecular Sciences 24, no. 14: 11263. https://doi.org/10.3390/ijms241411263
APA StyleZoroddu, S., Sanna, L., Bordoni, V., Lyu, W., Murineddu, G., Pinna, G. A., Forcales, S. V., Sala, A., Kelvin, D. J., & Bagella, L. (2023). RNAseq Analysis of Novel 1,3,4-Oxadiazole Chalcogen Analogues Reveals Anti-Tubulin Properties on Cancer Cell Lines. International Journal of Molecular Sciences, 24(14), 11263. https://doi.org/10.3390/ijms241411263







