Abstract
Aristolochia manshuriensis is a relic liana, which is widely used in traditional Chinese herbal medicine and is endemic to the Manchurian floristic region. Since this plant is rare and slow-growing, alternative sources of its valuable compounds could be explored. Herein, we established hairy root cultures of A. manshuriensis transformed with Agrobacterium rhizogenes root oncogenic loci (rol)B and rolC genes. The accumulation of nitrogenous secondary metabolites significantly improved in transgenic cell cultures. Specifically, the production of magnoflorine reached up to 5.72 mg/g of dry weight, which is 5.8 times higher than the control calli and 1.7 times higher than in wild-growing liana. Simultaneously, the amounts of aristolochic acids I and II, responsible for the toxicity of Aristolochia species, decreased by more than 10 fold. Consequently, the hairy root extracts demonstrated pronounced cytotoxicity against human glioblastoma cells (U-87 MG), cervical cancer cells (HeLa CCL-2), and colon carcinoma (RKO) cells. However, they did not exhibit significant activity against triple-negative breast cancer cells (MDA-MB-231). Our findings suggest that hairy root cultures of A. manshuriensis could be considered for the rational production of valuable A. manshuriensis compounds by the modification of secondary metabolism.
1. Introduction
Aristolochia L. belongs to a large genus of Aristolochiaceae, consisting of about 500 species, mainly distributed in tropical and subtropical regions [1]. Extensive research has been conducted on the unique pollination mechanisms employed by Aristolochia species, which involve a floral aroma and a perianth that entraps insects. Additionally, these species exhibit a species-specific relationship as host plants that serve as a habitat for swallowtail butterfly larvae [2,3,4]. The Aristolochia genus has garnered significant interest as a source of naturally occurring compounds with potential pharmaceutical properties. These compounds include nitrogen-containing secondary metabolites, flavonoids, lignans, terpenoids, and steroids. Consequently, Aristolochia plants are considered “natural factories” for producing these phytochemicals [5]. For centuries, Aristolochia species have been utilized in traditional medicine across Asia to treat various ailments, such as arthritis, rheumatism, angina, and myocardial infarction, alone or in combination with other herbs [6]. Studies have also suggested their potential as analgesics, antimalarials, antibacterial agents, and antidotes for snake bites [6,7]. However, certain Aristolochia species have been associated with an elevated risk of urinary tract cancer and renal interstitial fibrosis [7,8,9]. Consequently, their use in herbal medicinal products or dietary supplements has been banned [7,8].
The relict plant A. manshuriensis Kom. is an endemic of the Manchurian floristic region, native to Korea, China, and the southeastern part of Russia [10]. The dried stems of A. manshuriensis, known as Guan Mu Tong, are extensively used in traditional Chinese medicine [11]. Aristolochic acids (AAs) I, II, IIIa, IV, aristoloside, aristolamides, aristolactams, and alkaloids such as magnoflorine, manshurienine, tetrandrine, and aristopyridinone have been identified as the primary chemical constituents of A. manshuriensis [12,13,14]. These compounds are derived from L-tyrosine via (S)-norcoclaurine, with (S)-reticuline serving as the pivotal intermediate in the biosynthetic pathway for both phenanthrene derivatives and alkaloids [15]. Though A. manshuriensis plants contain AAs that cause nephrotoxicity and mutagenic and carcinogenic effects after prolonged use [16,17,18], it also accumulates various substances with excellent therapeutic potential. Aristolactam-type compounds from A. manshuriensis possess anti-tumor potential as inhibitors of cyclin-dependent kinase 2 [19]. Aristolamide II isolated from stems of A. manshuriensis showed an anti-inflammatory effect against superoxide anion production and human neutrophils releasing elastase. At the same time, AAs had no impact on either of the inflammatory mediators [13]. The polar quaternary aporphine magnoflorine is an important alkaloid with growing interest because of its broad spectrum of pharmacological properties, including anti-diabetic, anti-inflammatory, neuropsychopharmacological, immunomodulatory, hypotensive, antioxidant, and antifungal activities [20]. Magnoflorine reduced ischemia-induced neuronal damage in the cerebral cortex of rats, possibly accompanied by antioxidative stress, autophagy suppression, and Sirt1/AMPK pathway activation [21]. For practical purposes, it is crucial to obtain A. manshuriensis plant material containing high levels of beneficial compounds while minimizing harmful ones.
Propagation of Aristolochia is conducted through vegetative plant parts, seeds, or in vitro regeneration. These processes are time-consuming and labor-intensive. Though in vitro regeneration methods have been developed for A. indica [22], A. cathcartii [23], and A. tagala [24], pollination and fertilization in these plants cause weak fruit sets. Other limitations also include a small population, anthropogenic factors, and the absence of seed-dispersing agents [25,26]. Moreover, A. manshuriensis populations are affected by genetic drift due to a decline in the reproductive and effective population sizes, mainly linked to anthropogenic factors [27]. Therefore, developing an alternative plant biomass source is an exceedingly urgent task. In this regard, plant tissue culture techniques could become a potential tool for producing secondary metabolites of A. manshuriensis.
Plant cell cultures techniques, such as callus and cell suspension, have emerged as potential tools for producing secondary metabolites from medicinal plants. They can be used for the industrial and commercial production of essential plant chemicals because of their rapid growth and better productivity [28]. In vitro cultivation is an effective method for preserving rare and endangered natural species and protecting biodiversity. For example, callus cultures derived from leaf explants of A. indica and A. bracteolata have been shown to produce AA-I and AA-II [29]. Similarly, callus cultures of A. tuberosa have been found to produce 1.8 times more AAs than the original plant organs [30]. In addition, genetic transformation has been used to modify secondary metabolism and rationally produce valuable Aristolochia compounds. For instance, a suspension culture of A. manshuriensis transformed by wild-type Agrobacterium tumefaciens strain C58 with cardiotonic activity lost its ability to produce AAs [31]. Hairy roots, induced by the soil bacterium A. rhizogenes, are another promising approach for obtaining secondary metabolites, as they are fast-growing and higher-yielding compared to cell cultures [32,33]. However, there have been no studies on the development of hairy roots from Aristolochia species.
The present study establishes hairy root cultures of A. manshuriensis and investigates the effect of the rolB and rolC gene transfer on the metabolite profile of transformed cells. The anti-radical potential of the extracts and their cytotoxicity are also investigated.
2. Results and Discussion
2.1. Effect of Explants on Hairy Root Induction
Explants from the leaves, petioles, and stems of A. manshuriensis plants were infected with A. tumefaciens harboring rolA, rolB, and rolC genes to induce hairy root formation. They were cultured in a medium without hormones. Two weeks after infection, primary tumors were observed to be developing on rol-infected explants. After another week, adventitious roots emerged in the rolC- and rolB-infected stem and petiole-derived tumors, while no root growth was observed in rolA-transformed tumors. The induction of hairy roots in petiole explants infected with A. tumefaciens was lower compared to stems, while no induction effects were observed in the leaves (Table 1). In addition, it was observed that the development of adventitious roots was 2 times higher in stem and 1.3 times higher in petiole-type tumors infected with the rolB gene than those with the rolC gene (Table 1). The rolA calli grew very slowly as compact yellow globular aggregates, could not form roots, and were not included in further analysis. The control explants infected with A. tumefaciens bearing empty vectors (without rol genes) also showed no visible signs of root induction. Adventitious roots growing on stem-derived tumors were excised and transferred into the liquid media resulting in AC and AB hairy root cultures for the rolC and rolB transgenes, respectively. AC and AB cultures had yellow color and extensive lateral branching, typical for hairy root cultures (Figure 1A). The untransformed callus culture (A1) was previously derived from A. manshuriensis stems [34] and was taken as a control in this study.

Table 1.
The effectiveness of hairy root initiation from different explants, %.
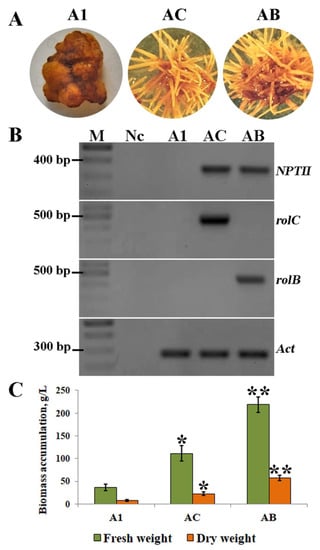
Figure 1.
(A) Phenotypes of the A. manshuriensis cell cultures. A1—untransformed callus line; AC—hairy root culture transformed with the rolC gene; AB—hairy root culture transformed with the rolB gene. (B) PCR products of nptII, rolC, rolB, and actin from the genomic DNA of A1, AC, and AB cell lines. M—DNA markers (100 bp + 1.5 kb ladder, SybEnzyme, Russia); Nc—negative control (no DNA added). The original image is shown in Figure S2. (C) Biomass accumulation in 4-week-old callus and hairy root cultures of A. manshuriensis. Data are presented as the mean values ± standard errors, * p < 0.05, ** p < 0.01 (Student’s t-test).
To prove the integration of the T-DNA region of pPCV002 plasmid into the hairy root cultures of A. manshuriensis, PCR analysis of the rol and nptII genes was performed. Using genomic DNA from A1, AB, and AC cell lines, fragments corresponding to nptII, rolC, and rolB with the predicted lengths were amplified, which confirmed the successful genetic transformation in AC and AB cultures (Figure 1B).
The accumulation of fresh and dry biomass of A. manshuriensis cell cultures was observed for five weeks. The exponential phase of the growth curve for all lines was observed from the third to the fourth week when a noticeably faster rate of biomass accumulation was detected (Figure S1). Comparatively, the four-week cultures showed the highest fresh and dry weights at 36.6 and 7.4 g/L for A1, 111.4 and 22.5 g/L for AC, and 219.0 and 57.5 g/L for AB, respectively (Figure 1C).
The efficacy of hairy root syndrome induction is influenced by various factors, including but not limited to the type of explant, co-cultivation conditions, the concentration of acetosyringone, and Agrobacterium strain, each exerting varying degrees of control [35,36]. In this investigation, previously optimized transformation conditions [37] were taken advantage of, and the effectiveness of hairy root induction on different types of explants by different rol genes was studied. Consistent with our findings, higher transformation rates from stem explants have also been documented in Trigonella foenum-graecum [38], Artemisia pallens [39], and Agastache rugosa [40]. Petiole explants are effective for inducing hairy roots in Withania somnifera [41], Isatis tinctoria [42], and Saussurea medusa [43]. In contrast, for many plant species, leaf explants have exhibited better performance [35,44,45,46]. This suggests that the availability of cells competent to serve as root initials in different types of explants can play a critical role in the successful induction of hairy roots. Additionally, it is essential to note that different Aristolochia species may have unique requirements for successful transformation. The results obtained for A. manshuriensis may not necessarily apply to other species. Therefore, a systematic approach should be taken to optimize transformation protocols for each species of interest.
As for individual rol genes, the higher root-inducing capacity of rolB is not surprising. Although all three loci (i.e., rolA, B, and C) could induce hairy root syndrome in tobacco, only the rolB gene induced root formation in Kalanchoe [47]. A rhizogenic effect was not observed for the rolA gene in either carrot discs or ginseng cell transformation [48,49]. Moreover, the rhizogenic effect of the rolA gene in tobacco was weaker than that of the rolC [47]. It is supposed that the RolB protein possesses the highest morphogenetic activity among the Agrobacterium oncogenes [50,51]. At the same time, RolC supports the growth potential by counteracting the negative effects of RolB (necrosis and cell death) [52,53]. The capacity of wild-type A. rhizogenes to trigger hairy roots on carrot disks has been considerably diminished by CRISPR/Cas9-mediated mutations in rolC and rolB genes [54]. Interestingly, according to our previous results, the rolC gene stimulates cell growth, while the rolB gene considerably suppresses biomass accumulation, especially at high levels of transgene expression [55]. Conversely, in A. manshuriensis hairy roots, the rolB gene provided better growth properties than rolC. AC and AB cell cultures grow much faster than the A1 callus line. Analogous to our results, hairy roots of many different plant species demonstrate superior growth potency over calli cells [32,33].
2.2. The Effect of rolC and rolB on Accumulation of Phenanthroic Acid Derivatives in Hairy Root Cultures of A. manshuriensis
Through HPLC-UV-MS analysis of the control culture A1, hairy roots (AC and AB), and stems of A. manshuriensis, seven main phenanthroic acid derivatives were identified, including six aristolochic acids (AAs) and one aporphine alkaloid (Figure 2). All compounds were identified by comparing their UV and MS data, including retention time, UV maxima, accurate mass, and fragmentation pattern, with those of the available standards and literature data [56,57,58]. The informative data obtained from the analysis are summarized in Table S1 and Supplementary HPLC-UV and HRMS data. Magnoflorine (2), an aporphine derivative, and two aristolochic acids (AA)-I (7) and AA-II (6) were identified by complete concurrence with standard compounds. The mass spectrometric behavior of AAs was studied using standard compounds (AA-I and AA-II) and previously published information [56,57,58]. It is noteworthy that the positive cluster ions with ammonia [M + NH4]+, as well as ions formed by elimination of neutral molecules with compositions [M + H-H2O]+, [M + H-NO2]+, and [M + H-CO2]+, were easily found in the positive ESI-MS full scan spectra of AAs. Additionally, the O-glucoside derivatives formed ions [M + H-Hex-H2O]+ and [M + H-Hex-NO2]+ due to the combined loss of the hexose moiety (162 Da) and the loss of the H2O (18 Da) or nitro group (46 Da), respectively. Furthermore, the negative ESI-MS full-scan spectra and MS2 fragmentation patterns of the defined compounds were studied and compared with the reference data (Table S1). Consequently, four aristolochic acid analogs were identified: AA-IIIa (4), AA-IVa/b (5), AA-IIIa O-glucoside (AA-IIIa-G) (1), and AA-IVa/b O-glucoside (AA-IVa/b-G) (3). The chemical structures of the detected compounds are provided in Figure 2.
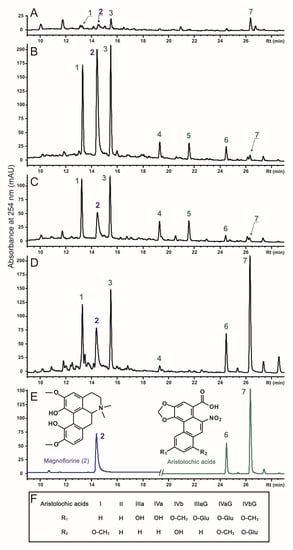
Figure 2.
The HPLC-UV separation of main phenanthrene derivatives from (A) control cell line A1, (B) rolC-transgenic cell line AC, (C) rolB-transgenic cell line AB, and (D) stems of A. manshuriensis recorded at 254 nm. Identified compounds: aristolochic acid IIIa O-glucoside (1), magnoflorine (2), aristolochic acid IVa/IVb O-glucoside (3), aristolochic acid IIIa (4), aristolochic acid IVa/IVb (5), aristolochic acids II (6), aristolochic acids I (7). Representative chromatogram of standard compounds and chemical structures of the identified phenanthrene derivatives are also shown (E,F).
The accumulation of total AAs in the AC and AB transgenic cell lines was 7–8-fold higher than that of the control A1 calli, but was 1.5–1.6 times less than that in the stems of A. manshuriensis (Table 2). While the effects of rolC and rolB were similar, they varied among different AA derivatives. The most significant increase (12–40 fold) was observed for AA-IIIa, AA-IVa/b, and AA-IIIa-G, whereas moderate effects (6–8 fold) were detected for AA-II and AA-IVa/b-G. In contrast, the content of AA-I was found to be reduced by 5.5 times. The content of magnoflorine was found to be maximum in rolC- and rolB-transformed hairy roots, exceeding its values in the A1 callus line by 5.8 and 2.8 times, respectively. Consequently, its production in four-week-old AC and AB lines reached 128.7 and 158.7 mg/L, respectively. Principal component analysis (PCA) revealed two significant components, where PC1 accounted for 87.81% of the variance, and PC2 accounted for 9.79%. The loading plot of PC1 was dominated by magnoflorine (Figure S3). At the same time, PC2 was mainly contributed by AA-I, AA-II, AA-IIIa-G, and AA-IVa/b-G, with other metabolites, including magnoflorine, having negative loading values. The samples were divided into four distinct zones based on PC1 and PC2 values (Figure S2), proving that each displayed substantial metabolic discrimination. The A1 and AC cell cultures exhibited the most significant metabolite dissimilarity, suggesting a substantial variance between these groups. Notably, the PCA conducted on the LC-MS and 1H NMR data of phytochemicals in 43 Aristolochia species indicated that magnoflorine was responsible for the positive PC1 values of the samples [57]. The investigation further demonstrated substantial diversity within discrete A. manshuriensis samples, which implies that numerous factors can influence the plant’s phytochemistry [57].

Table 2.
Aristolochic acids and magnoflorine content (mg/g of dry weight) in cell cultures and stems of A. manshuriensis measured by using HPLC-UV-MS.
It is well known that rol genes considerably alter the biosynthesis of secondary metabolites of the plant. For instance, they increase the content of alkaloids in Vinca minor and Catharanthus roseus [59,60], flavonoids in Artemisia annua [61], anthraquinones in Rubia cordifolia [55], and ginsenosides in Panax ginseng [37]. Although the rolB gene is known for its significant role in enhancing the secondary metabolism of transformed cells [62], its impact on the A. manshuriensis hairy roots was not observed to be as prominent as for the rolC gene. However, due to the excellent growth characteristics of the rolB-transformed hairy roots, the final magnoflorine yield reached the highest level reported in cell culture. Although magnoflorine is a naturally occurring alkaloid, its content in medicinal plants is typically less than 1 mg/g [20]. The yield of magnoflorine has been observed to be in the range of 0.17–0.27 mg/g in different tissues of A. fangchi [63]. Likewise, different tissues of Epimedium alpinum have been reported to accumulate 0.2–1.0 mg/g of magnoflorine [64]. In the bark of Ptychopetalum olacoides, magnoflorine was identified as a major compound, with a recorded concentration of up to 2.56 mg/g [65]. Previous attempts to produce magnoflorine in callus cultures of Papaver somniferum and Stephania glabra as alternative methods were insufficient in achieving maximum content [66,67]. A combination of transgenic Escherichia coli and Saccharomyces cerevisiae cells produced only 8.3 mg/L of culture medium [68]. Therefore, transformed hairy root cultures of A. manshuriensis can be deemed a promising and novel source of magnoflorine.
To safely use A. manshuriensis cell cultures, new strategies for metabolic engineering are needed to reduce AA contents while increasing the concentration of valuable phytochemicals like magnoflorine. A fascinating finding was that the A. manshuriensis cell suspension culture, which was transformed with the T-DNA from the wild-type A. tumefaciens, exhibited a loss of ability to produce AAs [31]. While the transformation of A. manshuriensis with rolC and rolB genes from A. rhizogenes T-DNA did not result in the complete elimination of all AAs, there was a significant reduction in the content of AA-I, which is primarily responsible for the nephrotoxic and carcinogenic effects of Aristolochia species [57].
The inactivation of key enzyme genes in the AAs biosynthetic pathway could open up a new possibility for using various AAs-containing plant sources. Developing novel techniques for CRISPR/Cas9-mediated modification of secondary metabolism in plants, calli, and hairy roots with improved traits has emerged as a promising research area [69,70,71,72,73]. Recent study has identified a total of 29 genes that are potentially involved in AA biosynthesis in the A. contorta genome [15]. Further investigation of these genes will enable the identification of suitable candidates for targeted manipulations in Aristolochia hairy roots.
2.3. Free Radical Scavenging Activity of A. manshuriensis Cell Cultures Extracts
DPPH assay evaluated the radical scavenging activity of A. manshuriensis plant and cell culture extracts. The results show that AC and AB hairy roots had an almost 2.8- and 2.9-fold higher antioxidant capacity than control calli, respectively, and were similar to those in A. manshuriensis stems (Table 3).

Table 3.
DPPH radical scavenging activity of cell cultures and stems of A. manshuriensis.
Antioxidant activities have been previously studied in extracts of A. indica [74,75], A. bracteata [76], A. longa [77], A. clematitis [78], and A. albida [79]. The results demonstrated that DPPH scavenging activity varied between 20 and 550 µg/mL and that roots have a higher capacity for free radicals than areal parts of the plants. The antioxidant activity of Aristolochia plants was primarily due to magnoflorine [80]. In particular, magnoflorine demonstrated remarkable antioxidant activity, as a dose of 50 μg/mL was found to scavenge approximately 70.8% of all the free radicals [81].
There has been limited research on the antioxidant properties of cell cultures derived from Aristolochia plants. Despite the accumulation of significant levels of magnoflorine in A1 calli, their extract demonstrated DPPH scavenging activity which was 37.9% of that found in the stem. This study is the first to demonstrate that rolC and rolB genes can effectively enhance the antioxidant activity of hairy roots to levels comparable to those found in wild A. manshuriensis liana stems, likely due to the higher concentration of magnoflorine. It is important to highlight that the DPPH-based measurements of antioxidant capacity should be utilized cautiously because they might not accurately represent the activity of extracts in vivo. Further investigations are necessary to fully elucidate the underlying mechanism of this phenomenon and explore its potential applications in fields such as medicine.
2.4. Cytotoxic Effect of Constituents from A. manshuriensis Cell Cultures
The cytotoxicity of A1, AC, AB, and stem extracts was examined by MTT assay on four cell lines: triple-negative breast cancer cells (MDA-MB-231), human glioblastoma cells (U-87 MG), cervical cancer cells (HeLa CCL-2), and human colon carcinoma cells (RKOs) (Figure 3). The extract from the A1 callus line reduced the activity of cancer cell lines HeLa CCL-2, U87MG, RKO, and MDA-MB-231 by 47%, 35%, 30%, and 19.2%, respectively. AC extracts reduced the metabolic activity of U-87 MG by 53% and HeLa CCL-2 cells by 36%, as well as RKO by 33% and MDA-MB-231 by 23.9%. AB extracts reduced the activity of HeLa CCL-2, U-87 MG, RKO, and MDA-MB-231 by 46%, 45%, 31%, and 13.8%, respectively. Stem extracts showed inhibitory effects on U-87 MG, MDA-MB-231, and HeLa CCL-2 cell growth at 48.4%, 35.4%, and 32.1%, respectively, while no cytotoxicity was observed with respect to RKO (Figure 3). In general, our findings indicate that extracts from hairy roots exhibited significant cytotoxic effects that differed across various cancer cell lines, implying that the impact was specific to each cell type.
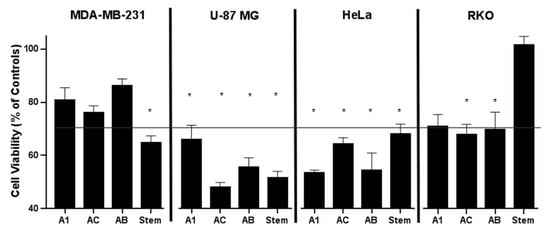
Figure 3.
Cell viability of MDA-MB-231, U-87 MG, HeLa CCL-2, and RKO cell lines in response to A1, AC, AB, and liana stems extracts, evaluated by MTT assay. Data are presented as the mean values ± standard errors. All data, with the exception of stem extract against RKO, are significantly different from the corresponding controls (set as 100%) at p < 0.01 (Student’s t-test). Asterisk designates viability inhibition greater than 30%.
The inhibitory properties of Aristolochia extracts may be attributed to the presence of magnoflorine [80]. Magnoflorine has been found to exhibit cytotoxic effects against several cancer cell lines, including those of brain, gastric, breast, and lung tumors, as well as hepatocellular carcinoma, among others [20,82,83,84,85,86]. The anti-tumor effects of magnoflorine have been linked to the induction of reactive oxygen species (ROS)-mediated apoptosis and autophagy through the AKT/mTOR and p38 signaling pathways [85,86]. However, some studies have reported that magnoflorine does not show cytotoxic effects against particular human cancer cell lines, such as KB, SiHa, A549, HaCaT, SK-OV-3, SK-MEL-2, XF498, HT-29, and HCT15 [81]. Moreover, magnoflorine is non-toxic to human embryonic kidney cells (HEK293) and normal gastric cells [81,85].
2.5. The Influence of A. manshuriensis Cell Cultures Extract on Cell Cycle of Cancer Cells
Upon observing the antiproliferative effects of A. manshuriensis cell culture extracts, cell cycle analysis was performed on the RKO cell line. Extracts obtained from AC and AB hairy roots induced a significant increase in the accumulation of G2/M phase cells compared to control calli (Figure 4A and Figure S4). Additionally, the treatment of RKO cancer cells with extracts from hairy roots resulted in a more than 10% increase in aneuploidy (Figure 4B and Figure S5). These findings suggest that the constituents of AC and AB extracts may trigger chromosome instability, leading to cell death and explaining the observed anti-proliferative effects of A. manshuriensis extracts via DNA damage. Previous studies have reported that magnoflorine induces ROS-induced apoptosis in cancer cells [85,86]. Other studies have demonstrated that ROS-induced apoptosis can be mediated by DNA double-strand breaks (DSBs) [87]. G2/M checkpoint arrest is a well-known mechanism that protects continuous cell cycles by preventing inaccurate chromosome segregation and leading to programmed cell death before critical genome instability can occur.
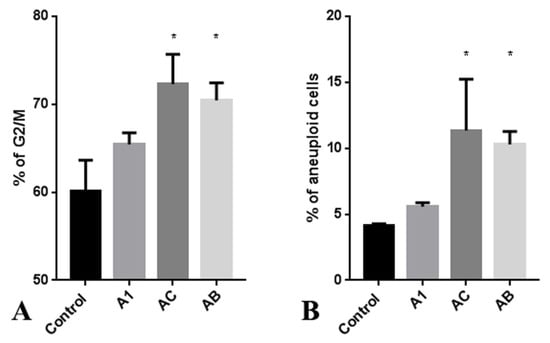
Figure 4.
Cell cycle analysis (A) and aneuploidy percentage (B) of RKO cancer cells after treatment with A1, AC, and AB extracts. Data are presented as the mean values ± standard errors, * p < 0.05 (Student’s t-test).
Studies have demonstrated that A. debilis extract induces sub-G1 arrest and apoptosis in HT-29 cells through changes in mitochondria-dependent apoptosis markers and the accumulation of ROS [88]. Similarly, magnoflorine treatment of gastric cancer cells has been shown to cause cell cycle arrest at the S/G2 phase via activation of ROS-dependent downstream signaling [84,85,86]. Another study revealed that extracts from A. clematitis, A. elegans, A. acuminate, and A. guentheri induced G2/M phase arrest and apoptosis in human kidney cells (HK-2) [57]. The observed levels of cytotoxicity, micronuclei induction, G2/M arrest, and apoptosis were found to be associated with aristolactam BI, AA-D, AA-IIIa, and AA-IIIa-G. Interestingly, the latter two metabolites were significantly upregulated in AC and AB hairy roots (Table 2). Surprisingly, the content of AA-I and AA-II, which are highly toxic agents found in Aristolochia species, did not correlate with the effects above [57]. These compounds were the most common aristolochic acid derivatives in A1 and stem, comprising 41–44% of total AAs. However, they only made up 3–5% of rol-transformed hairy roots of A. manshuriensis (Table 2). Thus, rolB and rolC genes induced alterations in the secondary metabolism of the hairy roots of A. manshuriensis that prevail in reducing toxicity and increasing valuable compounds. However, further research is still needed to validate the safety of obtained hairy roots, such as acute and long-term toxicology assessments on animals.
3. Materials and Methods
3.1. Plant Material and Tissue Cultures
In this study, we used parts of a wild-growing Aristolochia manshuriensis liana that had been collected in Nadegdinsky District (Nadegdinsk, Primorsky Krai, Russia). The control, untransformed callus culture (A1) was derived from 1-year-old A. manshuriensis stems as described in [34] and cultivated using W medium [89] supplemented with 1 mg/L 6-benzylaminopurine and 1 mg/L indole-3-acetic acid under darkness at 25 °C with a 30-day subculture cycle. During the period of observation (more than 10 years), A1 did not show any rhizogenic effects.
Petiole, leaf, and stem explants of 3-week-old clonally cultivated A. manshuriensis plantlets were inoculated with the GV3101 strains of Agrobacterium tumefaciens [90] carrying pPCV002-A, pPCV002-CaMVBT, and pPCV002-CaMVC binary vectors [48] (Figure S6). In these constructs, the rolA gene is driven by its native promoter, while the rolB and rolC genes are under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The T-DNA region also contains the gene for kanamycin resistance (nptII) under eukaryotic control sequences. As a control, we used A. thumefaciens carrying an empty pPCV002 vector (Figure S5). The optimized transformation protocol was previously described in detail [37]. In brief, the sterilized explants were cut into small pieces with the scalpel and dipped into the A. thumefaciens suspension in MES buffer (10 mM MES; pH, 5.6; 10 mM MgCl2; 100 μM acetosyringone) with OD600 = 0.5 for 10 min. After incubation, inoculated explants were blotted dry and then kept for 2 days on agar-solidified W medium containing 100 µM acetosyringone at 25 °C in the dark. Thereafter, explants were transferred to fresh medium supplemented with 250 mg/L cefotaxim and 50 mg/L kanamycin to produce lines of primary kanamycin-resistant tumors. After 3 weeks, rolC- and rolB-transformed primary tumors were able to spontaneously form adventitious roots. Root tips, isolated from adventitious roots were transferred into liquid W medium supplemented with 0.5 mg/L indole-3-butyric acid to further promote root growth. These cultures, designated as AC and AB, were cultivated in the dark at 25 °C in 500 mL Erlenmeyer flasks in an orbital shaker (100 rpm; 20 mm amplitude) and subcultured at 28-day intervals.
3.2. DNA Isolation and PCR Analysis
The isolation of genomic DNA from dry callus tissues was carried out with a CTAB-based method [91]. DNA quality was examined by electrophoresis on a 0.8% agarose gel. Spectrophotometric analysis using a BioSpec-nano spectrophotometer (Shimadzu, Kyoto, Japan) was employed to estimate the quantity and purity of extracted DNA.
We first aimed to amplify A. manshuriensis DNA sequences corresponding to the housekeeping actin gene. For this aim, PCRs were performed in 25 µL reaction volumes containing 50 ng of template DNA, 1× PCR buffer with 2.5 mM MgCl2, 200 μM of each deoxynucleotide, 1 μM of the primer pair (Table S2), and 1 U Taq polymerase (Evrogen, Russia). PCRs were performed in an iCycler (Bio-Rad Laboratories), and cycle conditions were as follows: 96 °C for 10 s, 50 °C for 30 s, and 72 °C for 1 min. DNA fragments of predicted lengths were obtained, isolated from agarose gels with a Glass Milk Kit (Sileks, Russia), and sequenced as described earlier [55] at the Instrumental Center of Biotechnology and Gene Engineering of FSCEATB FEB RAS using an ABI 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). This allowed identification of two actin isoforms, namely, AmACT1 (GenBank accession no.: OQ676410) and AmACT2a (GenBank accession no.: OQ676411), of which the latter sequence contains an 88 bp intron. To verify successful T-DNA integration, PCRs were performed with the above conditions and primer sets specific to the rolB, rolC, nptII, and AmACT1 genes (Table S2).
3.3. High-Performance Liquid Chromatography (HPLC) Analysis
For secondary metabolite analysis, A. manshuriensis calli were dried under hot air for 20 h and ground using a mortar and pestle.
3.3.1. Chemicals
Analytical standards (magnoflorine, AA-I, and AA-II) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Milli-Q water (Millipore, Bedford, MA, USA) was used for preparing standard solutions, extraction buffers, and eluents. Formic and acetic acids were of high-performance liquid chromatography (HPLC)-grade and obtained from Merck (Darmstadt, Germany). HPLC-grade acetonitrile (ACN) was obtained from PanReac AppliChem (Darmstadt, Germany). All the other chemicals were of analytical-grade.
3.3.2. Sample Preparation and HPLC Assays
Dried and ground samples (50 mg) were supplemented with 1 mL 70% ethanol, homogenized, sonicated for 30 min at 40 °C, and centrifuged at 15,000× g for 15 min at 4 °C. The supernatants were collected, filtered (0.45 μm nylon membrane, Millipore, Bedford, MA, USA), and analyzed by LC-UV-MS(MS2) as we described previously [92,93] with a few additions. Briefly, the HPLC-UV-MS studies were performed with an Agilent 1260 Infinity analytical HPLC system (Agilent Technologies, Santa Clara, CA, USA) equipped with a photodiode array detector, an analytical Zorbax C18 column (150 mm, 2.1 mm i.d., 3.5-μm part size, Agilent Technologies, USA), and interfaced with an ion trap mass spectrometer Bruker HCT Ultra PTM Discovery System (Bruker Daltonik GmbH, Bremen, Germany). High-resolution mass spectrometry (HRMS) was performed using a Shimadzu LCMS-IT-TOF instrument (Shimadzu, Kyoto, Japan) equipped with an ion trap time-of-flight mass spectrometer operating under electrospray ionization (ESI) conditions. All chromatographic and mass spectrometric conditions were the same [92]; detailed information is presented in Table S1 and Supplementary HPLC-UV and HRMS data. Chromatograms for quantification were recorded at 237 nm (for magnoflorine) and 254 nm (for AAs). Quantification of magnoflorine, AA-I, and AA-II was performed by corresponding peak areas using the absolute calibration method with commercially available standards. The amounts of AA-IIIa, AA-IVa/b, AA-IIIa-G, and AA-IVa/b-G, for which standards were not available, were calculated using the calibration curve of AA-I and corrected according to their molecular weight.
3.4. Antioxidant Activities of Extracts
The antioxidant activity of the extracts was determined using a 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging photometric assay [93]. A solution of 0.3 mM DPPH in ethanol was prepared, and 149 µL of this solution was mixed with 1 µL of extracts in ethanol at different concentrations (5–500 µg/mL). For comparison, ascorbic acid (vitamin C) was used. After incubation at room temperature, the absorbance was measured at 517 nm using a Benchmark Plus microplate spectrophotometer (Bio-Rad, Hercules, CA, USA) in 3 technical replicates and converted into percentage inhibition (%inh) using the following formula:
where AB is the absorbance of the negative control (equal amount of DPPH solution), and AS is the absorbance of the test sample. A similar amount of extract in ethanol was used as a blank. The IC50 values were calculated by using linear regression of plots, where the abscissa represented the concentration of tested extracts and the ordinate the average percent of antioxidant activity from three separate tests.
3.5. Anticancer Activities of Extracts
Extracts were tested on four cell lines using the MTT assay: triple-negative breast cancer cells (MDA-MB-231), human glioblastoma cells (U-87 MG), cervical cancer cells (HeLa CCL-2), human colon carcinoma cells (RKOs), all received from ATCC. Above cell cultures were cultivated in the Laboratory of Biomedical Cell Technologies at the Institute of Life Sciences and Biomedicine of Far Eastern Federal University.
Standard cell models were seeded onto a 96-well plate at a concentration of 5 × 103 cells per well in the appropriate culture medium. Since the extracts were dissolved in methanol (MeOH), we used 0.5% MeOH as the negative control added to the medium with cells (this concentration of the solvent is considered non-toxic). For convenience in testing and adding extracts during the experiment, we prepared master mixes with both the extracts and MeOH for the negative control at a concentration of 0.5%. At 24 h after seeding, the extracts from the prepared master mixes and MeOH as the control were added to the 96-well plate at a volume of 120 μL per well. The cells with added extracts were incubated for 72 h at 37 °C, 5% CO2, and 80% relative humidity. After incubation, cytotoxicity analysis was conducted using the MTT assay as previously described [94].
3.6. Cell Cycle Analysis
Cells (100 × 103 cells/well) were seeded in 6-well culture plates and treated for 5 days with different concentrations of extracts (up to a final methanol concentration of 0.5%) in three independent replicates. Pure methanol was added as a control up to a final concentration of 0.5%. After the incubation period, the cells were detached with a 0.5% trypsin solution and fixed with 4% paraformaldehyde. After a series of washes, the suspension was stained with the intercalating dye Hoechst 33342 at a concentration of 10 μg/mL for 20 min, washed, and analyzed using the BD FACS MoFlo Astrios cell sorter (BD Biosciences, Indianapolis, IN, USA) with a 355 nm ultraviolet laser. Emission was recorded at 488 nm. Results were analyzed using Kaluza 2.1 software with the cell cycle analysis function based on the Michael H. Fox algorithm.
3.7. Statistical Analysis
All values were expressed as the mean ± SE. For statistical evaluation, Student’s t-test was used to compare the two independent groups. For comparison among multiple data sets, analysis of variance (ANOVA) followed by multiple comparison procedures was employed. Fisher’s protected least significant difference (PLSD) post hoc test was employed for the intergroup comparison. PAST 4.03 software was used for principal component analysis (PCA) of secondary metabolites detected using HPLC-UV-MS [95].
4. Conclusions
This study reports the establishment of hairy root cultures of the rare and valuable medicinal plant A. manshuriensis through the genetic transfer of individual rolC and rolB genes. The results indicate that both rolC and rolB genes effectively induced the formation of roots on petiole and stem explants, with rolB having a more pronounced effect. The findings highlight the importance of selecting the appropriate explant and rol gene to induce hairy root syndrome in A. manshuriensis. Furthermore, it was found that both rolC and rolB genes led to a significant increase in AAs and magnoflorine in transformed A. manshuriensis cells, but to varying degrees. In the AC and AB cell lines, the accumulation of AA-I decreased considerably, while the contents of AA-IIIa, AA-IVa/b, and AA-IIIa-G increased up to 40 fold. The rolC gene had the most significant effect on the upregulation of magnoflorine quantity, with the AC cell line reaching 5.72 mg/g of dry weight. In addition, the results show that the hairy root extracts of A. manshuriensis exhibited significant cytotoxic effects on human glioblastoma cells (U-87 MG), cervical cancer cells (HeLa CCL-2), and colon carcinoma (RKO) cells. However, when tested on triple-negative breast cancer cells, the extracts did not show significant activity (MDA-MB-231). Further research is needed to explore the potential of A. manshuriensis hairy roots as a source of anti-cancer agents and to elucidate the underlying mechanisms of action.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms241411240/s1.
Author Contributions
Conceptualization, Y.N.S., V.V.K. and V.P.B.; methodology, Y.N.S., V.P.G., V.V.K. and V.P.B.; software, M.R.S.; validation, Y.N.S., Y.A.Y., V.P.G., V.V.K. and V.P.B.; formal analysis, T.Y.G., O.D.K. and A.I.D.; investigation, Y.N.S., G.K.T., Y.A.Y., V.P.G., M.R.S., M.S.O., N.A.S. and V.P.B.; resources, Y.N.S., V.V.K. and V.P.B.; data curation, Y.N.S., Y.A.Y. and V.V.K.; writing—original draft preparation, Y.N.S., Y.A.Y. and V.V.K.; writing—review and editing, Y.N.S.; visualization, Y.A.Y., M.S.O. and N.A.S.; supervision, Y.N.S., V.V.K. and V.P.B.; project administration, V.P.B.; funding acquisition, Y.N.S. and V.V.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Ministry of Science and Higher Education of the Russian Federation (theme no.: AAAA-A19-119012590040-4 and FZNS-2023-0017).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are contained within the article and Supplementary Materials. The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
This research was carried out within the state assignment of the Ministry of Science and Higher Education of the Russian Federation (theme no.: AAAA-A19-119012590040-4). The evaluation of cytotoxic activity and cell cycle assay was carried out within the state assignment of the Ministry of Science and Higher Education of the Russian Federation (theme no.: FZNS-2023-0017). The experiments described in this work were performed using equipment from the Instrumental Center for Biotechnology and Gene Engineering at the Federal Scientific Center of East Asia Terrestrial Biodiversity of the Far East Branch of the Russian Academy of Sciences. The authors are grateful to the Primorsky Aquarium Shared Equipment Facility at the A.V. Zhirmunsky National Scientific Center of Marine Biology of the Far Eastern Branch, the Russian Academy of Sciences (NSCMB FEB RAS), for providing equipment and resources.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Neinhuis, C.; Wanke, S.; Hilu, K.; Müller, K.; Borsch, T. Phylogeny of Aristolochiaceae based on parsimony, likelihood, and Bayesian analyses of trnL-trnF sequences. Plant. Syst. Evol. 2005, 250, 7–26. [Google Scholar] [CrossRef]
- Bänziger, H.; Disney, R.H.L. Scuttle flies (Diptera: Phoridae) imprisoned by Aristolochia baenzigeri (Aristolochiaceae) in Thailand. Mitt. Schweiz. entomol. Ges. 2006, 79, 29–61. [Google Scholar]
- Murugan, R.; Shivanna, K.R.; Rao, R.R. Pollination biology of Aristolochia tagala, a rare species of medicinal importance. Curr. Sci. 2006, 91, 795–798. [Google Scholar]
- Trujillo, C.G.; Sérsic, A.N. Floral biology of Aristolochia argentina (Aristolochiaceae). Flora Morphol. Distrib. Funct. Ecol. 2006, 201, 374–382. [Google Scholar] [CrossRef]
- Kuo, P.; Li, Y.; Wu, T. Chemical Constituents and Pharmacology of the Aristolochia species. J. Tradit. Complement. Med. 2012, 2, 249–266. [Google Scholar] [CrossRef]
- Lerma-Herrera, M.A.; Beiza-Granados, L.; Ochoa-Zarzosa, A.; López-Meza, J.E.; Navarro-Santos, P.; Herrera-Bucio, R.; Aviña-Verduzco, J.; García-Gutiérrez, H.A. Biological Activities of Organic Extracts of the Genus Aristolochia: A Review from 2005 to 2021. Molecules 2022, 27, 3937. [Google Scholar] [CrossRef] [PubMed]
- Grollman, A.P.; Marcus, D.M. Global hazards of herbal remedies: Lessons from Aristolochia: The lesson from the health hazards of Aristolochia should lead to more research into the safety and efficacy of medicinal plants. EMBO Rep. 2016, 17, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Martena, M.J.; van der Wielen, J.C.A.; van de Laak, L.F.J.; Konings, E.J.M.; de Groot, H.N.; Rietjens, I.M.C.M. Enforcement of the ban on aristolochic acids in Chinese traditional herbal preparations on the Dutch market. Anal. Bioanal. Chem. 2007, 389, 263–275. [Google Scholar] [CrossRef]
- Wu, L.; Sun, W.; Wang, B.; Zhao, H.; Li, Y.; Cai, S.; Xiang, L.; Zhu, Y.; Yao, H.; Song, J.; et al. An integrated system for identifying the hidden assassins in traditional medicines containing aristolochic acids. Sci. Rep. 2015, 5, 11318. [Google Scholar] [CrossRef]
- Nakonechnaya, O.V.; Koren, O.G.; Sidorenko, V.S.; Shabalin, S.A.; Markova, T.O. Poor fruit set due to lack of pollinators in Aristolochia manshuriensis (Aristolochiaceae). Plant. Ecol. Evol. 2021, 154, 39–48. [Google Scholar] [CrossRef]
- Cheung, T.P.; Xue, C.; Leung, K.; Chan, K.; Li, C.G. Aristolochic acids detected in some raw Chinese medicinal herbs and manufactured herbal products--a consequence of inappropriate nomenclature and imprecise labelling? Clin. Toxicol. 2006, 44, 371–378. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, J.X.; Gao, J.J.; Du, K.; Jia, Z.J. The antitumor constituents from stems of Aristolochia manshuriensis. J. Beijing Med. Univ. 2000, 32, 18–21. [Google Scholar]
- Chung, Y.M.; Chang, F.R.; Tseng, T.F.; Hwang, T.L.; Chen, L.C.; Wu, S.F.; Lee, C.L.; Lin, Z.Y.; Chuang, L.Y.; Su, J.H.; et al. A novel alkaloid, aristopyridinone A and anti-inflammatory phenanthrenes isolated from Aristolochia manshuriensis. Bioorg. Med. Chem. Lett. 2011, 21, 1792–1794. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, J. Alkaloids from Aristolochia manshuriensis (Aristolochiaceae). Helv. Chim. Acta 2006, 89, 2665–2670. [Google Scholar] [CrossRef]
- Cui, X.; Meng, F.; Pan, X.; Qiu, X.; Zhang, S.; Li, C.; Lu, S. Chromosome-level genome assembly of Aristolochia contorta provides insights into the biosynthesis of benzylisoquinoline alkaloids and aristolochic acids. Hortic. Res. 2022, 9, uhac005. [Google Scholar] [CrossRef] [PubMed]
- Arlt, V.M.; Stiborova, M.; Schmeiser, H.H. Aristolochic acid as a probable human cancer hazard in herbal remedies: A review. Mutagenesis 2002, 17, 265–277. [Google Scholar] [CrossRef]
- Zhang, Y.; Shao, Z.; Han, Y.; Wang, J.; Shi, Y. Study on the attenuation mechanism of Aristolochia manshuriensis compatibility with Zingiberis rhizoma based on different methods of boiling and blending. Int. J. Tradit. Chin. Med. 2019, 6, 1347–1352. [Google Scholar]
- Kang, Y.C.; Chen, M.H.; Lin, C.Y.; Lin, C.Y.; Chen, Y.T. Aristolochic acid-associated urinary tract cancers: An updated meta-analysis of risk and oncologic outcomes after surgery and systematic review of molecular alterations observed in human studies. Ther. Adv. Drug. Saf. 2021, 12, 1–25. [Google Scholar] [CrossRef]
- Hegde, V.R.; Borges, S.; Patel, M.; Das, P.R.; Wu, B.; Gullo, V.P.; Chan, T.M. New potential antitumor compounds from the plant Aristolochia manshuriensis as inhibitors of the CDK2 enzyme. Bioorg. Med. Chem. Lett. 2010, 20, 1344–1346. [Google Scholar] [CrossRef]
- Xu, T.; Kuang, T.; Du, H.; Li, Q.; Feng, T.; Zhang, Y.; Fan, G. Magnoflorine: A review of its pharmacology, pharmacokinetics and toxicity. Pharmacol. Res. 2020, 152, 104632. [Google Scholar] [CrossRef]
- Liang, H.; Chang, X.; Xia, R.N.; Wu, W.; Guo, H.J.; Yang, M. Magnoflorine attenuates cerebral ischemia-induced neuronal injury via autophagy/sirt1/AMPK signaling pathway. Evid. Based. Complement. Alternat. Med. 2022, 2131561. [Google Scholar] [CrossRef]
- Pattar, P.V.; Jayara, M. In vitro Regeneration of plantlets from leaf and nodal explants of Aristolochia indica L. – an important threatened medicinal plant. Asian Pac. J. Trop. Biomed. 2012, 2, S488–S493. [Google Scholar] [CrossRef]
- Sarma, B.; Tanti, B. In vitro regeneration of plantlets from nodal explants of Aristolochia saccata and Aristolochia cathcartii. Eur. J. Biol. Res. 2017, 7, 191–201. [Google Scholar] [CrossRef]
- Remya, M.; Narmatha Bai, V.; Murugesan, S.; Mutharaian, V.N. Changes in bioactive components of Aristolochia tagala. Cham, a rare species of medicinal importance during its in vitro development through direct regeneration. bioRxiv 2016, 037028. [Google Scholar] [CrossRef]
- Nakonechnaya, O.V.; Koren’, O.G.; Zhuravlev, Y.N. Allozyme variation of the relict plant Aristolochia manshuriensis Kom. (Aristolochiaceae). Russ. J. Genet. 2007, 43, 156–164. [Google Scholar] [CrossRef]
- Voronkova, N.M.; Kholina, A.B.; Koldaeva, M.N.; Nakonechnaya, O.V.; Nechaev, V.A. Morphophysiological dormancy, germination, and cryopreservation in Aristolochia contorta seeds. Plant Ecol. Evol. 2018, 151, 77–86. [Google Scholar] [CrossRef]
- Koren, O.G.; Nakonechnaya, O.V.; Zhuravlev, Y.N. Genetic structure of natural populations of the relict species Aristolochia manshuriensis (Aristolochiaceae) in disturbed and intact habitats. Russ. J. Genet. 2009, 45, 678–684. [Google Scholar] [CrossRef]
- Smetanska, I. Production of secondary metabolites using plant cell cultures. In Food Biotechnology; Stahl, U., Donalies, U., Nevoigt, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 111, pp. 187–228. [Google Scholar] [CrossRef]
- Prabha, A.L.; Nandagopalan, V.; Vasanth, K.; Jayabalan, N. Production of aristolochic acid (AA) I and II from Aristolochia indica and Aristolochia bracteolate through in vitro culture. Plant Cell Biotechnol. Mol. Biol. 2012, 13, 131–140. [Google Scholar]
- Zhang, H.; Wang, C. Callus formation of Aristolochia tuberosa and determination of aristolochic acid of callus by HPLC. J. West China Univ. Med. Sci. 1997, 28, 170–173. [Google Scholar]
- Bulgakov, V.P.; Zhuravlev, Y.N.; Fedoreyev, S.A.; Denisenko, V.A.; Veselova, M.; Kulesh, N.I.; Alshevskaya, E.K.; Radchenko, S.V. Constituents of Aristolochia manshuriensis cell suspension culture possessing cardiotonic activity. Fitoterapia 1996, 67, 238–240. [Google Scholar]
- Gutierrez-Valdes, N.; Häkkinen, S.T.; Lemasson, C.; Guillet, M.; Oksman-Caldentey, K.-M.; Ritala, A.; Cardon, F. Hairy root cultures – A versatile tool with multiple applications. Front. Plant Sci. 2020, 11, 33. [Google Scholar] [CrossRef]
- Stepanova, A.Y.; Malunova, M.V.; Gladkov, E.A.; Evsyukov, S.V.; Tereshonok, D.V.; Solov’eva, A.I.; Timiryazev, K.A. Collection of hairy roots as a basis for fundamental and applied research. Molecules 2022, 27, 8040. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Zhuravlev, Y.N. Generation of Aristolochia manshuriensis Kom. callus tissue cultures. Rastitel’nye Resur. 1989, 25, 266–270. [Google Scholar]
- Bansal, M.; Kumar, A.; Sudhakara Reddy, M. Influence of Agrobacterium rhizogenes strains on hairy root induction and ‘bacoside A’ production from Bacopa monnieri (L.) Wettst. Acta Physiol. Plant. 2014, 36, 2793–2801. [Google Scholar] [CrossRef]
- Panda, B.M.; Mehta, U.J.; Hazra, S. Optimizing culture conditions for establishment of hairy root culture of Semecarpus anacardium L. 3 Biotech 2017, 7, 21. [Google Scholar] [CrossRef]
- Degtyarenko, A.I.; Gorpenchenko, T.Y.; Grigorchuk, V.P.; Bulgakov, V.P.; Shkryl, Y.N. Optimization of the transient Agrobacterium-mediated transformation of Panax ginseng shoots and its use to change the profile of ginsenoside production. Plant Cell Tissue Organ Cult. 2021, 146, 357–373. [Google Scholar] [CrossRef]
- Shahabzadeh, Z.; Heidari, B.; Hafez, R.F. Induction of transgenic hairy roots in Trigonella foenum-graceum co-cultivated with Agrobacterium rhizogenes harboring a GFP gene. J. Crop Sci. Biotechnol. 2013, 16, 263–268. [Google Scholar] [CrossRef]
- Pala, Z.; Shukla, V.; Alok, A.; Kudale, S.; Desai, N. Enhanced production of an anti-malarial compound artesunate by hairy root cultures and phytochemical analysis of Artemisia pallens Wall. 3 Biotech 2016, 6, 182. [Google Scholar] [CrossRef]
- Lee, S.Y.; Xu, H.; Kim, Y.K.; Park, S.U. Rosmarinic acid production in hairy root cultures of Agastache rugosa Kuntze. World J. Microbiol. Biotechnol. 2008, 24, 969–972. [Google Scholar] [CrossRef]
- Saravanakumar, A.; Aslam, A.; Shajahan, A. Development and optimization of hairy root culture systems in Withania somnifera (L.) Dunal for withaferin-A production. Afr. J. Biotechnol. 2012, 11, 16412–16420. [Google Scholar]
- Gai, Q.Y.; Jiao, J.; Luo, M.; Wang, W.; Ma, W.; Zu, Y.G.; Fu, Y.J. Establishment of high-productive Isatis tinctoria L. hairy root cultures: A promising approach for efficient production of bioactive alkaloids. Biochem. Eng. J. 2015, 95, 37–47. [Google Scholar] [CrossRef]
- Zhao, D.; Fu, C.; Chen, Y.; Ma, F. Transformation of Saussurea medusa for hairy roots and jaceosidin production. Plant Cell Rep. 2004, 23, 468–474. [Google Scholar] [CrossRef]
- Sujatha, G.; Zdravković-Korać, S.; Ćalić, D.; Flamini, G.; Ranjitha Kumari, B.D. High-efficiency Agrobacterium rhizogenes-mediated genetic transformation in Artemisia vulgaris: Hairy root production and essential oil analysis. Ind. Crops Prod. 2013, 643–652. [Google Scholar] [CrossRef]
- Chung, I.M.; Rekha, K.; Rajakumar, G.; Thiruvengadam, M. Production of glucosinolates, phenolic compounds and associated gene expression profiles of hairy root cultures in turnip (Brassica rapa ssp. rapa). 3 Biotech 2016, 6, 175. [Google Scholar] [CrossRef] [PubMed]
- Mohebodini, M.; Fathi, R.; Mehri, N. Optimization of hairy root induction in chicory (Cichorium intybus L.) and effects of nanoparticles on secondary metabolites accumulation. Iran. J. Genet. Plant Breed. 2017, 6, 60–68. [Google Scholar] [CrossRef]
- Spena, A.; Schmülling, T.; Koncz, C.; Schell, J.S. Independent and synergistic activity of rol A, B and C loci in stimulating abnormal growth in plants. EMBO J. 1987, 6, 3891–3899. [Google Scholar] [CrossRef]
- Capone, I.; Spanò, L.; Cardarelli, M.; Bellincampi, D.; Petit, A.; Costantino, P. Induction and growth properties of carrot roots with different complements of Agrobacterium rhizogenes T-DNA. Plant Mol. Biol. 1989, 13, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Khodakovskaya, M.V.; Labetskaya, N.V.; Chernoded, G.K.; Zhuravlev, Y.N. The impact of plant rolC oncogene on ginsenoside production by ginseng hairy root cultures. Phytochemistry 1998, 49, 1929–1934. [Google Scholar] [CrossRef]
- Cardarelli, M.; Mariotti, D.; Pomponi, M.; Spanò, L.; Capone, I.; Costantino, P. Agrobacterium rhizogenes T-DNA genes capable of inducing hairy root phenotype. Mol. Gen. Genet. 1987, 209, 475–480. [Google Scholar] [CrossRef]
- Altamura, M.M. Agrobacterium rhizogenes rolB and rolD genes: Regulation and involvement in plant development. Plant Cell Tissue Organ Cult. 2004, 77, 89–101. [Google Scholar] [CrossRef]
- Schmülling, T.; Schell, J.; Spena, A. Single genes from Agrobacterium rhizogenes influence plant development. EMBO J. 1988, 7, 2621–2629. [Google Scholar] [CrossRef] [PubMed]
- Röder, F.T.; Schmülling, T.; Gatz, C. Efficiency of the tetracycline-dependent gene expression system: Complete suppression and efficient induction of the rolB phenotype in transgenic plants. Mol. Gen. Genet. 1994, 243, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.D.; Karimi, M.; Impens, L.; Van Lerberge, E.; Coussens, G.; Aesaert, S.; Rombaut, D.; Holtappels, D.; Ibrahim, H.M.M.; Van Montagu, M.; et al. Efficient CRISPR-mediated base editing in Agrobacterium spp. Proc. Natl. Acad. Sci. USA 2021, 118, e2013338118. [Google Scholar] [CrossRef] [PubMed]
- Shkryl, Y.N.; Veremeichik, G.N.; Bulgakov, V.P.; Tchernoded, G.K.; Mischenko, N.P.; Fedoreyev, S.A.; Zhuravlev, Y.N. Individual and combined effects of the rolA, B, and C genes on anthraquinone production in Rubia cordifolia transformed calli. Biotechnol. Bioeng. 2008, 100, 118–125. [Google Scholar] [CrossRef]
- Priestap, H.A.; Velandia, A.E.; Johnson, J.V.; Barbieri, M.A. Secondary metabolite uptake by the Aristolochia-feeding papilionoid butterfly Battus polydamas. Biochem. Syst. Ecol. 2012, 40, 126–137. [Google Scholar] [CrossRef]
- Michl, J.; Kite, G.C.; Wanke, S.; Zierau, O.; Vollmer, G.; Neinhuis, C.; Simmonds, M.S.J.; Heinrich, M. LC-MS- and 1H NMR-based metabolomic analysis and in vitro toxicological assessment of 43 Aristolochia species. J. Nat. Prod. 2016, 79, 30–37. [Google Scholar] [CrossRef]
- Yu, J.; Ma, C.M.; Wang, X.; Shang, M.Y.; Hattori, M.; Xu, F.; Jing, Y.; Dong, S.W.; Xu, Y.Q.; Zhang, C.Y.; et al. Analysis of aristolochic acids, aristololactams and their analogues using liquid chromatography tandem mass spectrometry. Chin. J. Nat. Med. 2016, 14, 626–640. [Google Scholar] [CrossRef]
- Verma, P.; Khan, S.A.; Masood, N.; Manika, N.; Sharma, A.; Verma, N.; Luqman, S.; Mathur, A.K. Differential rubisco content and photosynthetic efficiency of rol gene integrated Vinca minor transgenic plant: Correlating factors associated with morpho-anatomical changes, gene expression and alkaloid productivity. J. Plant Physiol. 2017, 219, 12–21. [Google Scholar] [CrossRef]
- Palazón, J.; Cusidó, R.M.; Gonzalo, J.; Bonfill, M.; Morales, C.; Piñol, M.T. Relation between the amount of rolC gene product and indole alkaloid accumulation in Catharanthus roseus transformed root cultures. J. Plant Physiol. 1998, 153, 712–718. [Google Scholar] [CrossRef]
- Dilshad, E.; Zafar, S.; Ismail, H.; Waheed, M.T.; Cusido, R.M.; Palazon, J.; Mirza, B. Effect of rol genes on polyphenols biosynthesis in Artemisia annua and their effect on antioxidant and cytotoxic potential of the plant. Appl. Biochem. Biotechnol. 2016, 179, 1456–1468. [Google Scholar] [CrossRef]
- Bulgakov, V.P. Functions of rol genes in plant secondary metabolism. Biotechnol. Adv. 2008, 26, 318–324. [Google Scholar] [CrossRef]
- Sim, H.J.; Kim, J.H.; Lee, K.R.; Hong, J. Simultaneous determination of structurally diverse compounds in different Fangchi species by UHPLC-DAD and UHPLC-ESI-MS/MS. Molecules 2013, 18, 5235–5250. [Google Scholar] [CrossRef]
- Canedo-Téxon, A.; Ramón-Farias, F.; Monribot-Villanueva, J.L. Novel findings to the biosynthetic pathway of magnoflorine and taspine through transcriptomic and metabolomic analysis of Croton draco (Euphorbiaceae). BMC Plant Biol. 2019, 19, 560. [Google Scholar] [CrossRef]
- Tian, X.; Guo, S.; He, K.; Roller, M.; Yang, M.; Liu, Q.; Zhang, L.; Ho, C.T.; Bai, N. Qualitative and quantitative analysis of chemical constituents of Ptychopetalum olacoides Benth. Nat. Prod. Res. 2018, 32, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.; Ikuta, A.; Syōno, K. Alkaloids from callus tissue of Papaver somniferum. Phytochemistry 1972, 11, 3041–3044. [Google Scholar] [CrossRef]
- Gorpenchenko, T.Y.; Grigorchuk, V.P.; Fedoreyev, S.A.; Tarbeeva, D.V.; Tchernoded, G.K.; Bulgakov, V.P. Stepharine production in morphogenic cell cultures of Stephania glabra (ROXB.) Miers. Plant Cell Tissue Organ Cult. 2017, 128, 67–76. [Google Scholar] [CrossRef]
- Minami, H.; Kim, J.S.; Ikezawa, N.; Takemura, T.; Katayama, T.; Kumagai, H.; Sato, F. Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. USA 2008, 105, 7393–7398. [Google Scholar] [CrossRef]
- Shkryl, Y.; Tsydeneshieva, Z.; Degtyarenko, A.; Yugay, Y.; Balabanova, L.; Rusapetova, T.; Bulgakov, V. Plant exosomal vesicles: Perspective information nanocarriers in biomedicine. Appl. Sci. 2022, 12, 8262. [Google Scholar] [CrossRef]
- Selma, S.; Ceulemans, E.; Goossens, A.; Lacchini, E. Clustered regularly interspaced short palindromic repeats tools for plant metabolic engineering: Achievements and perspectives. Curr. Opin. Biotechnol. 2023, 79, 102856. [Google Scholar] [CrossRef]
- Kowalczyk, T.; Merecz-Sadowska, A.; Picot, L.; Brčić Karačonji, I.; Wieczfinska, J.; Śliwiński, T.; Sitarek, P. Genetic manipulation and bioreactor culture of plants as a tool for industry and its applications. Molecules 2022, 27, 795. [Google Scholar] [CrossRef] [PubMed]
- Marone, D.; Mastrangelo, A.M.; Borrelli, G.M.; Mores, A.; Laidò, G.; Russo, M.A.; Ficco, D. Specialized metabolites: Physiological and biochemical role in stress resistance, strategies to improve their accumulation, and new applications in crop breeding and management. Plant Physiol. Biochem. 2022, 172, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Tan, H.; Li, Q.; Chen, J.; Gao, S.; Wang, Y.; Chen, W.; Zhang, L. CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry 2018, 148, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Subramaniyan, V.; Saravanan, R.; Baskaran, D.; Ramalalingam, S. In vitro free radical scavenging and anticancer potential of Aristolochia indica L. against MCF-7 cell line. Int. J. Pharm. 2015, 7, 392–396. [Google Scholar]
- Karan, S.K.; Mishra, S.K.; Pal, D.; Mondal, A. Isolation of β-sitosterol and evaluation of antidiabetic activity of Aristolochia indica in alloxan-induced diabetic mice with a reference to in vitro antioxidant activity. J. Med. Plant Res. 2012, 6, 1219–1223. [Google Scholar] [CrossRef]
- Jegadeeswari, P.; Daffodil, E.; Mohan, V.R. Quantification of total phenolics, flavonoid and in vitro antioxidant activity of Aristolochia bracteata Retz. Int. J. Pharm. Pharm. 2014, 6, 747–752. [Google Scholar]
- El Omari, N.; Sayah, K.; Fettach, S.; El Blidi, O.; Bouyahya, A.; Faouzi, M.E.A.; Kamal, R.; Barkiyou, M. Evaluation of in vitro antioxidant and antidiabetic activities of Aristolochia longa extracts. Evid. Based Complement. Altern. Med. 2019, 2019, 7384735. [Google Scholar] [CrossRef]
- Benmehdi, H.; Behilil, A.; Memmou, F.; Amrouche, A. Free radical scavenging activity, kinetic behaviour and phytochemical constituents of Aristolochia clematitis L. roots. Arab. J. Chem. 2017, 10, S1402–S1408. [Google Scholar] [CrossRef]
- Guinnin, F.F.D.; Sacramento, I.T.; Ategbo, J.M.; Agbangnan, C.D.P. Physico-chemical composition and radicalscavenging activity evaluation of the extracts of Aristolochia albida Duch. (Aristolochiaceae) of Benin. J. Appl. Biosci. 2016, 107, 10460–10470. [Google Scholar] [CrossRef]
- Okon, E.; Kukula-Koch, W.; Jarzab, A.; Halasa, M.; Stepulak, A.; Wawruszak, A. Advances in chemistry and bioactivity of magnoflorine and magnoflorine-containing extracts. Int. J. Mol. Sci. 2020, 21, 1330. [Google Scholar] [CrossRef]
- Li, C.; Wang, M.H. Potential biological activities of magnoflorine: A compound from Aristolochia debilis sieb. et Zucc. Korean J. Plant Res. 2014, 27, 223–228. [Google Scholar] [CrossRef]
- Mohamed, S.; Hassan, E.; Ibrahim, N. Cytotoxic and antiviral activities of aporphine alkaloids of Magnolia grandiflora L. Nat. Prod. Res. 2010, 24, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Ye, X.L.; Cui, X.L.; He, K.; Jin, Y.N.; Chen, Z.; Li, X.G. Cytotoxicity and antihyperglycemic effect of minor constituents from Rhizoma coptis in HepG2 cells. Fitoterapia 2012, 83, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Okon, E.; Kukula-Koch, W.; Halasa, M.; Jarzab, A.; Baran, M.; Dmoszynska-Graniczka, M.; Angelis, A.; Kalpoutzakis, E.; Guz, M.; Stepulak, A.; et al. Magnoflorine-isolation and the anticancer potential against NCI-H1299 lung, MDA-MB-468 breast, T98G glioma, and TE671 rhabdomyosarcoma cancer cells. Biomolecules 2020, 10, 1532. [Google Scholar] [CrossRef]
- Sun, X.L.; Zhang, X.W.; Zhai, H.J.; Zhang, D.; Ma, S.Y. Magnoflorine inhibits human gastric cancer progression by inducing autophagy, apoptosis and cell cycle arrest by JNK activation regulated by ROS. Biomed. Pharmacother. 2020, 125, 109118. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Xiaojun, X.; Peilong, C. Magnoflorine improves sensitivity to doxorubicin (DOX) of breast cancer cells via inducing apoptosis and autophagy through AKT/mTOR and p38 signaling pathways. Biomed. Pharmacother. 2020, 121, 109139. [Google Scholar] [CrossRef] [PubMed]
- Angelova, A.L.; Grekova, S.P.; Heller, A.; Kuhlmann, O.; Soyka, E.; Giese, T.; Aprahamian, M.; Bour, G.; Rüffer, S.; Cziepluch, C.; et al. Complementary induction of immunogenic cell death by oncolytic parvovirus H-1PV and gemcitabine in pancreatic cancer. J. Virol. 2014, 88, 5263–5276. [Google Scholar] [CrossRef] [PubMed]
- Chunmei, L.; Myeong-Hyeon, W. Aristolochia debilis Sieb. et Zucc. induces apoptosis and reactive oxygen species in the HT-29 human colon cancer cell line. Cancer Biother. Radiopharm. 2013, 28, 717–724. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Tchernoded, G.K.; Veselova, M.V.; Fedoreyev, S.A.; Muzarok, T.I.; Zhuravlev, Y.N. Catechin production in cultured cells of Taxus cuspidata and Taxus baccata. Biotechnol. Lett. 2011, 33, 1879–1883. [Google Scholar] [CrossRef]
- Hernalsteens, J.P.; Van Vliet, F.; De Beuckeleer, M.; Depicker, A.; Engler, G.; Lemmers, M.; Holsters, M.; Van Montagu, M.; Schell, J. The Agrobacterium tumefaciens Ti plasmid as a host vector system for introducing foreign DNA into plant cells. Nature 1980, 287, 654–656. [Google Scholar] [CrossRef]
- Echt, C.S.; Erdahl, L.A.; McCoy, T.J. Genetic segregation of random amplified polymorphic DNA in diploid cultivated alfalfa. Genome 1992, 35, 84–87. [Google Scholar] [CrossRef]
- Gorpenchenko, T.Y.; Grigorchuk, V.P.; Bulgakov, D.V.; Tchernoded, G.K.; Bulgakov, V.P. Tempo-spatial pattern of stepharine accumulation in Stephania glabra morphogenic tissues. Int. J. Mol. Sci. 2019, 20, 808. [Google Scholar] [CrossRef] [PubMed]
- Mensor, L.L.; Menezes, F.S.; Leitao, G.G.; Reis, A.S.; Santos, T.S.; Coube, C.S. Screening of Brazilian plant extracts for antioxidant activity by the use of DPPH free radical method. Phytother. Res. 2001, 15, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Shkryl, Y.; Rusapetova, T.; Yugay, Y.; Egorova, A.; Silant’ev, V.; Grigorchuk, V.; Karabtsov, A.; Timofeeva, Y.; Vasyutkina, E.; Kudinova, O.; et al. Biosynthesis and cytotoxic properties of Ag, Au, and bimetallic nanoparticles synthesized using Lithospermum erythrorhizon callus culture extract. Int. J. Mol. Sci. 2021, 22, 9305. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).