Molecular Mechanism of Zinc-Dependent Oligomerization of Alzheimer’s Amyloid-β with Taiwan (D7H) Mutation
Abstract
1. Introduction
2. Results
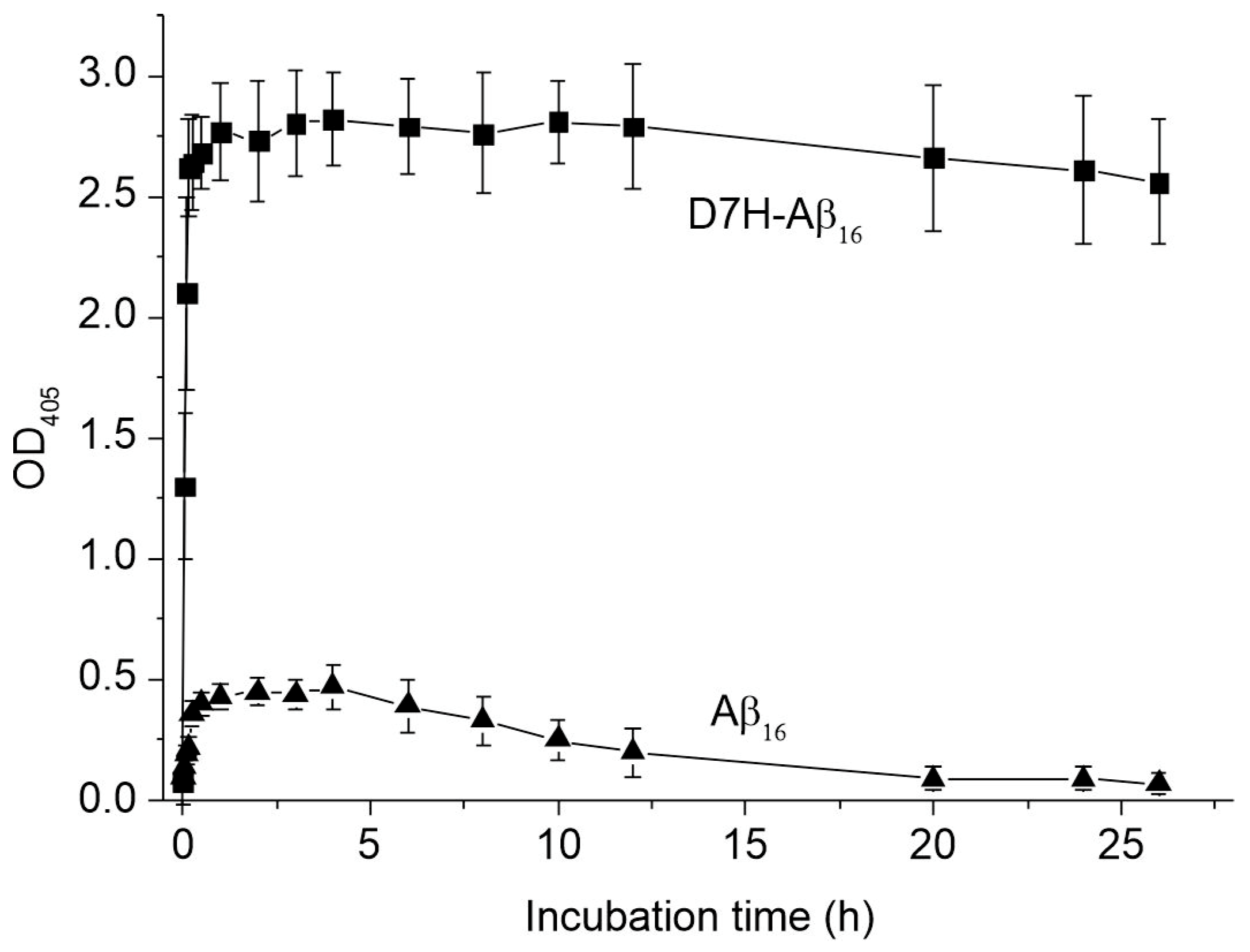
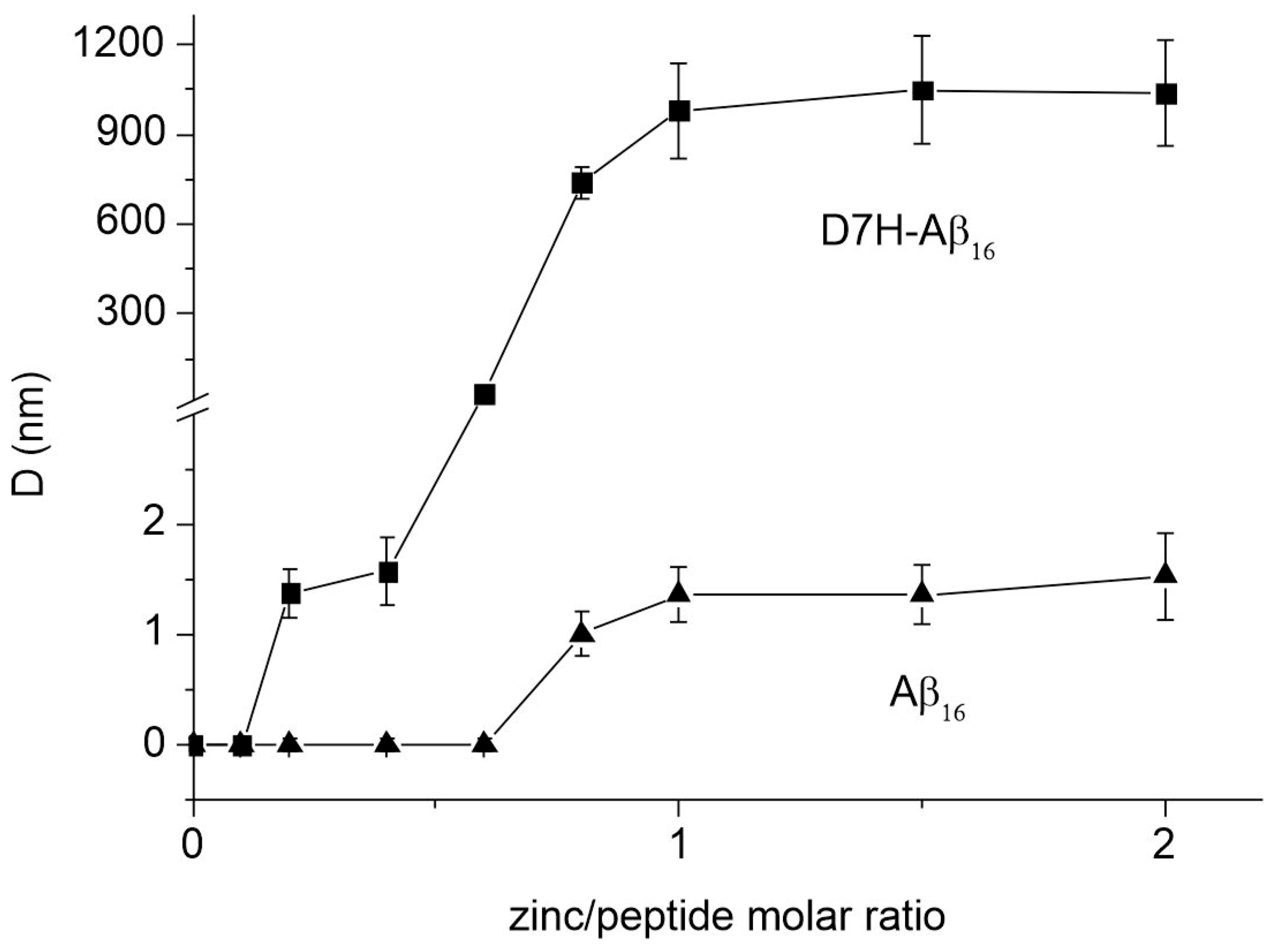
2.1. Aggregation States of Aβ16 and D7H-Aβ16 in the Presence of Zinc Ions
2.2. Determination of the Thermodynamic Parameters of the Formation of Zinc-Bound Complexes of Mutant Aβ16 Isoforms by Isothermal Titration Calorimetry (ITC)
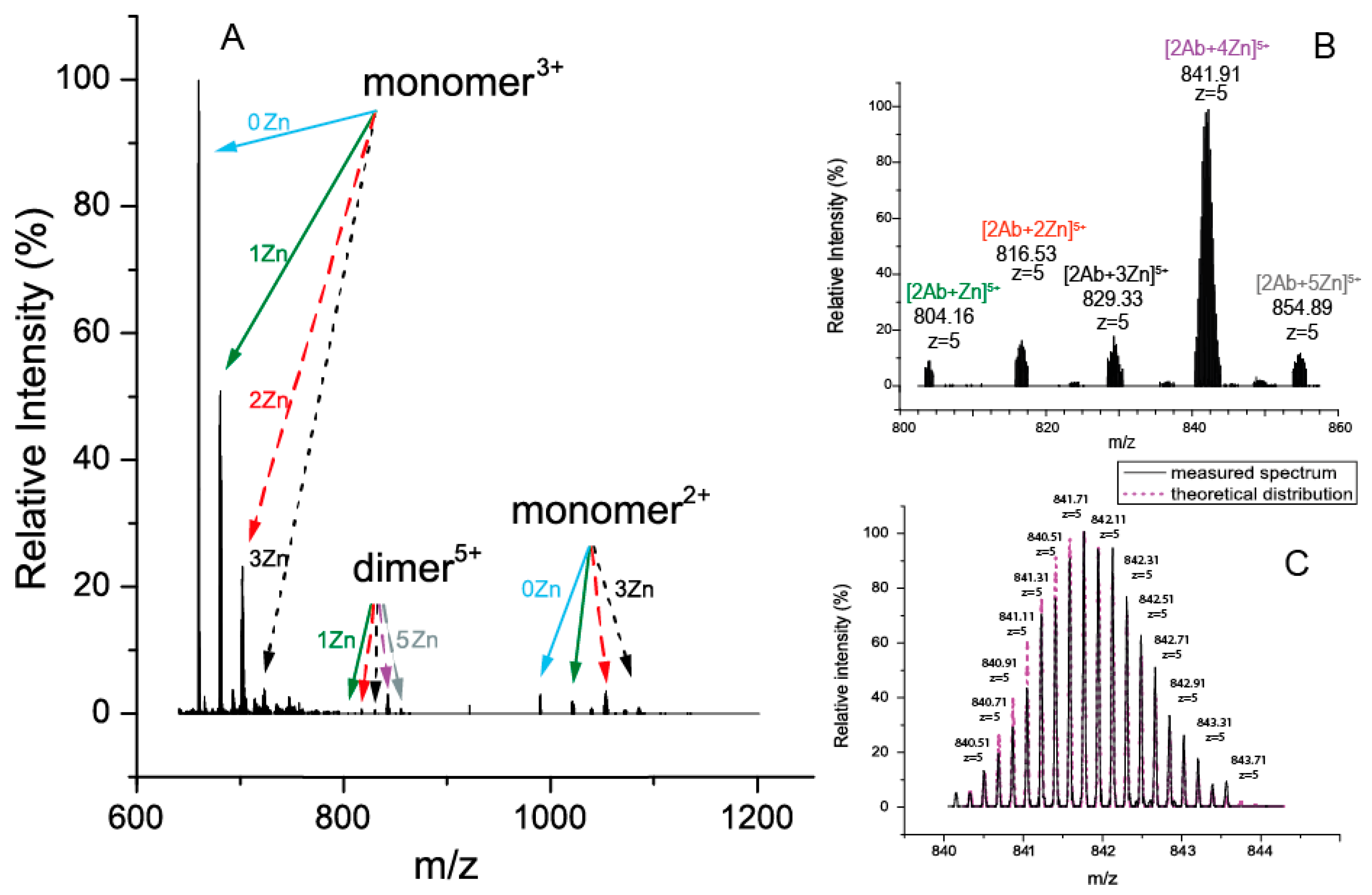
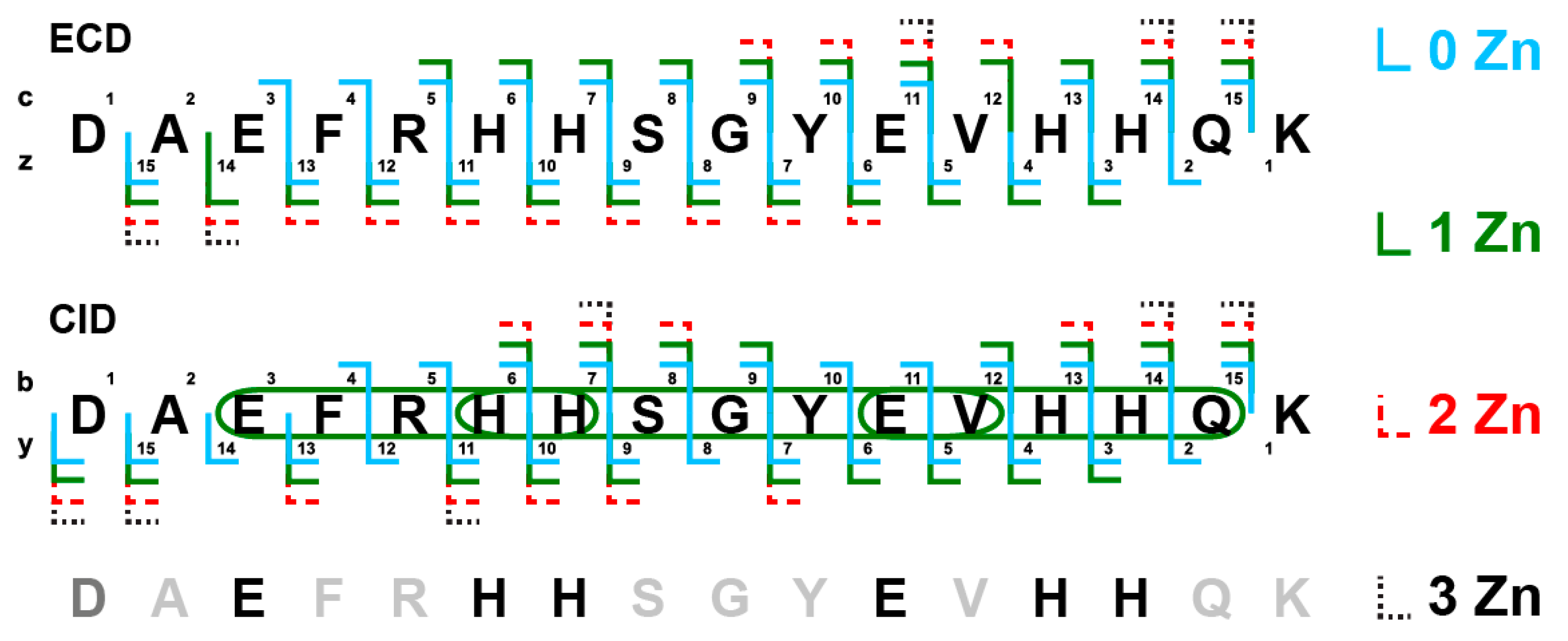
2.3. Detection of Zinc Ion Chelators in the D7H-Aβ16 Monomer and Dimer by Mass Spectrometry
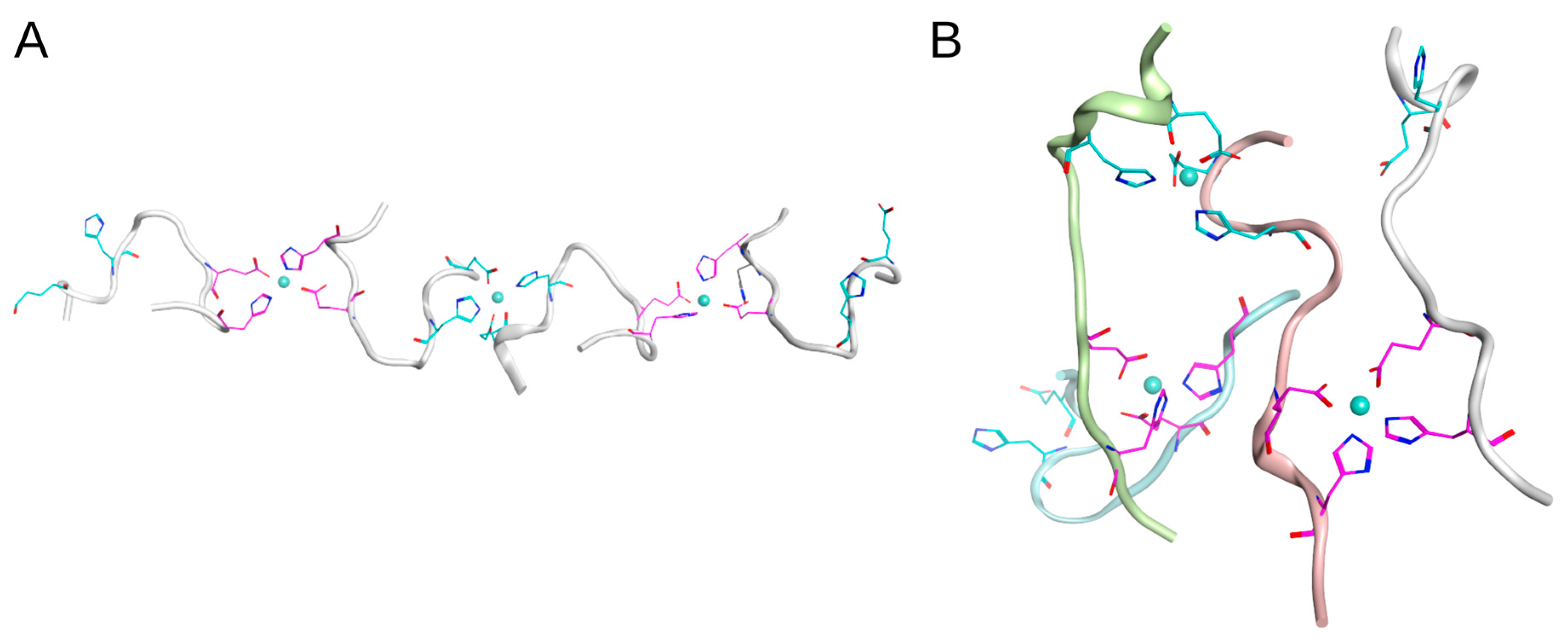
2.4. Molecular Modeling of Zinc-Induced Oligomers of D7H-Aβ16
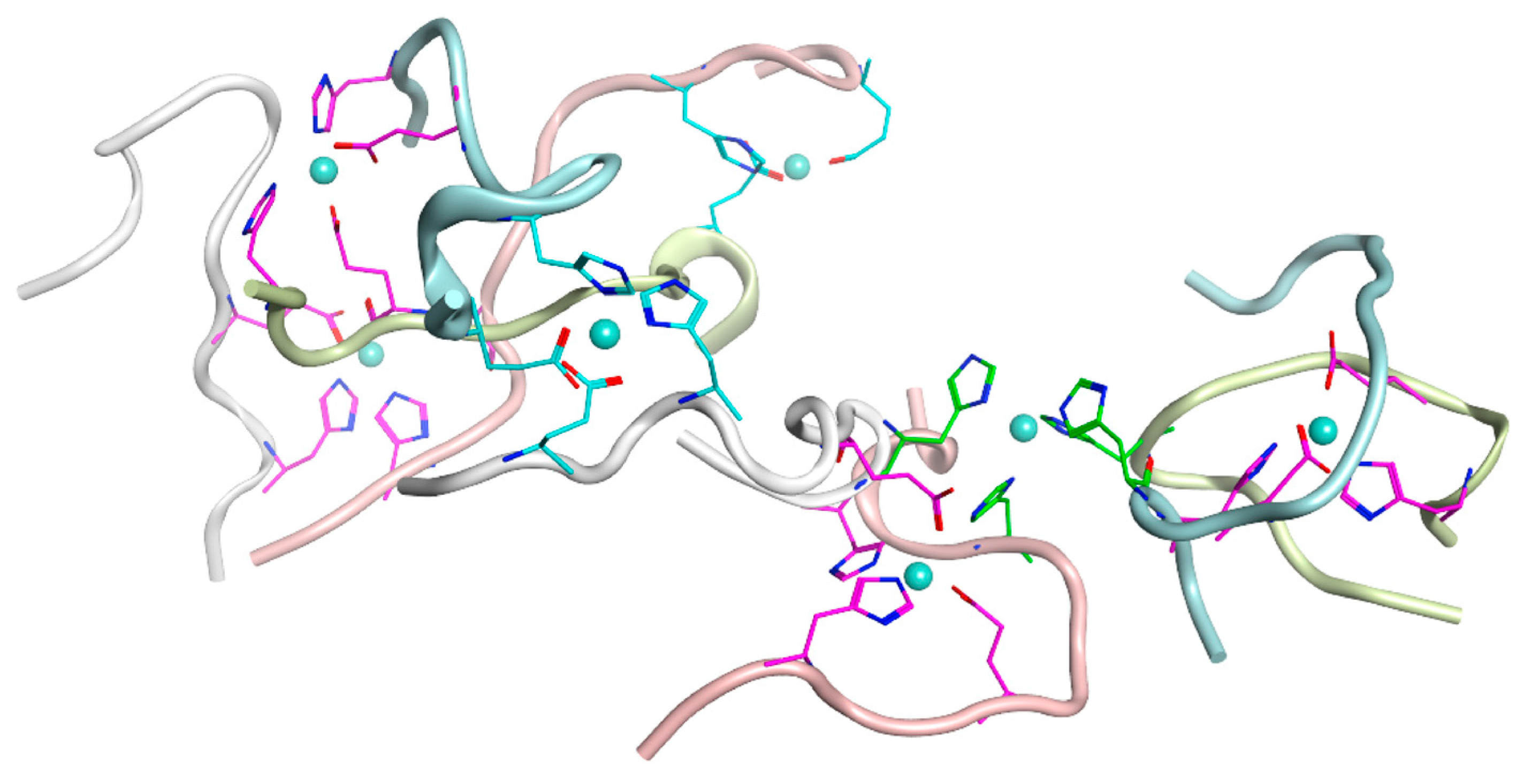
2.5. Molecular Modeling of Zinc-Bound Heterotrimers of Aβ16/Zn/D7H-Aβ16/Zn/Aβ16 and Aβ16/Zn/isoD7-Aβ16/Zn/Aβ16
3. Discussion
4. Materials and Methods
4.1. Amyloid Peptides
4.2. Turbidity and DLS Measurements
4.3. Isothermal Titration Calorimetry
4.4. Mass-Spectrometry
4.5. Molecular Modelling
4.5.1. Structure Preparation
4.5.2. Molecular Modeling Protocol
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Querfurth, H.W.; LaFerla, F.M. Alzheimer’s disease. N. Engl. J. Med. 2010, 362, 329–344. [Google Scholar] [CrossRef]
- Golde, T.E.; DeKosky, S.T.; Galasko, D. Alzheimer’s disease: The right drug, the right time. Science 2018, 362, 1250–1251. [Google Scholar] [CrossRef]
- Huang, L.K.; Chao, S.P.; Hu, C.J. Clinical trials of new drugs for Alzheimer disease. J. Biomed. Sci. 2020, 27, 18. [Google Scholar] [CrossRef]
- Sevigny, J.; Chiao, P.; Bussière, T.; Weinreb, P.H.; Williams, L.; Maier, M.; Dunstan, R.; Salloway, S.; Chen, T.; Ling, Y.; et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature 2016, 537, 50–56. [Google Scholar] [CrossRef]
- Perneczky, R.; Jessen, F.; Grimmer, T.; Levin, J.; Flöel, A.; Peters, O.; Froelich, L. Anti-amyloid antibody therapies in Alzheimer’s disease. Brain 2023, 146, 842–849. [Google Scholar] [CrossRef]
- Kozin, S.A.; Barykin, E.P.; Mitkevich, V.A.; Makarov, A.A. Anti-amyloid Therapy of Alzheimer’s Disease: Current State and Prospects. Biochemistry 2018, 83, 1057–1067. [Google Scholar] [CrossRef]
- Craddock, T.J.A.; Tuszynski, J.A.; Chopra, D.; Casey, N.; Goldstein, L.E.; Hameroff, S.R.; Tanzi, R.E. The Zinc Dyshomeostasis Hypothesis of Alzheimer’s Disease. PLoS ONE 2012, 7, e33552. [Google Scholar] [CrossRef]
- Lovell, M.A.; Robertson, J.D.; Teesdale, W.J.; Campbell, J.L.; Markesbery, W.R. Copper, iron and zinc in Alzheimer’s disease senile plaques. J. Neurol. Sci. 1998, 158, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Bush, A.I.; Pettingell, W.H.; Multhaup, G.; d Paradis, M.; Vonsattel, J.P.; Gusella, J.F.; Beyreuther, K.; Masters, C.L.; Tanzi, R.E. Rapid induction of Alzheimer A beta amyloid formation by zinc. Science 1994, 265, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Miller, Y.; Ma, B.; Nussinov, R. Zinc ions promote Alzheimer Aβ aggregation via population shift of polymorphic states. Proc. Natl. Acad. Sci. USA 2010, 107, 9490–9495. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.M.; Wang, Q.; Telivala, T.P.; Smith, R.J.; Lanzirotti, A.; Miklossy, J. Synchrotron-based infrared and X-ray imaging shows focalized accumulation of Cu and Zn co-localized with beta-amyloid deposits in Alzheimer’s disease. J. Struct. Biol. 2006, 155, 30–37. [Google Scholar] [CrossRef]
- Suh, S.W.; Jensen, K.B.; Jensen, M.S.; Silva, D.S.; Kesslak, P.J.; Danscher, G.; Frederickson, C.J. Histochemically-reactive zinc in amyloid plaques, angiopathy, and degenerating neurons of Alzheimer’s diseased brains. Brain Res. 2000, 852, 274–278. [Google Scholar] [CrossRef]
- Jucker, M.; Walker, L.C. Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1341–1349. [Google Scholar] [CrossRef]
- Kozin, S.A. Role of Interaction between Zinc and Amyloid Beta in Pathogenesis of Alzheimer’s Disease. Biochemistry 2023, 88, S75–S87. [Google Scholar] [CrossRef]
- Kozin, S.A.; Zirah, S.; Rebuffat, S.; Hoa, G.H.; Debey, P. Zinc binding to Alzheimer’s Abeta(1-16) peptide results in stable soluble complex. Biochem. Biophys. Res. Commun. 2001, 285, 959–964. [Google Scholar] [CrossRef]
- Zirah, S.; Kozin, S.A.; Mazur, A.K.; Blond, A.; Cheminant, M.; Segalas-Milazzo, I.; Debey, P.; Rebuffat, S. Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging. J. Biol. Chem. 2006, 281, 2151–2161. [Google Scholar] [CrossRef] [PubMed]
- Talmard, C.; Guilloreau, L.; Coppel, Y.; Mazarguil, H.; Faller, P. Amyloid-beta peptide forms monomeric complexes with Cu(II) and Zn(II) prior to aggregation. ChemBioChem 2007, 8, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Luehmann, M.; Coomaraswamy, J.; Bolmont, T.; Kaeser, S.; Schaefer, C.; Kilger, E.; Neuenschwander, A.; Abramowski, D.; Frey, P.; Jaton, A.L.; et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science 2006, 313, 1781–1784. [Google Scholar] [CrossRef]
- Tsvetkov, P.O.; Popov, I.A.; Nikolaev, E.N.; Archakov, A.I.; Makarov, A.A.; Kozin, S.A. Isomerization of the Asp7 residue results in zinc-induced oligomerization of Alzheimer’s disease amyloid beta(1-16) peptide. ChemBioChem 2008, 9, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Roher, A.E.; Palmer, K.C.; Yurewicz, E.C.; Ball, M.J.; Greenberg, B.D. Morphological and biochemical analyses of amyloid plaque core proteins purified from Alzheimer disease brain tissue. J. Neurochem. 1993, 61, 1916–1926. [Google Scholar] [CrossRef] [PubMed]
- Istrate, A.N.; Kozin, S.A.; Zhokhov, S.S.; Mantsyzov, A.B.; Kechko, O.I.; Pastore, A.; Makarov, A.A.; Polshakov, V.I. Interplay of histidine residues of the Alzheimer’s disease Aβ peptide governs its Zn-induced oligomerization. Sci. Rep. 2016, 6, 21734. [Google Scholar] [CrossRef] [PubMed]
- Kozin, S.A.; Cheglakov, I.B.; Ovsepyan, A.A.; Telegin, G.B.; Tsvetkov, P.O.; Lisitsa, A.V.; Makarov, A.A. Peripherally applied synthetic peptide isoAsp7-Abeta(1-42) triggers cerebral beta-amyloidosis. Neurotox. Res. 2013, 24, 370–376. [Google Scholar] [CrossRef]
- Kulikova, A.A.; Cheglakov, I.B.; Kukharsky, M.S.; Ovchinnikov, R.K.; Kozin, S.A.; Makarov, A.A. Intracerebral Injection of Metal-Binding Domain of Aβ Comprising the Isomerized Asp7 Increases the Amyloid Burden in Transgenic Mice. Neurotox. Res. 2016, 29, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Mitkevich, V.A.; Barykin, E.P.; Eremina, S.; Pani, B.; Katkova-Zhukotskaya, O.; Polshakov, V.I.; Adzhubei, A.A.; Kozin, S.A.; Mironov, A.S.; Makarov, A.A.; et al. Zn-dependent β-amyloid Aggregation and its Reversal by the Tetrapeptide HAEE. Aging Dis. 2023, 14, 309–318. [Google Scholar] [CrossRef]
- Barykin, E.P.; Mitkevich, V.A.; Kozin, S.A.; Makarov, A.A. Amyloid β Modification: A Key to the Sporadic Alzheimer’s Disease? Front. Genet. 2017, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Polshakov, V.I.; Mantsyzov, A.B.; Kozin, S.A.; Adzhubei, A.A.; Zhokhov, S.S.; van Beek, W.; Kulikova, A.A.; Indeykina, M.I.; Mitkevich, V.A.; Makarov, A.A. A Binuclear Zinc Interaction Fold Discovered in the Homodimer of Alzheimer’s Amyloid-β Fragment with Taiwanese Mutation D7H. Angew. Chem. Int. Ed. 2017, 56, 11734–11739. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.T.; Hong, C.J.; Lin, Y.T.; Chang, W.H.; Huang, H.T.; Liao, J.Y.; Chang, Y.J.; Hsieh, Y.F.; Cheng, C.Y.; Liu, H.C.; et al. Amyloid-beta (Aβ) D7H mutation increases oligomeric Aβ42 and alters properties of Aβ-zinc/copper assemblies. PLoS ONE 2012, 7, e35807. [Google Scholar] [CrossRef]
- Xu, L.; Chen, Y.; Wang, X. Dual effects of familial Alzheimer’s disease mutations (D7H, D7N, and H6R) on amyloid β peptide: Correlation dynamics and zinc binding. Proteins 2014, 82, 3286–3297. [Google Scholar] [CrossRef]
- Istrate, A.N.; Tsvetkov, P.O.; Mantsyzov, A.B.; Kulikova, A.A.; Kozin, S.A.; Makarov, A.A.; Polshakov, V.I. NMR solution structure of rat Aβ(1-16): Toward understanding the mechanism of rats’ resistance to Alzheimer’s disease. Biophys. J. 2012, 102, 136–143. [Google Scholar] [CrossRef]
- Kozin, S.A.; Kulikova, A.A.; Istrate, A.N.; Tsvetkov, P.O.; Zhokhov, S.S.; Mezentsev, Y.V.; Kechko, O.I.; Ivanov, A.S.; Polshakov, V.I.; Makarov, A.A. The English (H6R) familial Alzheimer’s disease mutation facilitates zinc-induced dimerization of the amyloid-β metal-binding domain. Metallomics 2015, 7, 422–425. [Google Scholar] [CrossRef]
- Kulikova, A.A.; Tsvetkov, P.O.; Indeykina, M.I.; Popov, I.A.; Zhokhov, S.S.; Golovin, A.V.; Polshakov, V.I.; Kozin, S.A.; Nudler, E.; Makarov, A.A. Phosphorylation of Ser8 promotes zinc-induced dimerization of the amyloid-β metal-binding domain. Mol. BioSyst. 2014, 10, 2590–2596. [Google Scholar] [CrossRef] [PubMed]
- Kozin, S.A.; Makarov, A.A. The Convergence of Alzheimer’s Disease Pathogenesis Concepts. Mol. Biol. 2019, 53, 896–903. [Google Scholar] [CrossRef]
- Radko, S.P.; Khmeleva, S.A.; Suprun, E.V.; Kozin, S.A.; Bodoev, N.V.; Makarov, A.A.; Archakov, A.I.; Shumyantseva, V.V. Physico-chemical methods for studying amyloid-β aggregation. Biochem. Suppl. Ser. B Biomed. Chem. 2015, 9, 258–274. [Google Scholar] [CrossRef]
- Kozin, S.A.; Mezentsev, Y.V.; Kulikova, A.A.; Indeykina, M.I.; Golovin, A.V.; Ivanov, A.S.; Tsvetkov, P.O.; Makarov, A.A. Zinc-induced dimerization of the amyloid-β metal-binding domain 1-16 is mediated by residues 11-14. Mol. BioSyst. 2011, 7, 1053–1055. [Google Scholar] [CrossRef] [PubMed]
- Mezentsev, Y.V.; Medvedev, A.E.; Kechko, O.I.; Makarov, A.A.; Ivanov, A.S.; Mantsyzov, A.B.; Kozin, S.A. Zinc-induced heterodimer formation between metal-binding domains of intact and naturally modified amyloid-beta species: Implication to amyloid seeding in Alzheimer’s disease? J. Biomol. Struct. Dyn. 2016, 34, 2317–2326. [Google Scholar] [CrossRef]
- Tsvetkov, P.O.; Kulikova, A.A.; Golovin, A.V.; Tkachev, Y.V.; Archakov, A.I.; Kozin, S.A.; Makarov, A.A. Minimal Zn(2+) binding site of amyloid-β. Biophys. J. 2010, 99, L84–L86. [Google Scholar] [CrossRef]
- Popov, I.A.; Indeikina, M.I.; Kononikhin, A.S.; Starodubtseva, N.L.; Kozin, S.A.; Makarov, A.A.; Nikolaev, E.N. ESI-MS identification of the minimal zinc-binding center in natural isoforms of β-amyloid domain 1–16. Mol. Biol. 2013, 47, 440–445. [Google Scholar] [CrossRef]
- Zirah, S.; Rebuffat, S.; Kozin, S.A.; Debey, P.; Fournier, F.; Lesage, D.; Tabet, J.-C. Zinc binding properties of the amyloid fragment Aβ(1–16) studied by electrospray-ionization mass spectrometry. Int. J. Mass Spectrom. 2003, 228, 999–1016. [Google Scholar] [CrossRef]
- Tolstova, A.P.; Makarov, A.A.; Adzhubei, A.A. Zinc Induced Aβ16 Aggregation Modeled by Molecular Dynamics. Int. J. Mol. Sci. 2021, 22, 12161. [Google Scholar] [CrossRef]
- Radko, S.P.; Khmeleva, S.A.; Kaluzhny, D.N.; Kechko, O.I.; Kiseleva, Y.Y.; Kozin, S.A.; Mitkevich, V.A.; Makarov, A.A. The English (H6R) Mutation of the Alzheimer’s Disease Amyloid-β Peptide Modulates Its Zinc-Induced Aggregation. Biomolecules 2020, 10, 961. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Macchiagodena, M.; Pagliai, M.; Andreini, C.; Rosato, A.; Procacci, P. Upgraded AMBER Force Field for Zinc-Binding Residues and Ligands for Predicting Structural Properties and Binding Affinities in Zinc-Proteins. ACS Omega 2020, 5, 15301–15310. [Google Scholar] [CrossRef] [PubMed]





| N b | Ka c × 10−4, M−1 | ΔH d kcal mol−1 | |
|---|---|---|---|
| Aβ16 e | 1.1 | 1.8 | −4 |
| D7H-Aβ16 | 1 | 2.3 | −10.3 |
| 0.35 | 48 | −8.0 | |
| E11A-D7H-Aβ16 | 1.1 | 2.1 | −8.7 |
| H13A-D7H-Aβ16 | 1 | 1.7 | −8.7 |
| 0.3 | 20 | −6.0 | |
| H14A-D7H-Aβ16 | 1 | 1.8 | −9.0 |
| D7H-Aβ10 f | 1 | 4.1 | −10.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kechko, O.I.; Adzhubei, A.A.; Tolstova, A.P.; Indeykina, M.I.; Popov, I.A.; Zhokhov, S.S.; Gnuchev, N.V.; Mitkevich, V.A.; Makarov, A.A.; Kozin, S.A. Molecular Mechanism of Zinc-Dependent Oligomerization of Alzheimer’s Amyloid-β with Taiwan (D7H) Mutation. Int. J. Mol. Sci. 2023, 24, 11241. https://doi.org/10.3390/ijms241411241
Kechko OI, Adzhubei AA, Tolstova AP, Indeykina MI, Popov IA, Zhokhov SS, Gnuchev NV, Mitkevich VA, Makarov AA, Kozin SA. Molecular Mechanism of Zinc-Dependent Oligomerization of Alzheimer’s Amyloid-β with Taiwan (D7H) Mutation. International Journal of Molecular Sciences. 2023; 24(14):11241. https://doi.org/10.3390/ijms241411241
Chicago/Turabian StyleKechko, Olga I., Alexei A. Adzhubei, Anna P. Tolstova, Maria I. Indeykina, Igor A. Popov, Sergey S. Zhokhov, Nikolay V. Gnuchev, Vladimir A. Mitkevich, Alexander A. Makarov, and Sergey A. Kozin. 2023. "Molecular Mechanism of Zinc-Dependent Oligomerization of Alzheimer’s Amyloid-β with Taiwan (D7H) Mutation" International Journal of Molecular Sciences 24, no. 14: 11241. https://doi.org/10.3390/ijms241411241
APA StyleKechko, O. I., Adzhubei, A. A., Tolstova, A. P., Indeykina, M. I., Popov, I. A., Zhokhov, S. S., Gnuchev, N. V., Mitkevich, V. A., Makarov, A. A., & Kozin, S. A. (2023). Molecular Mechanism of Zinc-Dependent Oligomerization of Alzheimer’s Amyloid-β with Taiwan (D7H) Mutation. International Journal of Molecular Sciences, 24(14), 11241. https://doi.org/10.3390/ijms241411241







