Quantification of Empty, Partially Filled and Full Adeno-Associated Virus Vectors Using Mass Photometry
Abstract
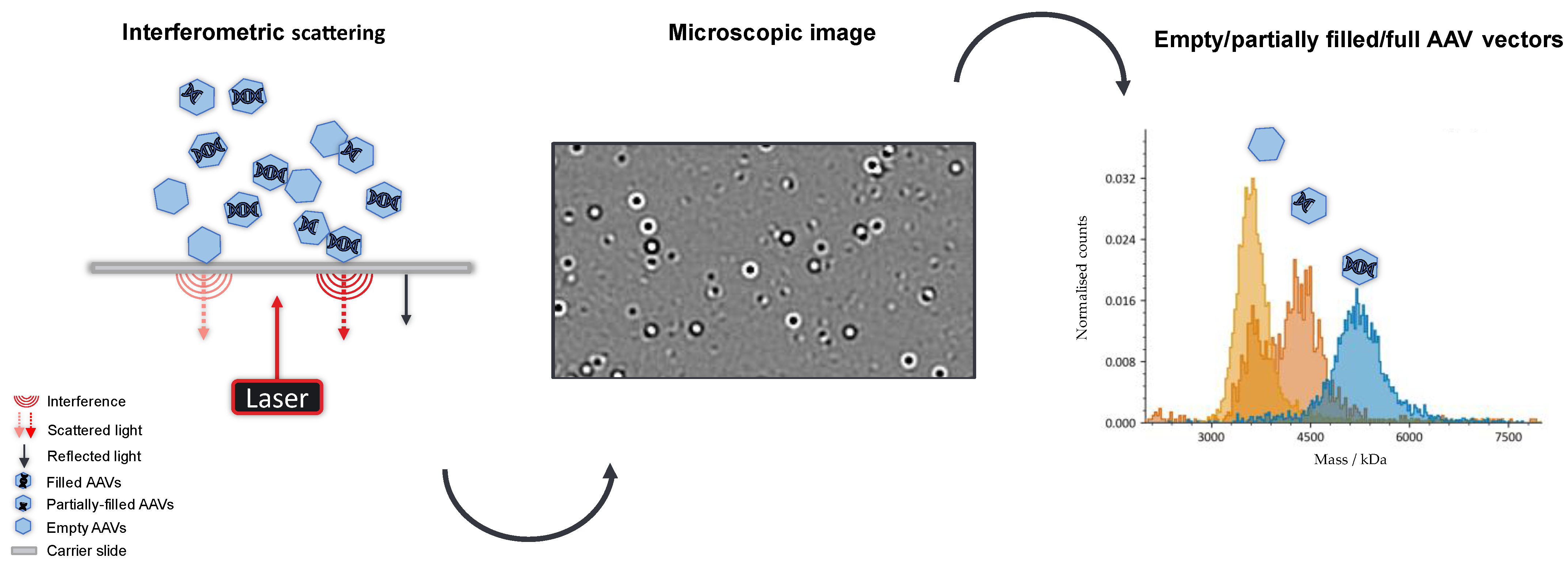
1. Introduction
2. Results and Discussion
2.1. Comparison of % Filled AAV Capsids by Orthogonal Methods
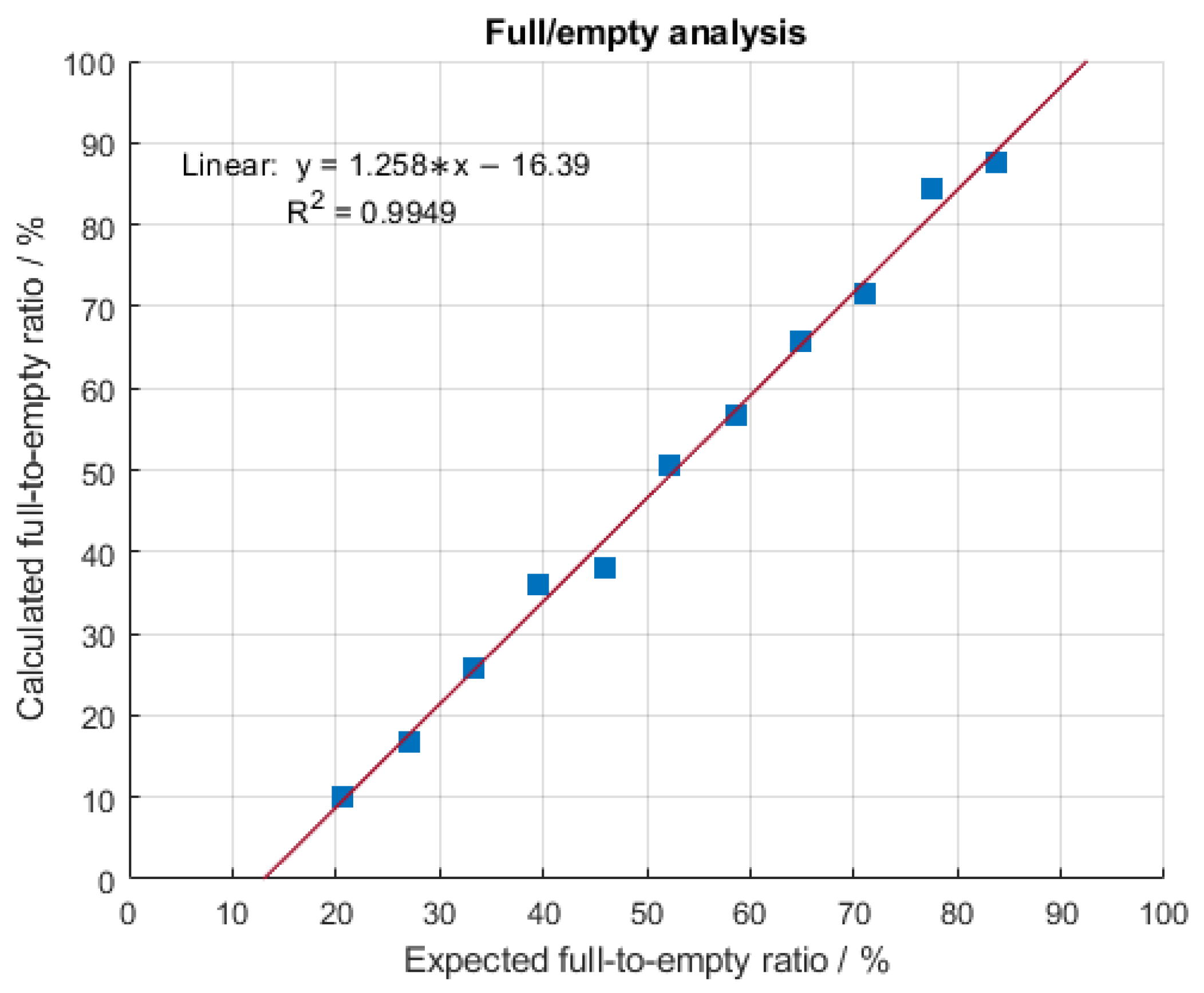
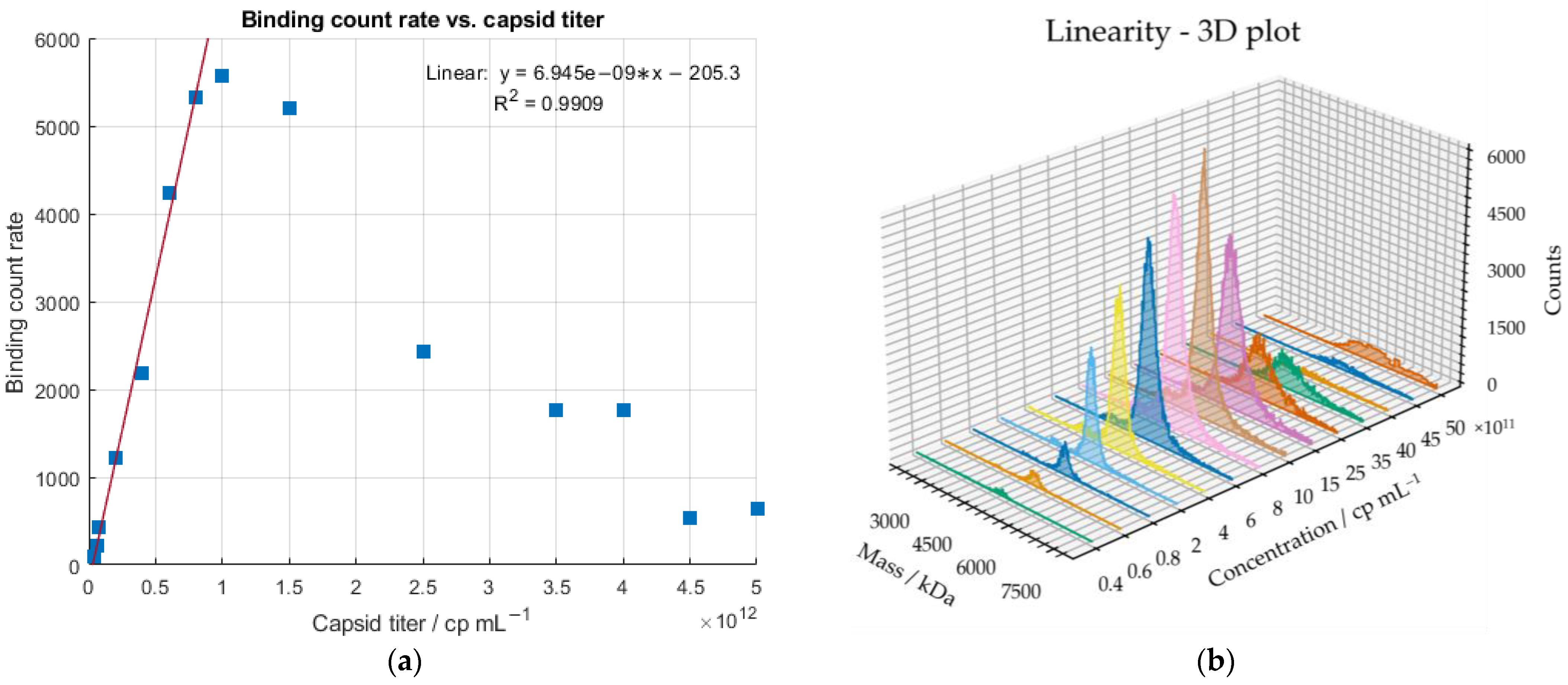
2.2. Linearity of the Mass Photometer
2.3. Precision of the Mass Photometer
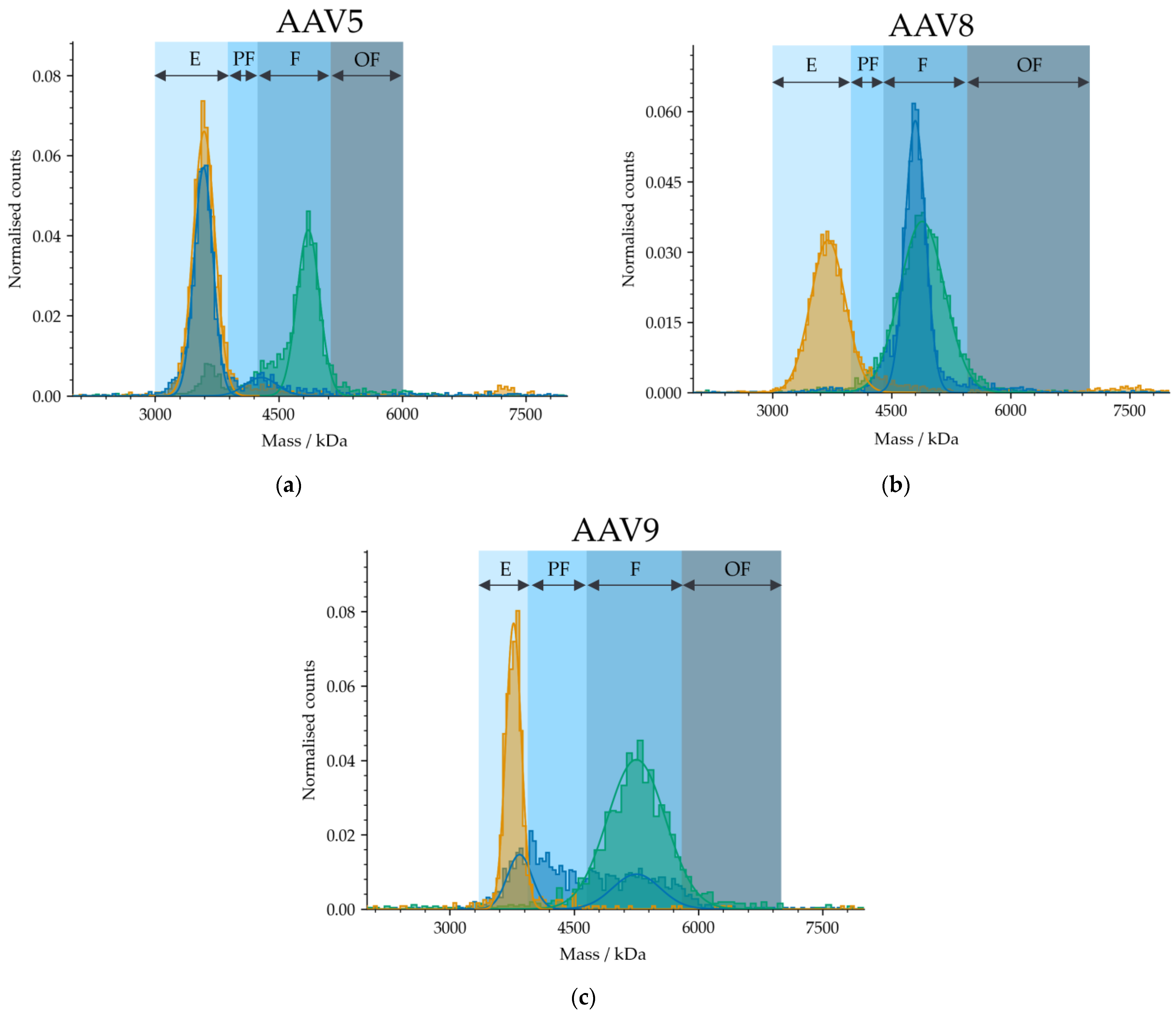
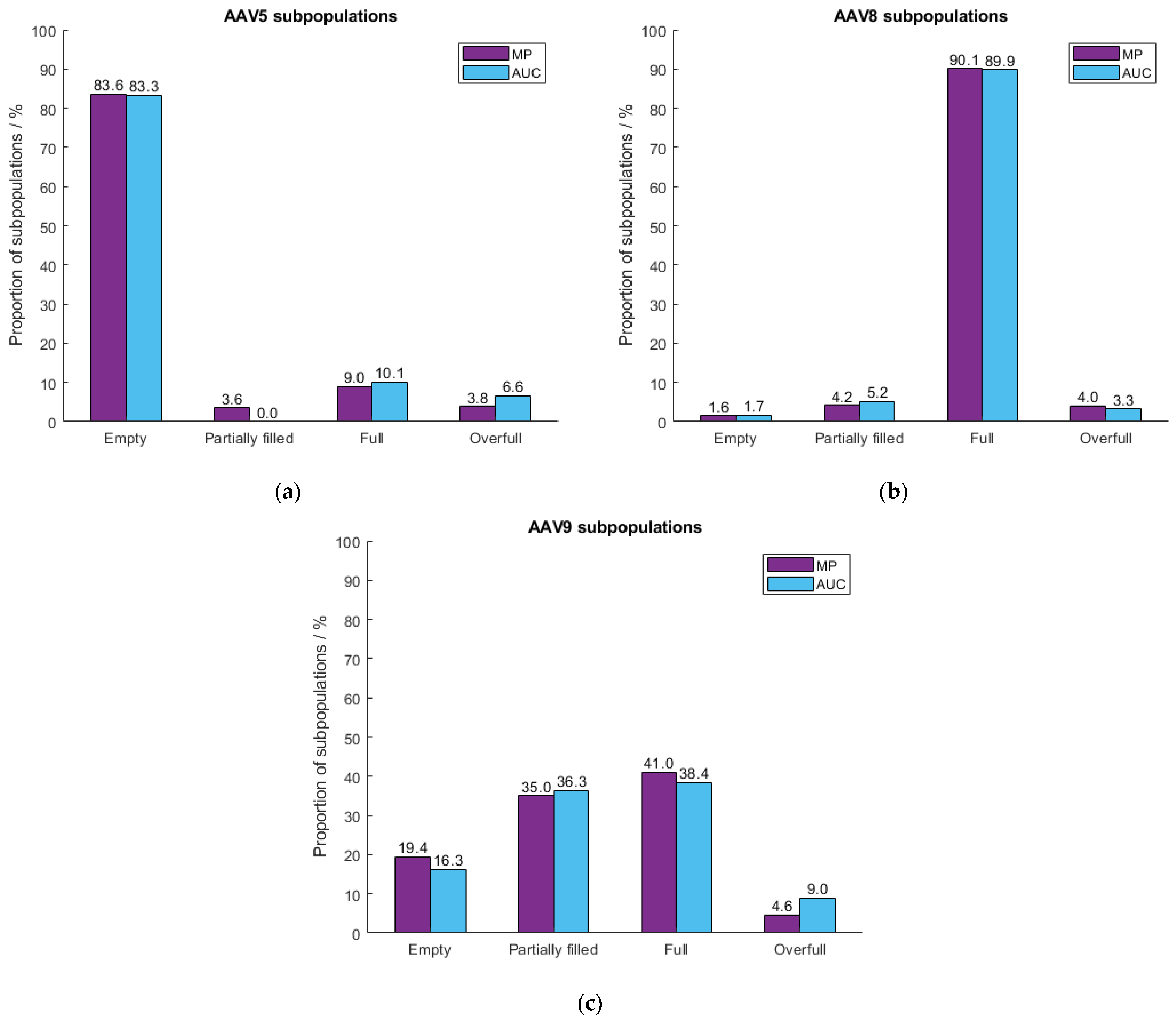
2.4. Measurement of Different Serotypes
2.5. Confirmation of Transgene Size
3. Materials and Methods
3.1. Sample Preparation
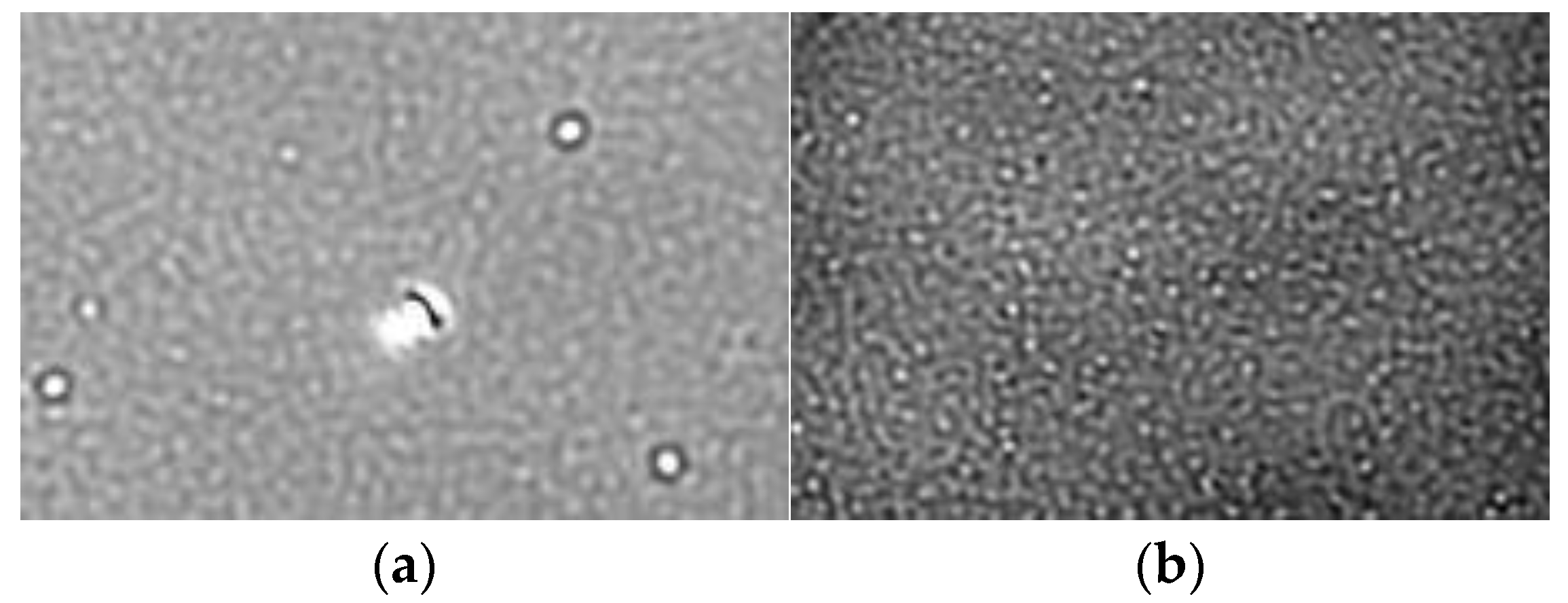
3.2. Measurements and Experimental Setup
3.3. Data Collection and Processing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Chandler, R.; Sands, M.S.; Venditti, C.P. Recombinant Adeno-Associated Viral Integration and Genotoxicity: Insights from Animal Models. Hum. Gene Ther. 2017, 28, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Maestro, S.; Weber, N.D.; Zabaleta, N.; Aldabe, R.; Gonzalez-Aseguinolaza, G. Novel vectors and approaches for gene therapy in liver diseases. JHEP Rep. 2021, 3, 100300. [Google Scholar] [CrossRef]
- Aponte-Ubillus, J.J.; Barajas, D.; Peltier, J.; Bardliving, C.; Shamlou, P.; Gold, D. Molecular design for recombinant adeno-associated virus (rAAV) vector production. Appl. Microbiol. Biotechnol. 2018, 102, 1045–1054. [Google Scholar] [CrossRef]
- Wileys, J. Gene Therapy Clinical Trials Worldwide. Available online: https://a873679.fmphost.com/fmi/webd/GTCT (accessed on 12 April 2023).
- EU/3/12/1091: Orphan Designation for the Treatment of Beta Thalassaemia Intermedia and Major. Available online: https://www.ema.europa.eu/en/medicines/human/orphan-designations/eu3121091 (accessed on 12 April 2023).
- FDA Approves First Cell-Based Gene Therapy to Treat Adult and Pediatric Patients with Beta-thalassemia Who Require Regular Blood Transfusions. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-cell-based-gene-therapy-treat-adult-and-pediatric-patients-beta-thalassemia-who (accessed on 12 April 2023).
- Wang, C.; Mulagapati, S.H.R.; Chen, Z.; Du, J.; Zhao, X.; Xi, G.; Chen, L.; Linke, T.; Gao, C.; Schmelzer, A.E.; et al. Developing an Anion Exchange Chromatography Assay for Determining Empty and Full Capsid Contents in AAV6.2. Mol. Ther. Methods Clin. Dev. 2019, 15, 257–263. [Google Scholar] [CrossRef]
- Geoffroy, M.-C.; Salvetti, A. Helper Functions Required for Wild Type and Recombinant Adeno- Associated Virus Growth. Curr. Gene Ther. 2005, 5, 265–271. [Google Scholar] [CrossRef]
- Buller, R.M.L.; Janik, J.E.; Sebring, E.D.; Rose, J.A. Herpes Simplex Virus Types 1 and 2 Completely Help Adenovirus-Associated Virus Replication. J. Virol. 1981, 40, 241–247. [Google Scholar] [CrossRef]
- Venkatakrishnan, B.; Yarbrough, J.; Domsic, J.; Bennett, A.; Bothner, B.; Kozyreva, O.G.; Samulski, R.J.; Muzyczka, N.; McKenna, R.; Agbandje-McKenna, M. Structure and Dynamics of Adeno-Associated Virus Serotype 1 VP1-Unique N-Terminal Domain and Its Role in Capsid Trafficking. J. Virol. 2013, 87, 4974–4984. [Google Scholar] [CrossRef]
- Rose, J.A.; Maizel, J.V., Jr.; Inman, J.K.; Shatkin, A.J. Structural Proteins of Adenovirus-Associated Viruses. J. Virol. 1971, 8, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Barnes, L.F.; Draper, B.E.; Kurian, J.; Chen, Y.-T.; Shapkina, T.; Powers, T.W.; Jarrold, M.F. Analysis of AAV-Extracted DNA by Charge Detection Mass Spectrometry Reveals Genome Truncations. Anal. Chem. 2023, 95, 4310–4316. [Google Scholar] [CrossRef] [PubMed]
- Wörner, T.P.; Bennett, A.; Habka, S.; Snijder, J.; Friese, O.; Powers, T.; Agbandje-McKenna, M.; Heck, A.J.R. Adeno-associated virus capsid assembly is divergent and stochastic. Nat. Commun. 2021, 12, 1642. [Google Scholar] [CrossRef] [PubMed]
- Oyama, H.; Ishii, K.; Maruno, T.; Torisu, T.; Uchiyama, S.; Troxell, B.; Tsai, I.-W.; Shah, K.; Knuckles, C.I.; Shelton, S.; et al. Characterization of Adeno-Associated Virus Capsid Proteins with Two Types of VP3-Related Components by Capillary Gel Electrophoresis and Mass Spectrometry. Hum. Gene Ther. 2021, 32, 1403–1416. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Brown, A.M.; Jenkins, C.; Campbell, K. Viral Vector Systems for Gene Therapy: A Comprehensive Literature Review of Progress and Biosafety Challenges. Appl. Biosaf. 2020, 25, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016, 21, 75–80. [Google Scholar] [CrossRef]
- Wright, J.F. Product-Related Impurities in Clinical-Grade Recombinant AAV Vectors: Characterization and Risk Assessment. Biomedicines 2014, 2, 80–97. [Google Scholar] [CrossRef]
- Li, T.; Gao, T.; Chen, H.; Demianova, Z.; Wang, F.; Malik, M.; Luo, J.; Yowanto, H.; Mollah, S. Determination of Full, Partial and Empty Capsid Ratios for Adeno-Associated Virus (AAV) Analysis; SCIEX: Framingham, MA, USA, 2020. [Google Scholar]
- Fu, X.; Chen, W.C.; Argento, C.; Clarner, P.; Bhatt, V.; Dickerson, R.; Bou-Assaf, G.; Bakhshayeshi, M.; Lu, X.; Bergelson, S.; et al. Analytical Strategies for Quantification of Adeno-Associated Virus Empty Capsids to Support Process Development. Hum. Gene Ther. Methods 2019, 30, 144–152. [Google Scholar] [CrossRef]
- Gimpel, A.L.; Katsikis, G.; Sha, S.; Maloney, A.J.; Hong, M.S.; Nguyen, T.N.; Wolfrum, J.; Springs, S.L.; Sinskey, A.J.; Manalis, S.R.; et al. Analytical methods for process and product characterization of recombinant adeno-associated virus-based gene therapies. Mol. Ther. Methods Clin. Dev. 2021, 20, 740–754. [Google Scholar] [CrossRef]
- Pierson, E.E.; Keifer, D.Z.; Asokan, A.; Jarrold, M.F. Resolving Adeno-Associated Viral Particle Diversity With Charge Detection Mass Spectrometry. Anal. Chem. 2016, 88, 6718–6725. [Google Scholar] [CrossRef]
- Werle, A.K.; Powers, T.W.; Zobel, J.F.; Wappelhorst, C.N.; Jarrold, M.F.; Lyktey, N.A.; Sloan, C.D.; Wolf, A.J.; Adams-Hall, S.; Baldus, P.; et al. Comparison of analytical techniques to quantitate the capsid content of adeno-associated viral vectors. Mol. Ther. Methods Clin. Dev. 2021, 23, 254–262. [Google Scholar] [CrossRef]
- Takeda, K.; Noda, M.; Maruno, T.; Uchiyama, S. Critical Calibration of Mass Photometry for Higher-Mass Samples Such as Adeno-Associated Virus Vectors. J. Pharm. Sci. 2022, 112, 1145–1150. [Google Scholar] [CrossRef]
- Wu, D.; Piszczek, G. Standard protocol for mass photometry experiments. Eur. Biophys J. 2021, 50, 403–409. [Google Scholar] [CrossRef]
- Sonn-Segev, A.; Belacic, K.; Bodrug, T.; Young, G.; VanderLinden, R.T.; Schulman, B.A.; Schimpf, J.; Friedrich, T.; Dip, P.V.; Schwartz, T.U.; et al. Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat. Commun. 2020, 11, 1772. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, P.J. Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta 1993, 272, 1–40. [Google Scholar] [CrossRef]
- Wagner, C.; Innthaler, B.; Lemmerer, M.; Pletzenauer, R.; Birner-Gruenberger, R. Biophysical Characterization of Adeno-Associated Virus Vectors Using Ion-Exchange Chromatography Coupled to Light Scattering Detectors. Int. J. Mol. Sci. 2022, 23, 12715. [Google Scholar] [CrossRef] [PubMed]
- Benefits and Applications of Analytical Ultracentrifugation (AUC). Available online: https://www.nanolytics.com/ (accessed on 14 June 2023).
- Saleun, S.; Mas, C.; Le Roy, A.; Penaud-Budloo, M.; Adjali, O.; Blouin, V.; Ebel, C. Analytical ultracentrifugation sedimentation velocity for the characterization of recombinant adeno-associated virus vectors sub-populations. Eur. Biophys. J. 2023, 52, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Refeyn Ltd. Quantifying Heterogeneous AAV Capsid Loading Using Mass Photometry. [White Paper]. 2022. Available online: https://info.refeyn.com/app-note-aav-loading (accessed on 14 June 2023).

| Empty AAVs | Full AAVs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Average Mass/kDa | RSD/% | Average Mass/kDa | RSD/% | Calculated Transgene Size/kDa | Average Transgene Size/kDa | RSD/% | Expected Transgene Size/kDa | Error/% | |
| Day 1 | 3583 | 2.2 | 4987 | 2.2 | 1405 | 1447 | 2.5 | 1400 | 3.3 |
| Day 2 | 3746 | 0.9 | 5217 | 0.9 | 1471 | ||||
| Day 3 | 3789 | 1.3 | 5253 | 1.3 | 1464 | ||||
| MP | AUC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Average E/% | RSD/% | Average PF/% | RSD/% | Average F/% | RSD/% | Average OF/% | RSD/% | E/% | PF/% | F/% | OF/% | |
| Day 1 | 39.7 | 5.4 | 7.1 | 8.7 | 49.7 | 2.2 | 3.5 | 9.8 | 33.1 | 6.3 | 52.3 | 8.4 |
| Day 2 | 40.0 | 6.8 | 8.8 | 9.8 | 47.8 | 8.6 | 2.8 | 5.4 | ||||
| Day 3 | 40.4 | 3.8 | 8.3 | 1.8 | 48.1 | 3.1 | 2.3 | 8.7 | ||||
| Serotype | Empty AAVs/kDa | Full AAVs/kDa | Calculated Transgene Size/kDa | Expected Transgene Size/kDa | Error/% |
|---|---|---|---|---|---|
| AAV9_a | 3750 | 5253 | 1503 | 1400 | 7.4 |
| AAV9_b | 3992 | 5319 | 1327 | 1370 | 3.1 |
| AAV9_c | 3754 | 4644 | 890 | 890 | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, C.; Fuchsberger, F.F.; Innthaler, B.; Lemmerer, M.; Birner-Gruenberger, R. Quantification of Empty, Partially Filled and Full Adeno-Associated Virus Vectors Using Mass Photometry. Int. J. Mol. Sci. 2023, 24, 11033. https://doi.org/10.3390/ijms241311033
Wagner C, Fuchsberger FF, Innthaler B, Lemmerer M, Birner-Gruenberger R. Quantification of Empty, Partially Filled and Full Adeno-Associated Virus Vectors Using Mass Photometry. International Journal of Molecular Sciences. 2023; 24(13):11033. https://doi.org/10.3390/ijms241311033
Chicago/Turabian StyleWagner, Christina, Felix F. Fuchsberger, Bernd Innthaler, Martin Lemmerer, and Ruth Birner-Gruenberger. 2023. "Quantification of Empty, Partially Filled and Full Adeno-Associated Virus Vectors Using Mass Photometry" International Journal of Molecular Sciences 24, no. 13: 11033. https://doi.org/10.3390/ijms241311033
APA StyleWagner, C., Fuchsberger, F. F., Innthaler, B., Lemmerer, M., & Birner-Gruenberger, R. (2023). Quantification of Empty, Partially Filled and Full Adeno-Associated Virus Vectors Using Mass Photometry. International Journal of Molecular Sciences, 24(13), 11033. https://doi.org/10.3390/ijms241311033






