Antibodies Capable of Enhancing SARS-CoV-2 Infection Can Circulate in Patients with Severe COVID-19
Abstract
1. Introduction
2. Results
2.1. mAb RS2 Selection, Its Neutralization Activity, and Affinity
2.2. Epitope Mapping
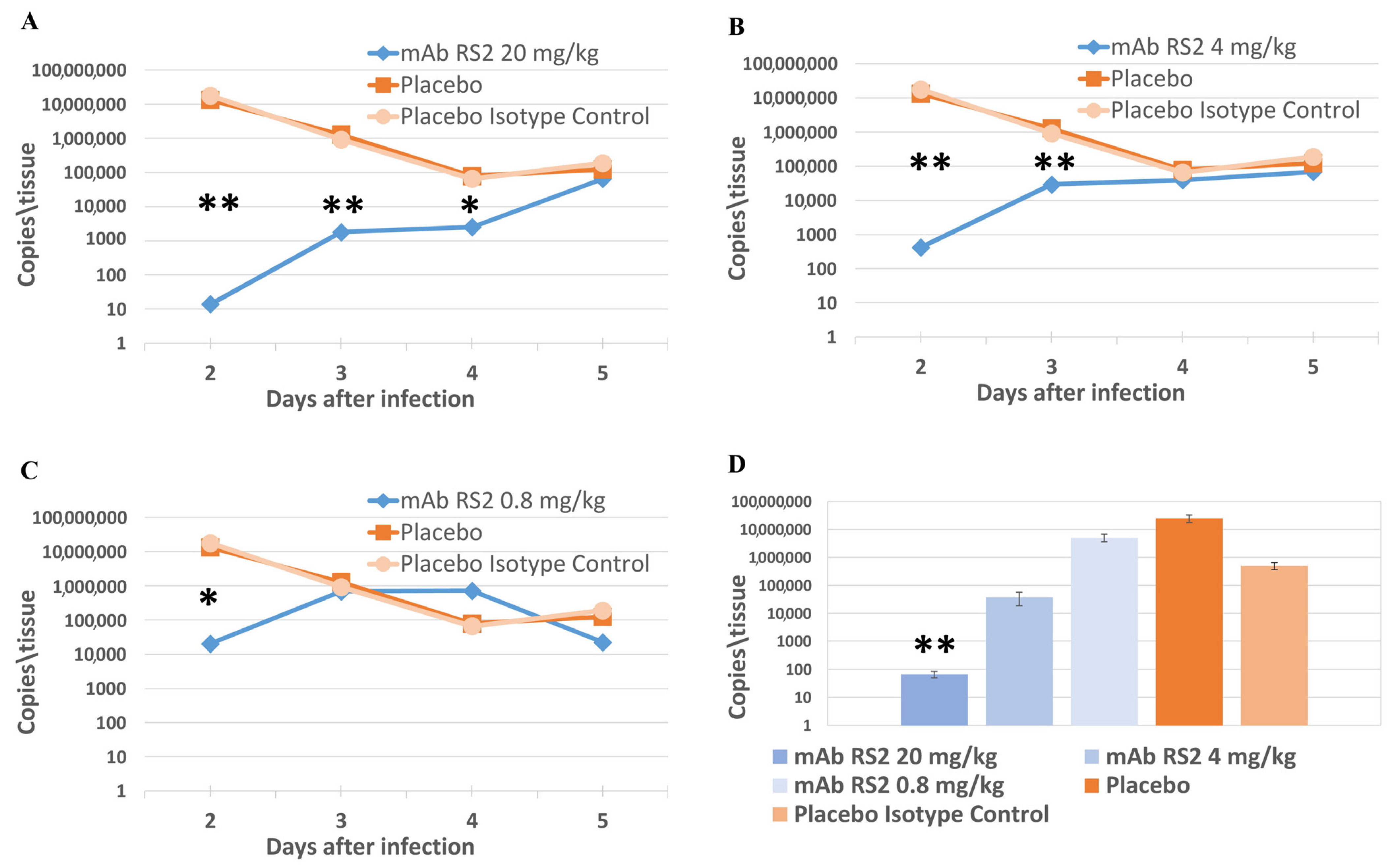
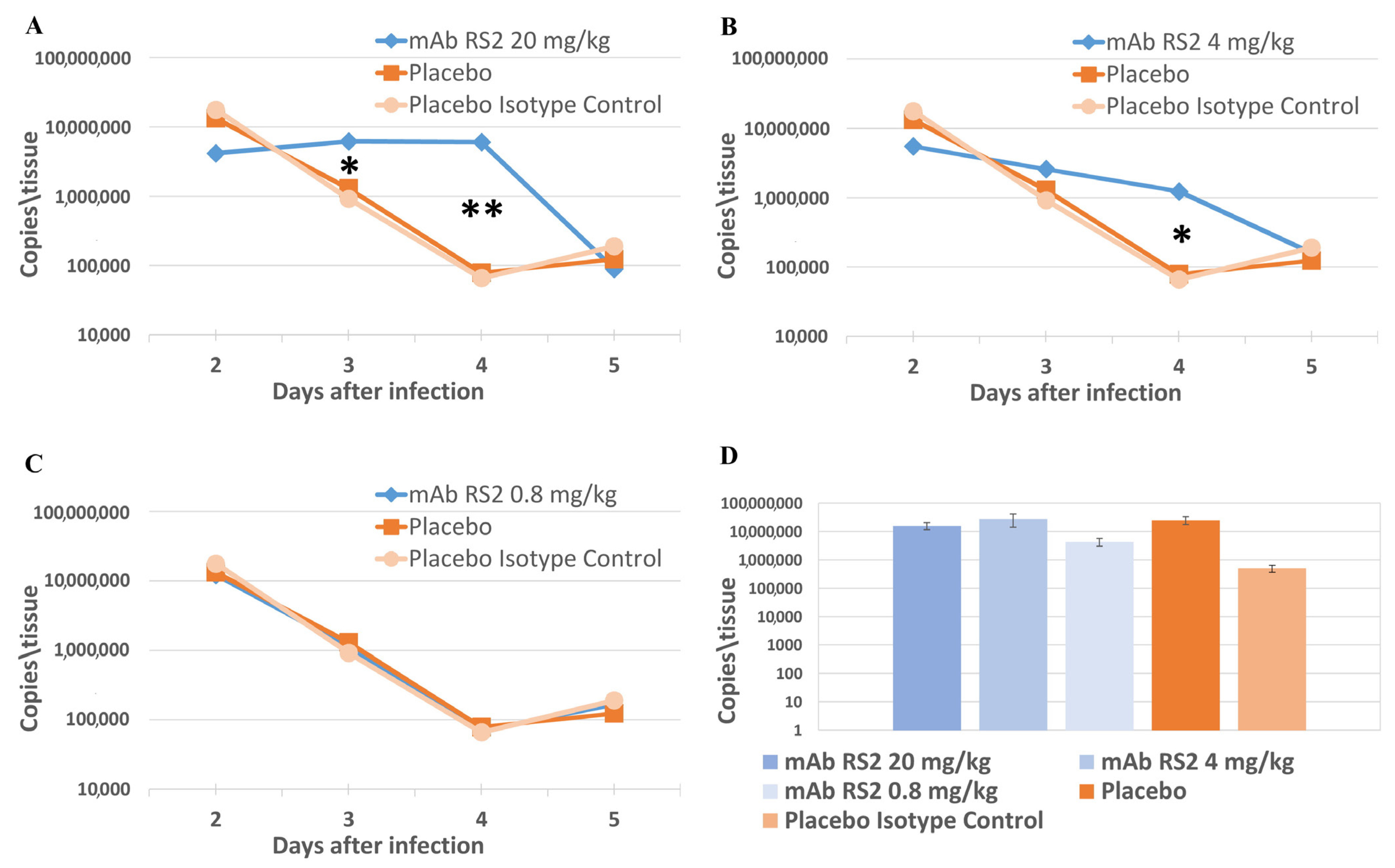
2.3. Pre- and Post-Exposure Administration of mAb RS2
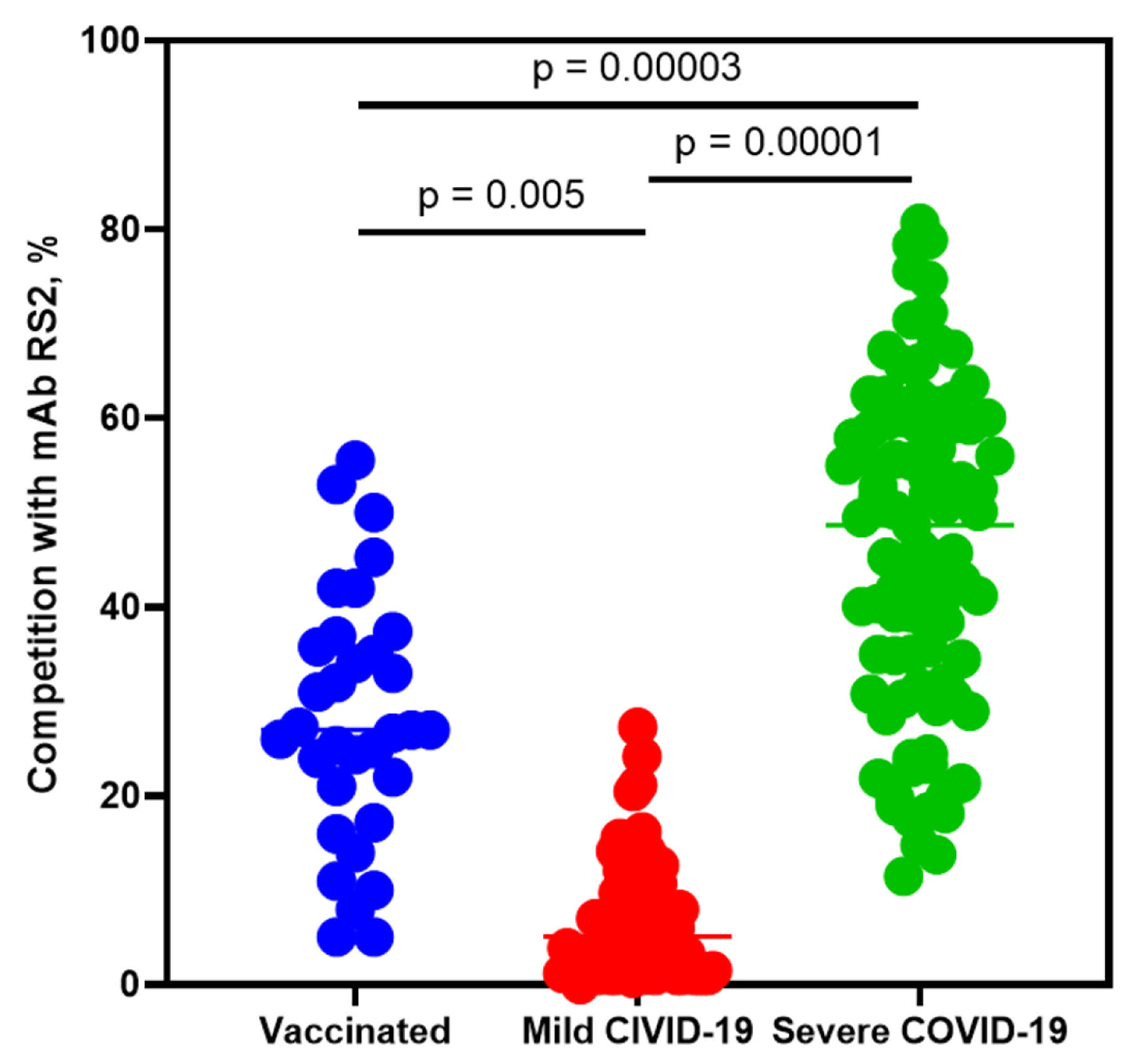
2.4. Competitive Analysis of Human Sera Antibodies with mAb RS2
3. Discussion
4. Materials and Methods
4.1. Animals, Virus, Sera, and Cells
4.2. Antigens Production
4.3. Mouse Immunization and mAbs Selection
4.4. Production, Purification, and Evaluation of the mAb RS2
4.5. Assessment of Neutralization Activity In Vitro
4.6. Animal Studies
4.7. Competitive ELISA
4.8. Epitope Mapping
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Coronavirus (COVID-19) statistics. Available online: https://covid19.who.int/ (accessed on 5 May 2023).
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.J.; Bickerton, E.; Britton, P. Coronaviruses: Methods and Protocols; Humana: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281.e6–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, L.; Zhang, G.; Yao, Y.; Zhou, H.; Shen, S.; Shen, B.; Li, B.; Li, X.; Zhang, Q.; et al. A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes. Nat. Commun. 2021, 12, 2623. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H.; et al. Aptamer Blocking Strategy Inhibits SARS-CoV-2 Virus Infection. Angew. Chem. 2021, 60, 10266–10272. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Silva Andrade, B.; Giovanetti, M.; Almeida Costa, E.; Kumavath, R.; Ghosh, P.; Góes-Neto, A.; Alcantara, L.C.J.; Azevedo, V. Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain. F1000Research 2020, 9, 576. [Google Scholar] [CrossRef]
- Chaouat, A.E.; Achdout, H.; Kol, I.; Berhani, O.; Roi, G.; Vitner, E.B.; Melamed, S.; Politi, B.; Zahavy, E.; Brizic, I.; et al. SARS-CoV-2 receptor binding domain fusion protein efficiently neutralizes virus infection. PLoS Pathog. 2021, 17, e1010175. [Google Scholar] [CrossRef]
- Nassar, M.; Nso, N.; Gonzalez, C.; Lakhdar, S.; Alshamam, M.; Elshafey, M.; Abdalazeem, Y.; Nyein, A.; Punzalan, B.; Durrance, R.J.; et al. COVID-19 vaccine-induced myocarditis: Case report with literature review. Diabetes Metab. Syndr. 2021, 15, 102205. [Google Scholar] [CrossRef]
- Morgan, M.C.; Atri, L.; Harrell, S.; Al-Jaroudi, W.; Berman, A. COVID-19 vaccine-associated myocarditis. World J. Cardiol. 2022, 14, 382–391. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 585, 174–177. [Google Scholar] [CrossRef]
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.; Yang, F.; Tan, W.; Wu, G.; Xu, M.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2020, 22, 196–208. [Google Scholar] [CrossRef]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Lambe, T.; Spencer, A.; Belij-Rammerstorfer, S.; Purushotham, J.N.; Port, J.R.; Avanzato, V.A.; Bushmaker, T.; Flaxman, A.; Ulaszewska, M.; et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020, 586, 578–582. [Google Scholar] [CrossRef]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Baum, A.; Ajithdoss, D.; Copin, R.; Zhou, A.; Lanza, K.; Negron, N.; Ni, M.; Wei, Y.; Mohammadi, K.; Musser, B.; et al. REGN-COV2 antibody cocktail prevents and treats SARS-CoV-2 infection in rhesus macaques and hamsters. Nature 2020, 370, 1110–1115. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Du, S.; Cao, Y.; Zhu, Q.; Yu, P.; Qi, F.; Wang, G.; Du, X.; Bao, L.; Deng, W.; Zhu, H.; et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell 2020, 183, 1013–1023. [Google Scholar] [CrossRef]
- Graham, B.S. Vaccines against respiratory syncytial virus: The time has finally come. Vaccine 2016, 34, 3535–3541. [Google Scholar] [CrossRef]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef]
- Wang, T.T.; Sewatanon, J.; Memoli, M.J.; Wrammert, J.; Bournazos, S.; Bhaumik, S.K.; Pinsky, B.A.; Chokephaibulkit, K.; Onlamoon, N.; Pattanapanyasat, K.; et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science 2017, 355, 395–398. [Google Scholar] [CrossRef]
- Liu, L.; Wei, Q.; Lin, Q.; Fang, J.; Wang, H.; Kwok, H.; Tang, H.; Nishiura, K.; Peng, J.; Tan, Z.; et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019, 4, e123158. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.S.; Tao, X.; Algaissi, A.; Garron, T.; Narayanan, K.; Peng, B.H.; Couch, R.B.; Tseng, C.-T.K. Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum. Vaccin. Immunother. 2016, 12, 2351–2356. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020, 5, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Matveev, A.L.; Krylov, V.B.; Khlusevich, Y.A.; Baykov, I.K.; Yashunsky, D.V.; Emelyanova, L.A.; Tsvetkov, Y.E.; Karelin, A.A.; Bardashova, A.V.; Wong, S.S.W.; et al. Novel mouse monoclonal antibodies specifically recognizing β-(1→3)-D-glucan antigen. PLoS ONE 2019, 14, e0215535. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Sedykh, S.E.; Ermakov, E.A.; Matveev, A.L.; Odegova, E.I.; Sedykh, T.A.; Shcherbakov, D.N.; Merkuleva, I.A.; Volosnikova, E.A.; Nesmeyanova, V.S.; et al. Natural IgG against S-Protein and RBD of SARS-CoV-2 Do Not Bind and Hydrolyze DNA and Are Not Autoimmune. Int. J. Mol. Sci. 2022, 23, 13681. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Sun, S.; Tai, W.; Chen, J.; Geng, Q.; He, L.; Chen, Y.; Wu, J.; Shi, Z.; et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020, 94, e02015-19. [Google Scholar] [CrossRef]
- Houser, K.V.; Broadbent, A.J.; Gretebeck, L.; Vogel, L.; Lamirande, E.W.; Sutton, T.; Bock, K.W.; Minai, M.; Orandle, M.; Moore, I.N.; et al. Enhanced inflammation in New Zealand white rabbits when MERS-CoV reinfection occurs in the absence of neutralizing antibody. PLoS Pathog. 2017, 13, e1006565. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Yu, X.; Jiang, W.; Chen, S.; Wang, R.; Wang, M.; Jiao, S.; Yang, Y.; Wang, W.; et al. Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires FcγRIIB and virus-antibody complex with bivalent interaction. Commun Biol. 2022, 5, 262. [Google Scholar] [CrossRef]
- Shen, X.R.; Li, Q.; Li, H.L.; Wang, X.; Wang, Q.; Zheng, X.S.; Geng, R.; Zhang, Y.-L.; Li, B.; Jiang, R.-D.; et al. Antibody-Dependent Enhancement of SARS-CoV-2 Infection of Human Immune Cells: In Vitro Assessment Provides Insight in COVID-19 Pathogenesis. Viruses 2021, 13, 2483. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef]
- Fan, W.; Renhong, Y.; Mei, L.; Zezhong, L.; Yingdan, W.; Die, L. Antibody-dependent enhancement (ADE) of SARS-CoV-2 infection in recovered COVID-19 patients: Studies based on cellular and structural biology analysis. medRxiv 2020, 1–52. [Google Scholar] [CrossRef]
- Kreil, T.R.; Eibl, M.M. Pre- and postexposure protection by passive immunoglobulin but no enhancement of infection with a flavivirus in a mouse model. J. Virol. 1997, 71, 2921–2927. [Google Scholar] [CrossRef]
- Haslwanter, D.; Blaas, D.; Heinz, F.X.; Stiasny, K. A novel mechanism of antibody-mediated enhancement of flavivirus infection. PLoS Pathog. 2017, 13, e1006643. [Google Scholar] [CrossRef]
- Barba-Spaeth, G.; Dejnirattisai, W.; Rouvinski, A.; Vaney, M.C.; Medits, I.; Sharma, A.; Simon-Lorière, E.; Sakuntabhai, A.; Cao-Lormeau, V.-M.; Haouz, A.; et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 2016, 536, 48–53. [Google Scholar] [CrossRef]
- Bröker, M.; Kollaritsch, H. After a tick bite in a tick-borne encephalitis virus endemic area: Current positions about post-exposure treatment. Vaccine 2007, 26, 863–868. [Google Scholar] [CrossRef]
- Levanov, L.N.; Matveev, L.E.; Goncharova, E.P.; Lebedev, L.R.; Ryzhikov, A.B.; Yun, T.E.; Batanova, T.A.; Shvalov, A.N.; Baykov, I.K.; Shingarova, L.N.; et al. Chimeric antibodies against tick-borne encephalitis virus. Vaccine 2010, 28, 5265–5271. [Google Scholar] [CrossRef]
- Borgoyakova, M.B.; Karpenko, L.I.; Rudometov, A.P.; Shanshin, D.V.; Isaeva, A.A.; Nesmeyanova, V.S.; Volkova, N.V.; Belenkaya, S.V.; Murashkin, D.E.; Shcherbakov, D.N.; et al. Immunogenic Properties of the DNA Construct Encoding the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein. Mol. Biol. 2021, 55, 889–898. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Shipovalov, A.V.; Vanin, A.F.; Pyankov, O.V.; Bagryanskaya, E.G.; Mikoyan, V.D.; Tkachev, N.A.; Asanbaeva, N.A.; Popkova, V.Y. Antiviral Activity of Nitrosonium Cations against SARS-CoV-2 on a Syrian Hamster Model. Biophysics 2022, 67, 785–795. [Google Scholar] [CrossRef]
- Matveev, A.L.; Kozlova, I.V.; Stronin, O.V.; Khlusevich, Y.A.; Doroshchenko, E.K.; Baykov, I.K.; Lisak, O.V.; Emelyanova, L.A.; Suntsova, O.V.; Matveeva, V.A.; et al. Post-exposure administration of chimeric antibody protects mice against European, Siberian, and Far-Eastern subtypes of tick-borne encephalitis virus. PLoS ONE 2019, 14, e0215075. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matveev, A.; Pyankov, O.; Khlusevich, Y.; Tyazhelkova, O.; Emelyanova, L.; Timofeeva, A.; Shipovalov, A.; Chechushkov, A.; Zaitseva, N.; Kudrov, G.; et al. Antibodies Capable of Enhancing SARS-CoV-2 Infection Can Circulate in Patients with Severe COVID-19. Int. J. Mol. Sci. 2023, 24, 10799. https://doi.org/10.3390/ijms241310799
Matveev A, Pyankov O, Khlusevich Y, Tyazhelkova O, Emelyanova L, Timofeeva A, Shipovalov A, Chechushkov A, Zaitseva N, Kudrov G, et al. Antibodies Capable of Enhancing SARS-CoV-2 Infection Can Circulate in Patients with Severe COVID-19. International Journal of Molecular Sciences. 2023; 24(13):10799. https://doi.org/10.3390/ijms241310799
Chicago/Turabian StyleMatveev, Andrey, Oleg Pyankov, Yana Khlusevich, Olga Tyazhelkova, Lyudmila Emelyanova, Anna Timofeeva, Andrey Shipovalov, Anton Chechushkov, Natalia Zaitseva, Gleb Kudrov, and et al. 2023. "Antibodies Capable of Enhancing SARS-CoV-2 Infection Can Circulate in Patients with Severe COVID-19" International Journal of Molecular Sciences 24, no. 13: 10799. https://doi.org/10.3390/ijms241310799
APA StyleMatveev, A., Pyankov, O., Khlusevich, Y., Tyazhelkova, O., Emelyanova, L., Timofeeva, A., Shipovalov, A., Chechushkov, A., Zaitseva, N., Kudrov, G., Yusubalieva, G., Yussubaliyeva, S., Zhukova, O., Baklaushev, V., Sedykh, S., Lifshits, G., Tikunov, A., & Tikunova, N. (2023). Antibodies Capable of Enhancing SARS-CoV-2 Infection Can Circulate in Patients with Severe COVID-19. International Journal of Molecular Sciences, 24(13), 10799. https://doi.org/10.3390/ijms241310799






