Transcriptome Landscape Analyses of the Regulatory Network for Zygotic Embryo Development in Paeonia ostii
Abstract
1. Introduction
2. Results
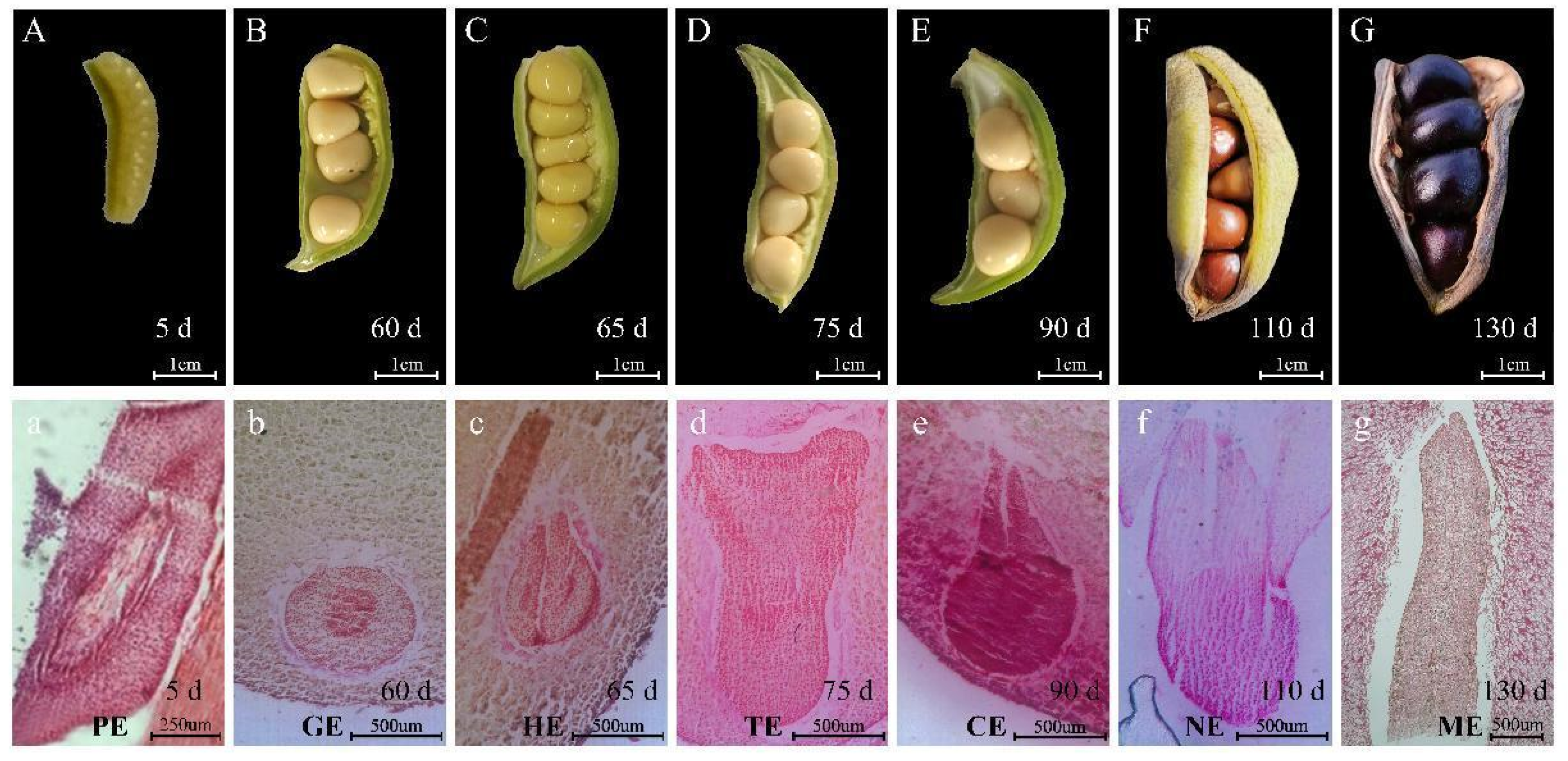
2.1. Morphological Observation of Tree Peony Pod Development after Pollination
2.2. Anatomical Observation of Embryo Development of Tree Peony
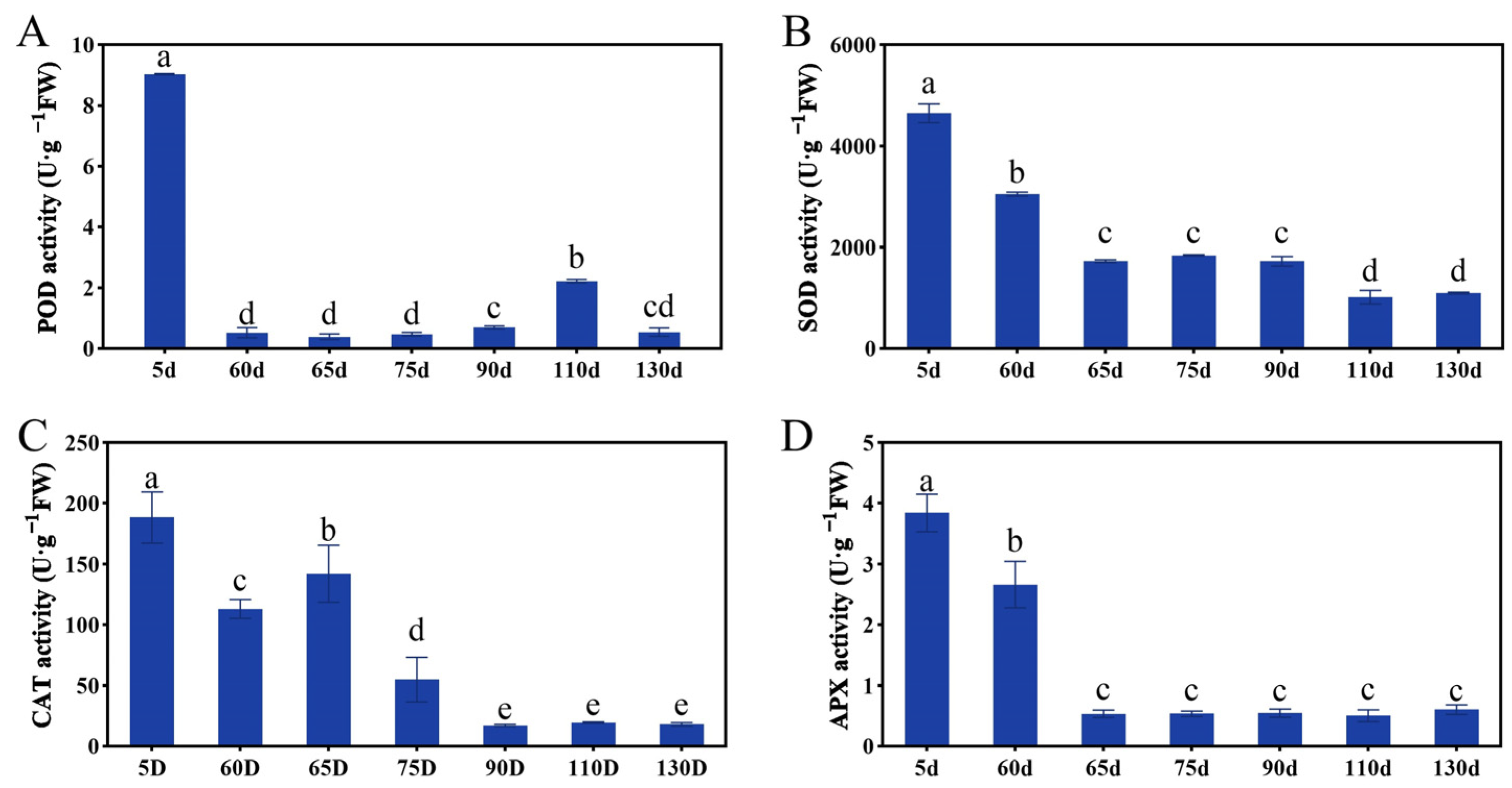
2.3. Antioxidant Enzymes Activities during the Seed Embryo Development of P. ostii
2.4. Global Analysis of the Time-Course Transcriptome Data in Embryo Development
2.5. Gene Expression during Different Embryo Development Stages
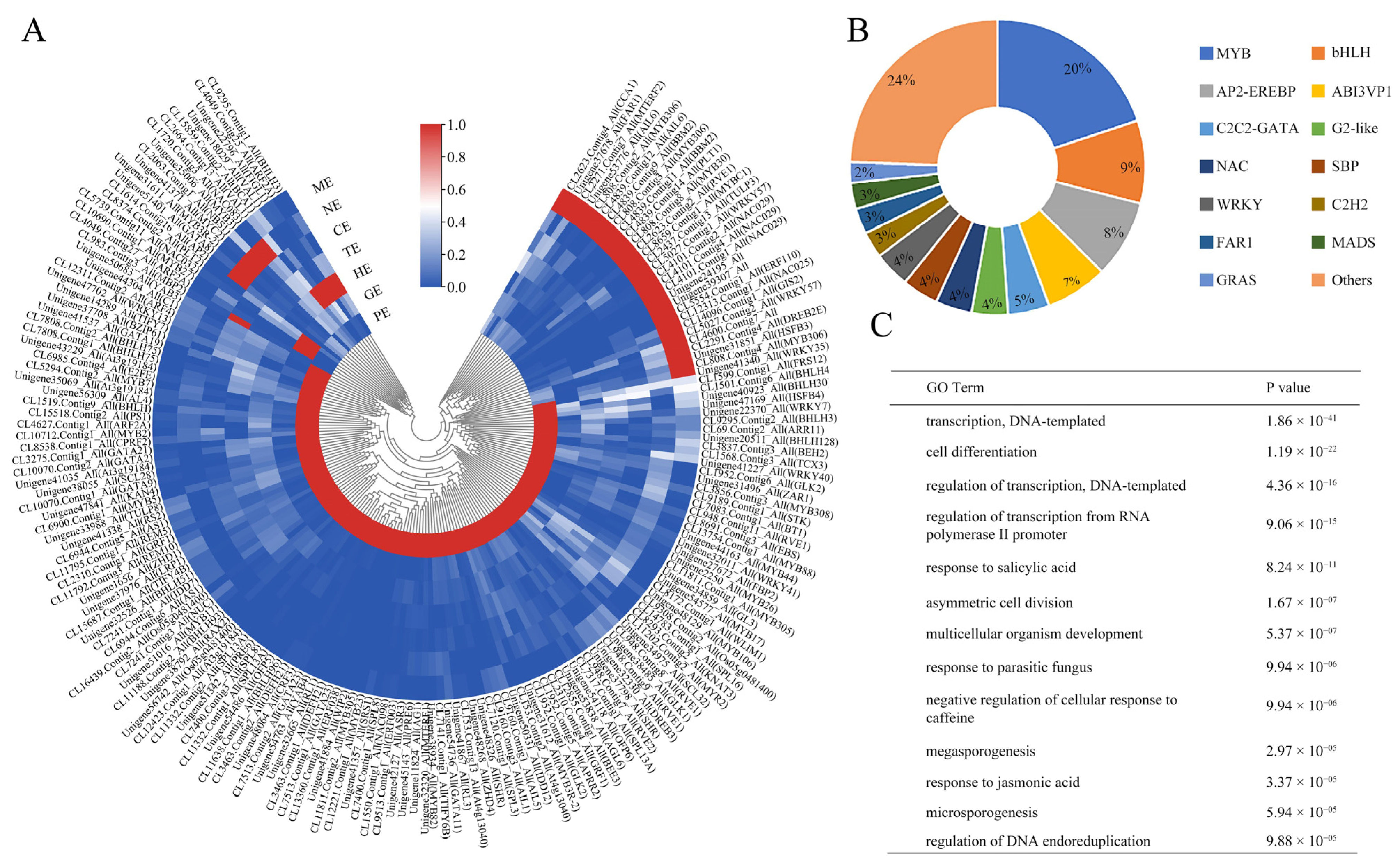
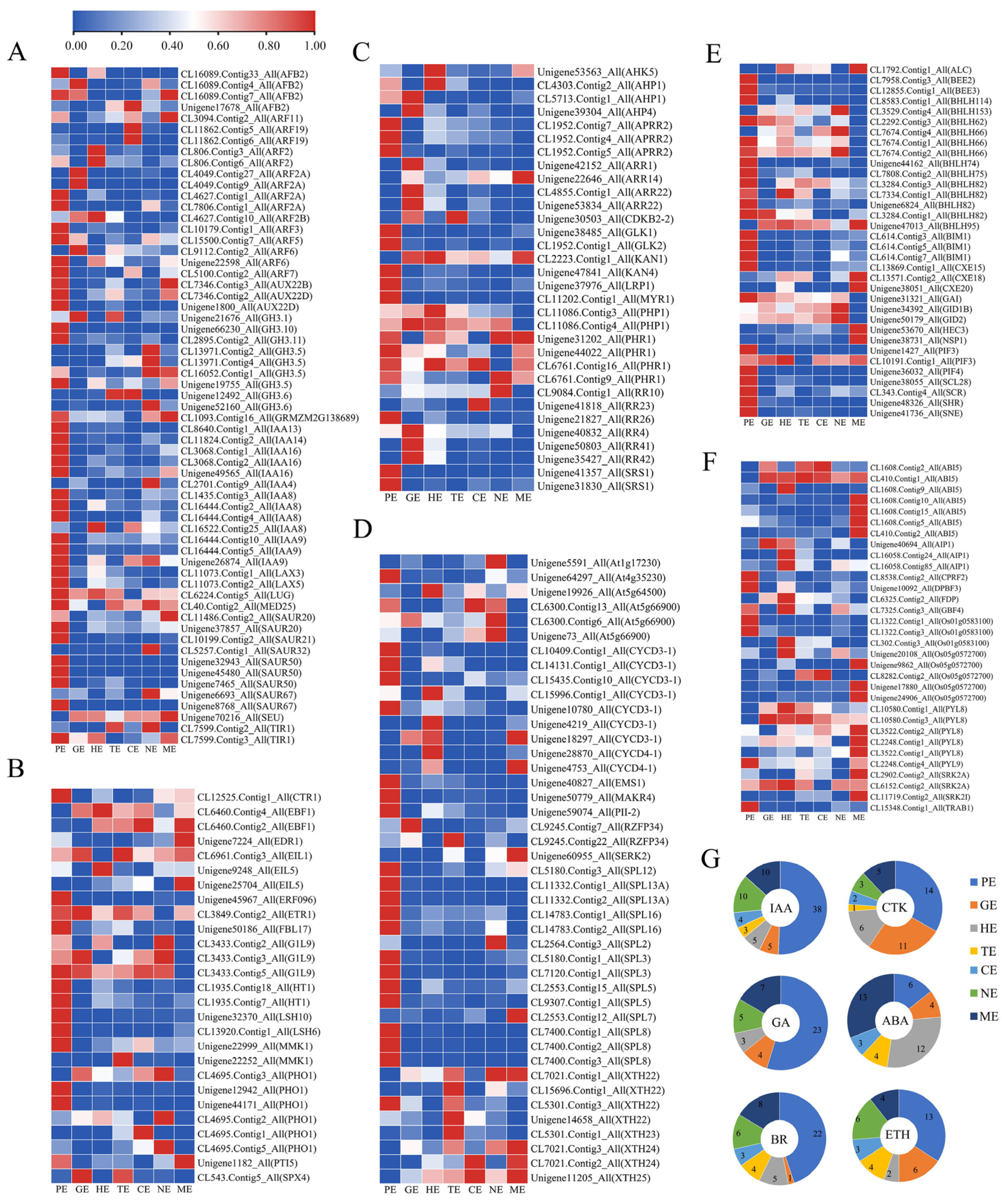
2.6. Spatial and Temporal Expression Pattern of Transcription Factors
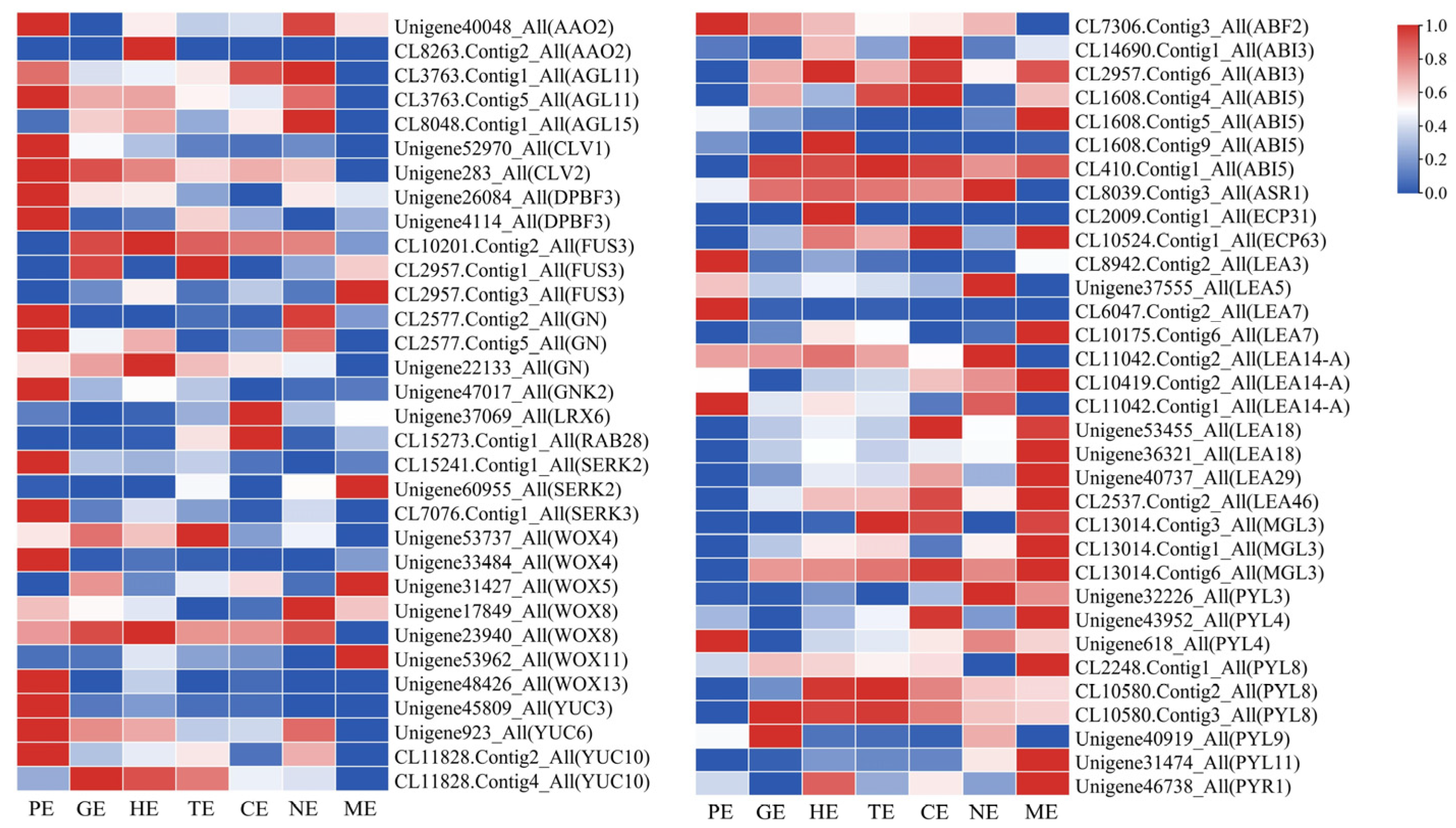
2.7. Expression Profiles of Key TFs Associated with Somatic Embryogenesis during Embryo Development of P. ostii
2.8. Analysis of Antioxidant Enzyme-Related Genes in Embryo Development
2.9. DEGs Analysis Involved in Plant Hormones Signal Transduction and Biosynthesis during Embryo Development
2.10. Expression Patterns of Endosperm-Related Genes during Different Embryo Development Stages
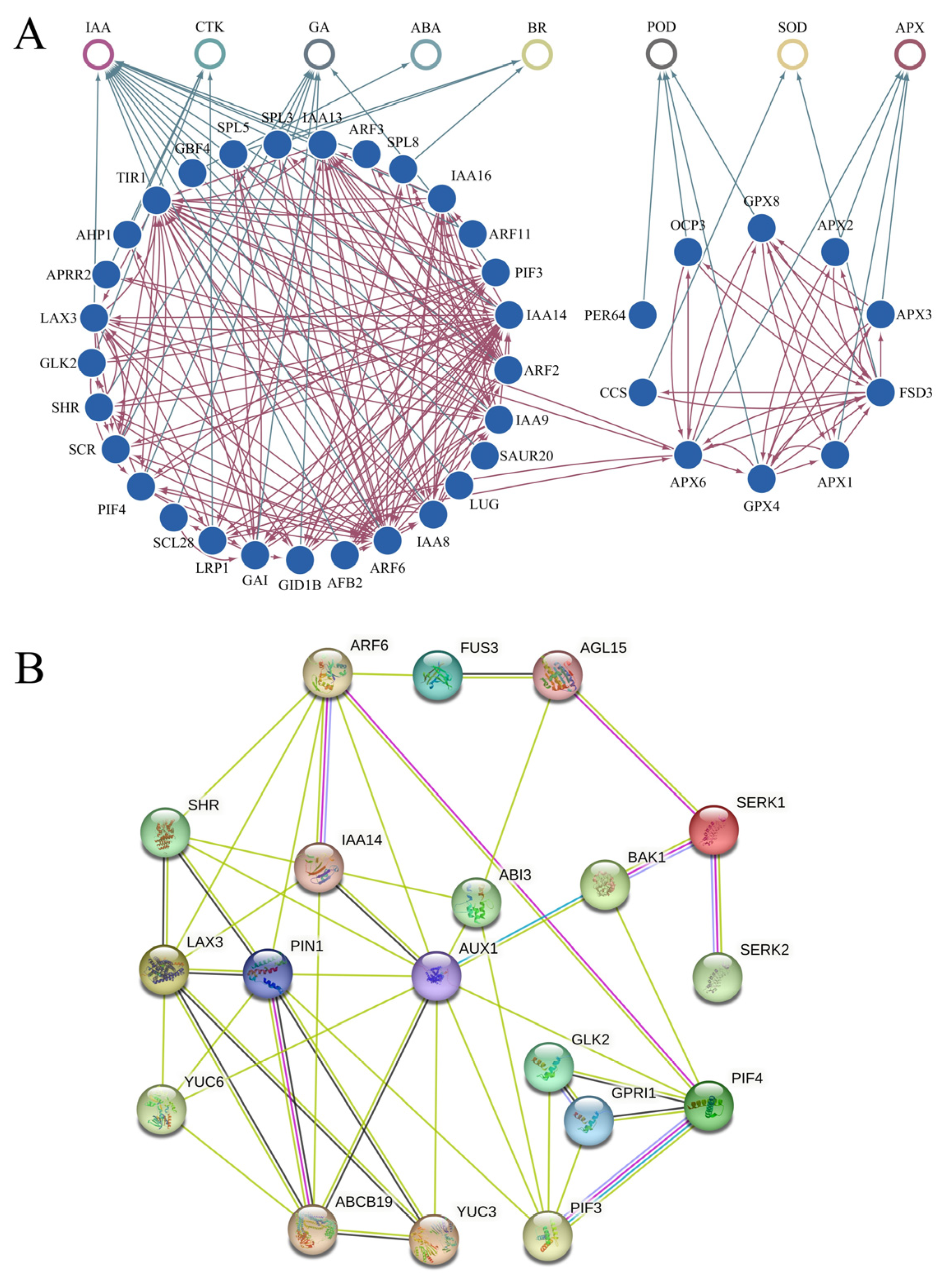
2.11. Prediction of the Protein-Protein Interaction Network in the Proembryo Development of Tree Peony
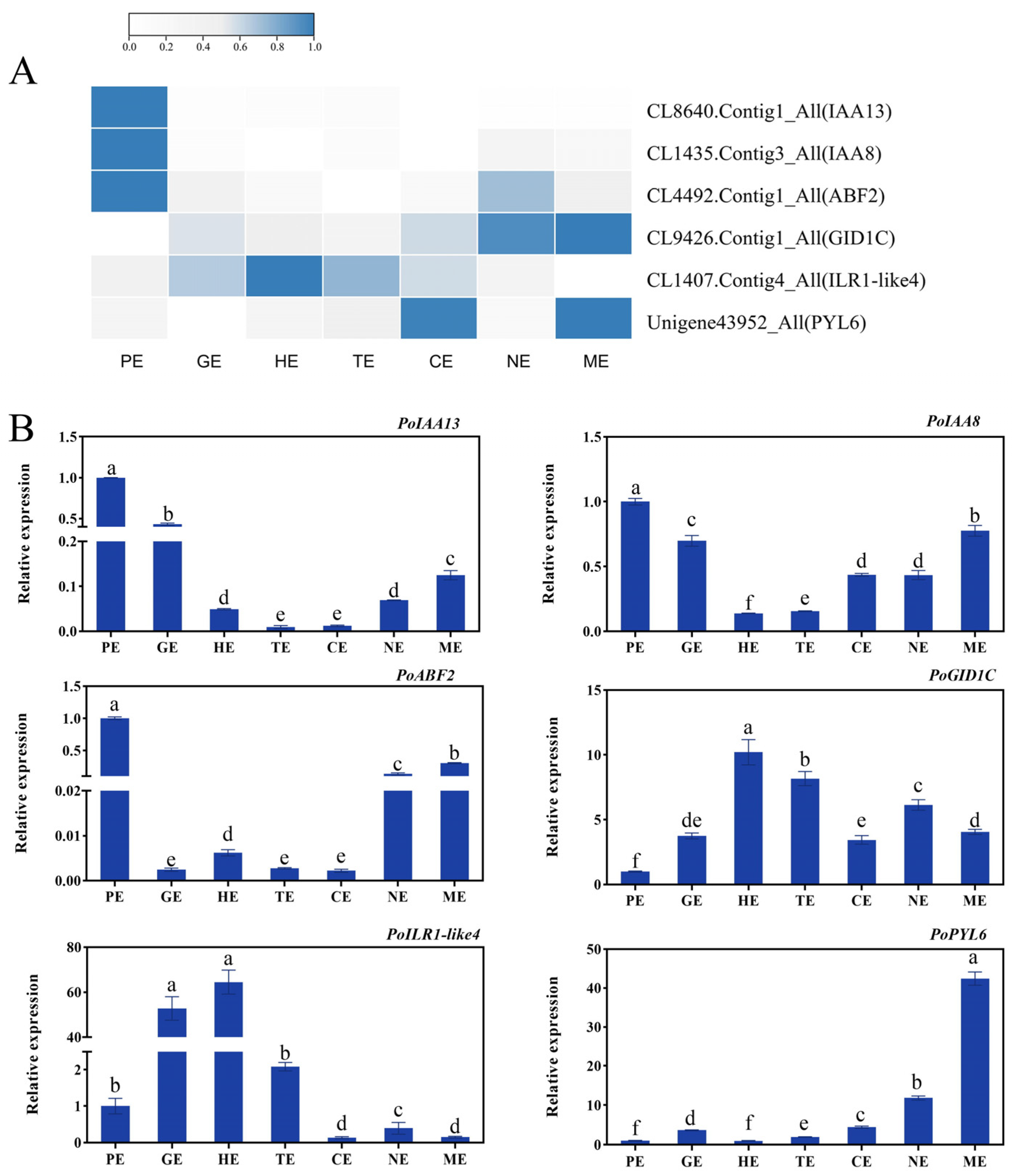
2.12. The Validation of RNA-Seq Data by qRT-PCR
3. Discussion
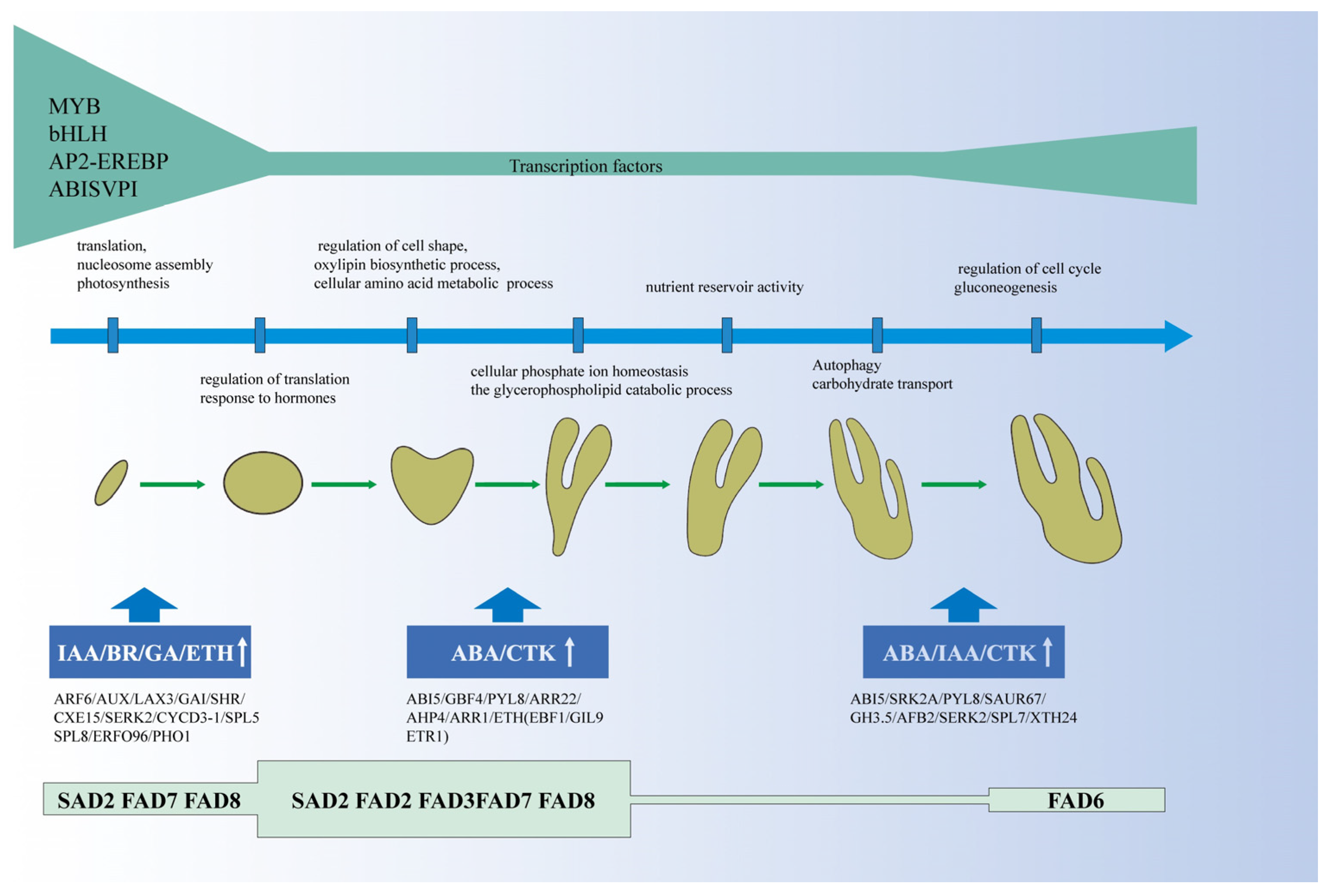
3.1. Growth and Development of P. ostii Seed
3.2. Genes Related to Plant Hormones and Enzymes Were Important for the Embryo Development of Tree Peony
3.3. The Molecular Study on Embryo Development Provided the Basis for Somatic Embryo Culture Technology in Tree Peony
4. Materials and Methods
4.1. Plant Material
4.2. Histology Observation
4.3. Measurement of Physiological Data
4.4. Total RNA Extraction and RNA-Seq Library Preparation
4.5. Transcriptome Analysis and Genes Expression Patterns Analysis
4.6. The Prediction of Protein-Protein Interaction Network
4.7. Validation of Transcriptome Data by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savadi, S. Molecular regulation of seed development and strategies for engineering seed size in crop plants. Plant. Growth Regul. 2018, 84, 401–422. [Google Scholar] [CrossRef]
- Locascio, A.; Roig-Villanova, I.; Bernardi, J.; Varotto, S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: A focus on auxin. Front. Plant. Sci. 2014, 5, 412. [Google Scholar] [CrossRef]
- Bian, J.X.; Deng, P.C.; Zhan, H.S.; Wu, X.T.; Nishantha, M.D.; Yan, Z.G.; Du, X.H.; Nie, X.J.; Song, W.N. Transcriptional Dynamics of Grain Development in Barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2019, 20, 962. [Google Scholar] [CrossRef]
- Juarez-Escobar, J.; Bojórquez-Velázquez, E.; Elizalde-Contreras, J.M.; Guerrero-Analco, J.A.; Loyola-Vargas, V.M.; Mata-Rosas, M.; Ruiz-May, E. Current Proteomic and Metabolomic Knowledge of Zygotic and Somatic Embryogenesis in Plants. Int. J. Mol. Sci. 2021, 22, 11807. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Lv, Q.Y.; Deng, J.; Huang, J.; Cai, F.; Liang, C.G.; Chen, Q.J.; Wang, Y.; Zhu, L.W.; Zhang, X.N.; et al. Transcriptome Analysis Reveals Key Seed-Development Genes in Common Buckwheat (Fagopyrum esculentum). Int. J. Mol. Sci. 2019, 20, 4303. [Google Scholar] [CrossRef] [PubMed]
- Ci, H.T.; Li, C.Y.; Aung, T.T.; Wang, S.L.; Yun, C.; Wang, F.; Ren, X.X.; Zhang, X.X. A Comparative Transcriptome Analysis Reveals the Molecular Mechanisms That Underlie Somatic Embryogenesis in Peaonia ostii ‘Fengdan’. Int. J. Mol. Sci. 2022, 23, 10595. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Hernández, H.A.; Ledezma-Rodríguez, M.; Avilez-Montalvo, R.N.; Juárez-Gómez, Y.L.; Skeete, A.; Avilez-Montalvo, J.; De-la-Peña, C.; Loyola-Vargas, V.M. Signaling Overview of Plant Somatic Embryogenesis. Front. Plant. Sci. 2019, 10, 77. [Google Scholar] [CrossRef]
- Alemanno, L.; Devic, M.; Niemenak, N.; Sanier, C.; Guilleminot, J.; Rio, M.; Verdeil, J.L.; Montoro, P. Characterization of leafy cotyledon1-like during embryogenesis in Theobroma cacao L. Planta 2008, 227, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Salvo, S.A.; Hirsch, C.N.; Buell, C.R.; Kaeppler, S.M.; Kaeppler, H.F. Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS ONE 2014, 9, e111407. [Google Scholar] [CrossRef] [PubMed]
- Boulard, C.; Fatihi, A.; Lepiniec, L.; Dubreucq, B. Regulation and evolution of the interaction of the seed B3 transcription factors with NF-Y subunits. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 1069–1078. [Google Scholar] [CrossRef]
- Paul, P.; Joshi, S.; Tian, R.; Diogo Junior, R.; Chakrabarti, M.; Perry, S.E. The MADS-domain factor AGAMOUS-Like18 promotes somatic embryogenesis. Plant. Physiol. 2022, 188, 1617–1631. [Google Scholar] [CrossRef] [PubMed]
- Luan, A.P.; He, Y.H.; Xie, T.; Chen, C.J.; Mao, Q.; Wang, X.S.; Li, C.H.; Ding, Y.Q.; Lin, W.Q.; Liu, C.Y.; et al. Identification of an Embryonic Cell-Specific Region within the Pineapple SERK1 Promoter. Genes 2019, 10, 883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Z.; Liu, X.G.; Lin, Y.A.; Xie, G.N.; Fu, F.L.; Liu, H.L.; Wang, J.; Gao, S.B.; Lan, H.; Rong, T.Z. Characterization of a ZmSERK gene and its relationship to somatic embryogenesis in a maize culture. Plant Cell Tissue Organ Cult. 2011, 105, 29–37. [Google Scholar] [CrossRef]
- Baudino, S.; Hansen, S.; Brettschneider, R.; Hecht, V.F.; Dresselhaus, T.; Lörz, H.; Dumas, C.; Rogowsky, P.M. Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta 2001, 213, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Khurana, P. Ectopic expression of Triticum aestivum SERK genes (TaSERKs) control plant growth and development in Arabidopsis. Sci. Rep. 2017, 7, 12368. [Google Scholar] [CrossRef]
- Zheng, Y.M.; Ren, N.; Wang, H.; Stromberg, A.J.; Perry, S.E. Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 2009, 21, 2563–2577. [Google Scholar] [CrossRef]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.; Opsahl-Sorteberg, H.G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef]
- Cao, J.S.; Li, G.J.; Qu, D.J.; Li, X.; Wang, Y.N. Into the Seed: Auxin Controls Seed Development and Grain Yield. Int. J. Mol. Sci. 2020, 21, 1662. [Google Scholar] [CrossRef]
- Verma, S.; Attuluri, V.P.S.; Robert, H.S. An Essential Function for Auxin in Embryo Development. Cold Spring Harb. Perspect. Biol. 2021, 13, a039966. [Google Scholar] [CrossRef]
- Robert, H.S.; Park, C.; Gutièrrez, C.L.; Wójcikowska, B.; Pěnčík, A.; Novák, O.; Chen, J.Y.; Grunewald, W.; Dresselhaus, T.; Friml, J.; et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 2018, 4, 548–553. [Google Scholar] [CrossRef]
- Pérez, M.; Viejo, M.; LaCuesta, M.; Toorop, P.; Cañal, M.J. Epigenetic and hormonal profile during maturation of Quercus Suber L. somatic embryos. J. Plant Physiol. 2015, 173, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Ugartechea-Chirino, Y.; Swarup, R.; Swarup, K.; Péret, B.; Whitworth, M.; Bennett, M.; Bougourd, S. The AUX1 LAX family of auxin influx carriers is required for the establishment of embryonic root cell organization in Arabidopsis thaliana. Ann. Bot. 2010, 105, 277–289. [Google Scholar] [CrossRef] [PubMed]
- De Vega-Bartol, J.J.; Simões, M.; Lorenz, W.W.; Rodrigues, A.S.; Alba, R.; Dean, J.F.; Miguel, C.M. Transcriptomic analysis highlights epigenetic and transcriptional regulation during zygotic embryo development of Pinus pinaster. BMC Plant Biol. 2013, 13, 123. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.F.; Dai, X.H.; Zhao, Y.D. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Braybrook, S.A.; Paula, S.L.; Kwong, L.W.; Meuser, J.; Pelletier, J.; Hsieh, T.-F.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3151–3156. [Google Scholar] [CrossRef]
- Wang, H.A.; Caruso, L.V.; Downie, A.B.; Perry, S.E. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 2004, 16, 1206–1219. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Tsuchiya, Y.; Lumba, S.; Okamoto, M.; McCourt, P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 2004, 7, 373–385. [Google Scholar] [CrossRef]
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004, 136, 3660–3669. [Google Scholar] [CrossRef]
- Libik, M.; Konieczny, R.; Pater, B.; Slesak, I.; Miszalski, Z. Differences in the activities of some antioxidant enzymes and in H2O2 content during rhizogenesis and somatic embryogenesis in callus cultures of the ice plant. Plant Cell Rep. 2005, 23, 834–841. [Google Scholar] [CrossRef]
- Bahmankar, M.; Mortazavian, S.M.M.; Tohidfar, M.; Sadat Noori, S.A.; Izadi Darbandi, A.; Corrado, G.; Rao, R. Chemical Compositions, Somatic Embryogenesis, and Somaclonal Variation in Cumin. Biomed. Res. Int. 2017, 2017, 7283806. [Google Scholar] [CrossRef]
- Yoshida, K.; Kaothien, P.; Matsui, T.; Kawaoka, A.; Shinmyo, A. Molecular biology and application of plant peroxidase genes. Appl. Microbiol. Biotechnol. 2003, 60, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Zhao, Z.Q.; Si, J.; Di, C.X.; Han, J.; An, L.Z. Brassinosteroids alleviate chilling-induced oxidative damage by enhancing antioxidant defense system in suspension cultured cells of Chorispora bungeana. Plant Growth Regul. 2009, 59, 207–214. [Google Scholar] [CrossRef]
- Chou, C.H.; Huang, C.H.; Liu, Z.H. Peroxidase genes differentially respond to auxin during the formation of adventitious roots in soybean hypocotyl. Plant Growth Regul. 2010, 60, 151–161. [Google Scholar] [CrossRef]
- Kibinza, S.; Bazin, J.; Bailly, C.; Farrant, J.M.; Corbineau, F.; El-Maarouf-Bouteau, H. Catalase is a key enzyme in seed recovery from ageing during priming. Plant Sci. 2011, 181, 309–315. [Google Scholar] [CrossRef]
- Zi, J.; Zhang, J.Y.; Wang, Q.H.; Zhou, B.J.; Zhong, J.Y.; Zhang, C.L.; Qiu, X.M.; Wen, B.; Zhang, S.Y.; Fu, X.Q.; et al. Stress responsive proteins are actively regulated during rice (Oryza sativa) embryogenesis as indicated by quantitative proteomics analysis. PLoS ONE 2013, 8, e74229. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.C.; Caixeta, F.; Clemente, A.C.; Pinho, E.V.; Rosa, S.D. Gene expression of antioxidant enzymes and coffee seed quality during pre- and post-physiological maturity. Genet. Mol. Res. 2014, 13, 10983–10993. [Google Scholar] [CrossRef]
- Xie, D.M.; Yu, N.J.; Huang, L.Q.; Peng, D.Y.; Liu, C.B.; Zhu, Y.J.; Huang, H. Next generation sequencing and transcriptome analysis of root bark from Paeonia suffruticosa cv. Feng Dan. Zhong Yao Za Zhi 2017, 42, 2954–2961. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, L.; Xu, Z.; Yu, X. Transcriptome sequencing of Paeonia suffruticosa ‘Shima Nishiki’ to identify differentially expressed genes mediating double-color formation. Plant Physiol. Biochem. 2018, 123, 114–124. [Google Scholar] [CrossRef]
- Wang, S.L.; Gao, J.; Xue, J.; Xue, Y.Q.; Li, D.D.; Guan, Y.R.; Zhang, X.X. De novo sequencing of tree peony (Paeonia suffruticosa) transcriptome to identify critical genes involved in flowering and floral organ development. BMC Genom. 2019, 20, 572. [Google Scholar] [CrossRef]
- Gao, J.; Xue, J.Q.; Xue, Y.Q.; Liu, R.; Ren, X.X.; Wang, S.L.; Zhang, X.X. Transcriptome sequencing and identification of key callus browning-related genes from petiole callus of tree peony (Paeonia suffruticosa cv. Kao) cultured on media with three browning inhibitors. Plant Physiol. Biochem. 2020, 149, 36–49. [Google Scholar] [CrossRef]
- Yan, J.; Buer, H.; Wang, Y.P.; Zhula, G.; Bai, Y.E. Transcriptomic Time-Series Analyses of Gene Expression Profile During Zygotic Embryo Development in Picea mongolica. Front. Genet. 2021, 12, 738649. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Peng, P.; Duan, G.; Lin, T.; Bai, Y.E. Multiple analyses of various factors affecting the plantlet regeneration of Picea mongolica (H. Q. Wu) W.D. Xu from somatic embryos. Sci. Rep. 2021, 11, 6694. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, N.; Altschmied, L.; Radchuk, V.; Gubatz, S.; Wobus, U.; Weschke, W. Transcript profiles and deduced changes of metabolic pathways in maternal and filial tissues of developing barley grains. Plant J. 2004, 37, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Wang, L.S.; Shu, Q.Y.; Wu, J.; Chen, L.G.; Shao, S.; Yin, D.D. Fatty acid composition of developing tree peony (Paeonia section Moutan DC.) seeds and transcriptome analysis during seed development. BMC Genom. 2015, 16, 208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Yu, R.; Sun, D.Y.; Rahman, M.M.; Xie, L.H.; Hu, J.Y.; He, L.X.; Kilaru, A.; Niu, L.X.; Zhang, Y.L. Comparative Transcriptome Analysis Reveals an Efficient Mechanism of α-Linolenic Acid in Tree Peony Seeds. Int. J. Mol. Sci. 2018, 20, 65. [Google Scholar] [CrossRef]
- Wang, X.J.; Liang, H.Y.; Guo, D.L.; Duan, X.G.; Jia, Q.S.; Hou, X.G. Integrated analysis of transcriptomic and proteomic data from tree peony (P. ostii) seeds reveals key developmental stages and candidate genes related to oil biosynthesis and fatty acid metabolism. Hortic. Res. 2019, 6, 111. [Google Scholar] [CrossRef]
- Chang, M.; Wang, Z.T.; Zhang, T.; Wang, T.; Liu, R.J.; Wang, Y.; Jin, Q.Z.; Wang, X.G. Characterization of fatty acids, triacylglycerols, phytosterols and tocopherols in peony seed oil from five different major areas in China. Food Res. Int. 2020, 137, 109416. [Google Scholar] [CrossRef]
- He, C.L.; Zhang, K.Y.; Hou, X.G.; Han, D.B.; Wang, S.B. Foraging Behavior and Pollination Efficiency of Apis mellifera L. on the Oil Tree Peony ‘Feng Dan’ (Paeonia ostii T. Hong et J.X. Zhang). Insects 2019, 10, 116. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, D.L.; Guo, L.L.; Guo, Q.; Wang, H.F.; Hou, X.G. Construction of a high-density genetic map and QTLs mapping with GBS from the interspecific F1 population of P. ostii ‘Fengdan Bai’ and P. suffruticosa ‘Xin Riyuejin’. Sci. Hortic. 2019, 246, 190–200. [Google Scholar] [CrossRef]
- Tian, D.; Tilt, K.M.; Dane, F.; Woods, F.M.; Sibley, J.L. Comparison of shoot induction ability of different explants in herbaceous peony (Paeonia lactiflora Pall.). Sci. Hortic. 2010, 123, 385–389. [Google Scholar] [CrossRef]
- Ferreira, J.C.B.; de Araújo Silva-Cardoso, I.M.; de Oliveira Meira, R.; Scherwinski-Pereira, J.E.S. Somatic embryogenesis and plant regeneration from zygotic embryos of the palm tree Euterpe precatoria Mart. Plant Cell Rep. 2022, 148, 667–686. [Google Scholar] [CrossRef]
- Du, Y.M.; Cheng, F.Y.; Zhong, Y. Induction of direct somatic embryogenesis and shoot organogenesis and histological study in tree peony (Paeonia sect. Moutan). Plant Cell Tiss. Organ. Cult. 2020, 141, 557–570. [Google Scholar] [CrossRef]
- Ikeda, M.; Umehara, M.; Kamada, H. Embryogenesis-related genes; Its expression and roles during somatic and zygotic embryogenesis in carrot and Arabidopsis. Plant Biotechnol. 2006, 23, 153–161. [Google Scholar] [CrossRef]
- Elbl, P.; Lira, B.S.; Andrade, S.C.S.A.; Jo, L.; Santos, A.L.W.D.; Coutinho, L.L.; Floh, E.I.S.; Rossi, M. Comparative transcriptome analysis of early somatic embryo formation and seed development in Brazilian pine, Araucaria angustifolia (Bertol.) Kuntze. Plant Cell Tiss. Organ. Cult. 2015, 120, 903–915. [Google Scholar] [CrossRef]
- Armenta-Medina, A.; Gillmor, C.S.; Gao, P.; Mora-Macias, J.; Kochian, L.V.; Xiang, D.; Datla, R. Developmental and genomic architecture of plant embryogenesis: From model plant to crops. Plant Commun. 2020, 2, 100136. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pastrana, J.; Testillano, P.S.; Barany, I.; Canto-Flick, A.; Álvarez-López, D.; Pijeira-Fernández, G.; Avilés-Viñas, S.A.; Peña-Yam, L.; Muñoz-Ramírez, L.; Nahuat-Dzib, S. Endogenous auxin accumulation/localization during zygotic and somatic embryogenesis of Capsicum chinense Jacq. J. Plant Physiol. 2021, 258–259, 153333. [Google Scholar] [CrossRef]
- Wang, X.D.; Song, Y.H.; Sheahan, M.B.; Garg, M.L.; Rose, R.J. From embryo sac to oil and protein bodies: Embryo development in the model legume Medicago truncatula. New. Phytol. 2012, 193, 327–338. [Google Scholar] [CrossRef]
- Zhang, K.; Cao, W.; Baskin, J.M.; Baskin, C.C.; Sun, J.; Yao, L.; Tao, J. Seed development in Paeonia ostii (Paeoniaceae), with particular reference to embryogeny. BMC Plant Biol. 2021, 21, 603. [Google Scholar] [CrossRef]
- Xiu, Y.; Wu, G.; Tang, W.; Peng, Z.; Bu, X.; Chao, L.; Yin, X.; Xiong, J.N.; Zhang, H.W.; Zhao, X.Q.; et al. Oil biosynthesis and transcriptome profiles in developing endosperm and oil characteristic analyses in Paeonia ostii var. lishizhenii. J. Plant Physiol. 2018, 228, 121–133. [Google Scholar] [CrossRef]
- Radoeva, T.; Lokerse, A.S.; Llavata-Peris, C.I.; Wendrich, J.R.; Xiang, D.Q.; Liao, C.Y.; Vlaar, L.; Boekschoten, M.; Hooiveld, G.; Datla, R.; et al. A Robust Auxin Response Network Controls Embryo and Suspensor Development through a Basic Helix Loop Helix Transcriptional Module. Plant Cell 2019, 31, 52–67. [Google Scholar] [CrossRef]
- Robert, H.S.; Grunewald, W.; Sauer, M.; Cannoot, B.; Soriano, M.; Swarup, R.; Weijers, D.; Bennett, M.; Boutilier, K.; Friml, J. Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 2015, 142, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Winkelmann, T. Somatic Versus Zygotic Embryogenesis: Learning from Seeds. Methods Mol. Biol. 2016, 1359, 25–46. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Zhang, Z.J.; Laux, T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 2004, 20, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Breuninger, H.; Rikirsch, E.; Hermann, M.; Ueda, M.; Laux, T. Differential expression of WOX genes mediates apical–basal axis formation in the Arabidopsis embryo. Dev. Cell 2008, 14, 867–876. [Google Scholar] [CrossRef]
- Gambino, G.; Minuto, M.; Boccacci, P.; Perrone, I.; Vallania, R.; Gribaudo, I. Characterization of expression dynamics of WOX homeodomain transcription factors during somatic embryogenesis in Vitis vinifera. J. Exp. Bot. 2011, 62, 1089–1101. [Google Scholar] [CrossRef]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Schmidt, E.D.L.; Boutilier, K.; Grossniklaus, U.; de Vries, S.C. The Arabidopsis Somatic Embryogenesis Receptor Kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 2001, 127, 803–816. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L.; Yang, Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta 2005, 222, 107–117. [Google Scholar] [CrossRef]
- He, K.; Gou, X.P.; Yuan, T.; Lin, H.H.; Asami, T.; Yoshida, S.; Russell, S.D.; Li, J. BAK1 and BKK1 Regulate Brassinosteroid-Dependent Growth and Brassinosteroid-Independent Cell-Death Pathways. Curr. Biol. 2007, 17, 1109–1115. [Google Scholar] [CrossRef]
- Karlova, R.; Boeren, S.; Russinova, E.; Aker, J.; Vervoort, J.; de Vries, S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 2006, 18, 626–638. [Google Scholar] [CrossRef]
- Blakeslee, J.J.; Bandyopadhyay, A.; Lee, O.R.; Mravec, J.; Titapiwatanakun, B.; Sauer, M.; Makam, S.N.; Cheng, Y.; Bouchard, R.; Adamec, J.; et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 2007, 19, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Piya, S.; Shrestha, S.K.; Binder, B.; Stewart, C.N., Jr.; Hewezi, T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front. Plant Sci. 2014, 5, 744. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, M.; Qiu, Z.; Wang, K.; Du, Y.; Gu, L.; Cui, X. Spatiotemporal transcriptome provides insights into early fruit development of tomato (Solanum lycopersicum). Sci. Rep. 2016, 6, 23173. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.L.; Wang, S.; Tong, R.R.; Wang, S.; Chen, Y.H.; Wu, W.J.; He, F.Z.; Wan, R.; Jian, Z.H.; Hu, Q.X.; et al. Widely targeted secondary metabolomics explored pomegranate aril browning during cold storage. Postharvest Biol. Technol. 2022, 186, 111839. [Google Scholar] [CrossRef]
- Chotikakham, S.; Pany, A.; Saengnil, K. Methyl salicylate treatment alleviates peel spotting of ‘Sucrier’ banana by improving mitochondrial physiological properties and functions. Postharvest Biol. Technol. 2022, 186, 111832. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, 14; R Foundation for Statistical Computing: Vienna, Austria, 2014; pp. 12–21. Available online: https://www.r-project.org/ (accessed on 14 March 2019).
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fan, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.N.; Sun, D.Y.; Wang, S.; Luo, S.; Fu, Y.Q.; Niu, L.X.; Shi, Q.Q.; Zhang, Y.L. Integrating full-length transcriptomics and metabolomics reveals the regulatory mechanisms underlying yellow pigmentation in tree peony (Paeonia suffruticosa Andr.) flowers. Hortic. Res. 2021, 8, 235. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Shang, W.; Li, L.; Song, Y.; Wang, G.; Shi, L.; Shen, Y.; Sun, Y.; He, S.; Wang, Z. Transcriptome Landscape Analyses of the Regulatory Network for Zygotic Embryo Development in Paeonia ostii. Int. J. Mol. Sci. 2023, 24, 10715. https://doi.org/10.3390/ijms241310715
Xu Y, Shang W, Li L, Song Y, Wang G, Shi L, Shen Y, Sun Y, He S, Wang Z. Transcriptome Landscape Analyses of the Regulatory Network for Zygotic Embryo Development in Paeonia ostii. International Journal of Molecular Sciences. 2023; 24(13):10715. https://doi.org/10.3390/ijms241310715
Chicago/Turabian StyleXu, Yufeng, Wenqian Shang, Linda Li, Yinglong Song, Guiqing Wang, Liyun Shi, Yuxiao Shen, Yuke Sun, Songlin He, and Zheng Wang. 2023. "Transcriptome Landscape Analyses of the Regulatory Network for Zygotic Embryo Development in Paeonia ostii" International Journal of Molecular Sciences 24, no. 13: 10715. https://doi.org/10.3390/ijms241310715
APA StyleXu, Y., Shang, W., Li, L., Song, Y., Wang, G., Shi, L., Shen, Y., Sun, Y., He, S., & Wang, Z. (2023). Transcriptome Landscape Analyses of the Regulatory Network for Zygotic Embryo Development in Paeonia ostii. International Journal of Molecular Sciences, 24(13), 10715. https://doi.org/10.3390/ijms241310715





