Single-Cell RNA-Seq Identifies Pathways and Genes Contributing to the Hyperandrogenemia Associated with Polycystic Ovary Syndrome
Abstract
1. Introduction
2. Results
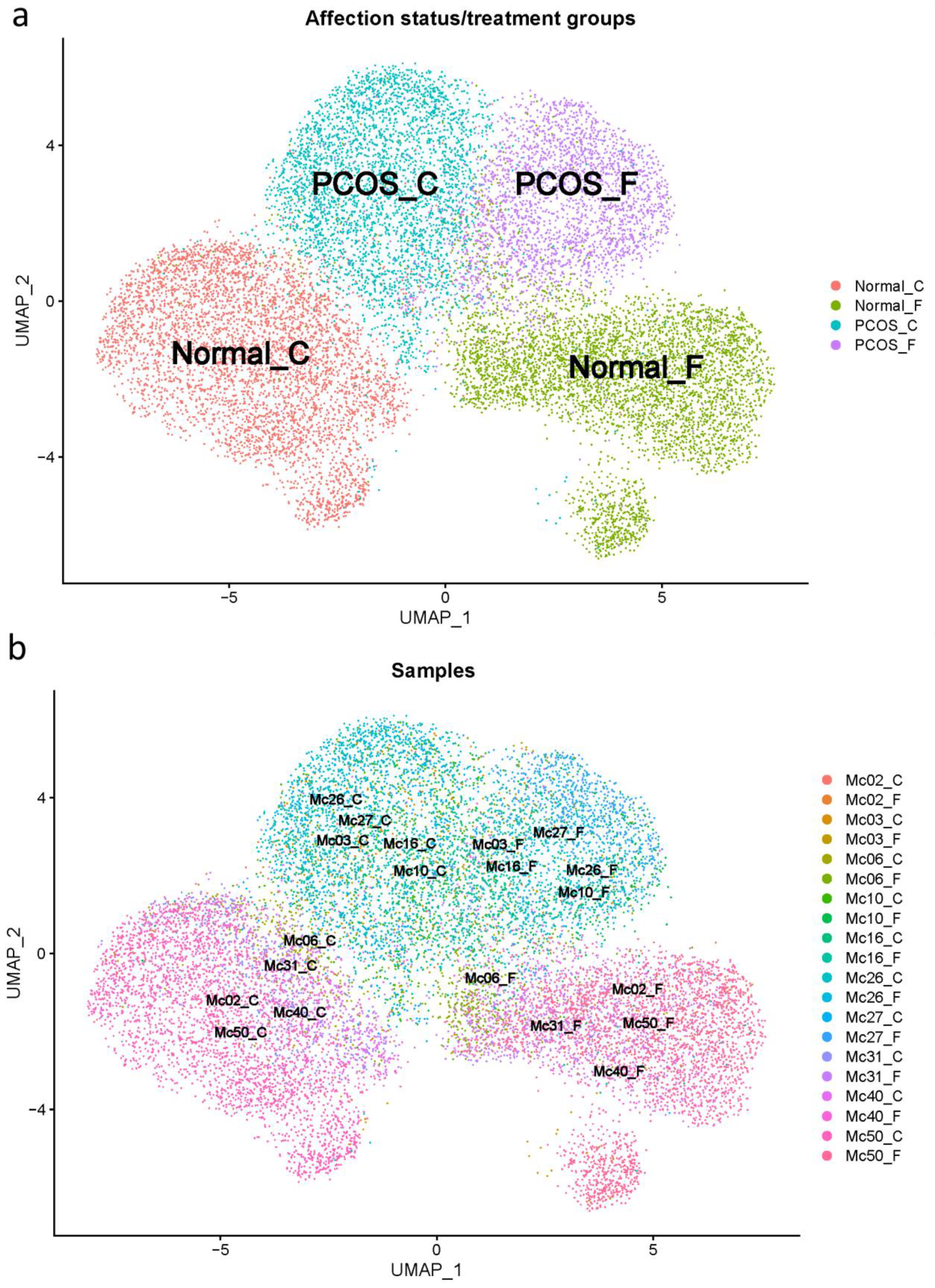
2.1. scRNA-Seq Reveals That PCOS Theca Cells Have Distinctive Gene Expression Profiles
2.2. Bulk RNA-Seq Confirms a Distinctive Repertoire of Gene Expression in PCOS Theca Cells
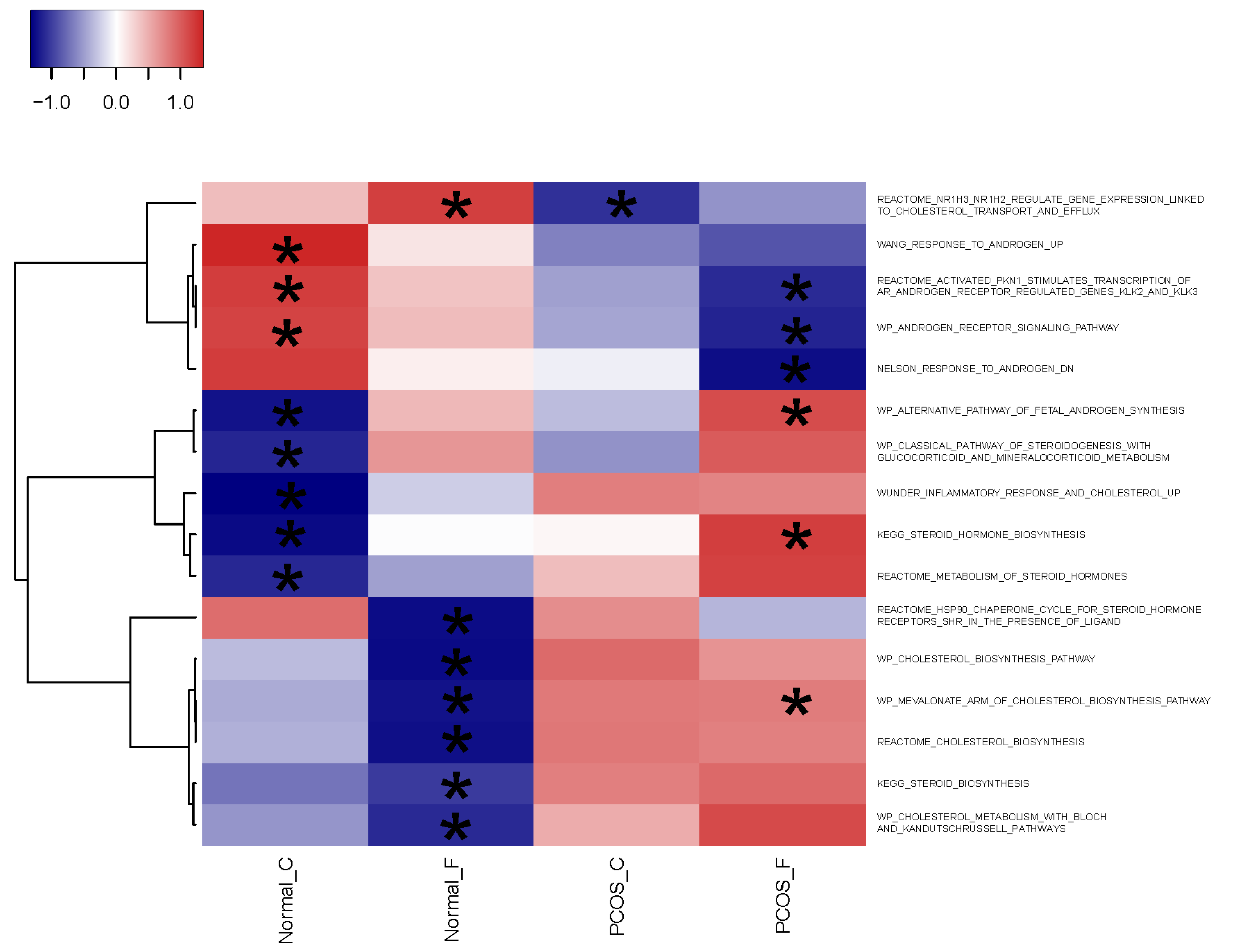
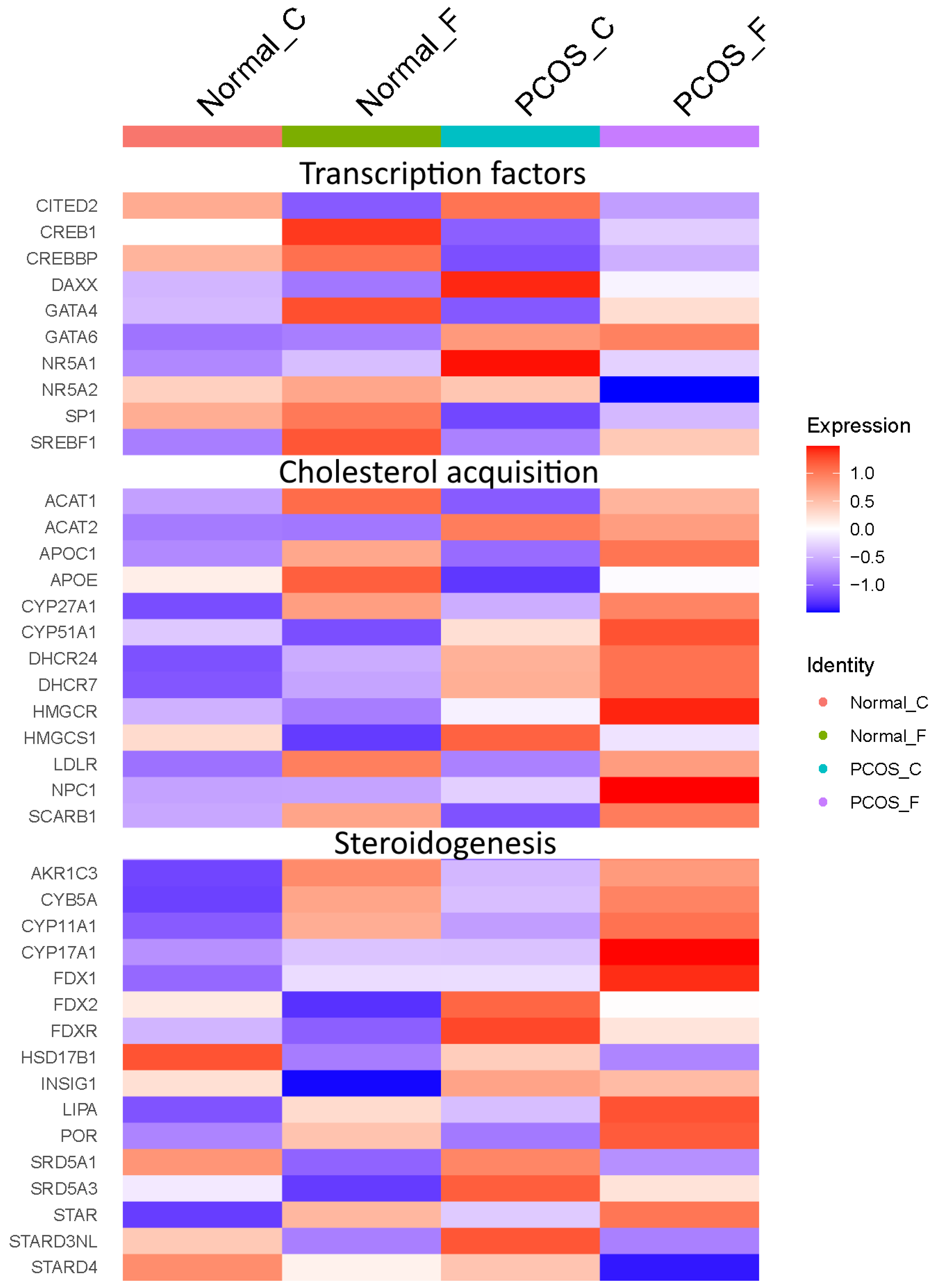
2.3. Differential Gene Expression Identified by scRNA-Seq Is Consistent with the PCOS Molecular Signature Detected by Microarrays
3. Discussion
4. Materials and Methods
4.1. Theca Cell Cultures
4.2. Single-Cell RNA Sequencing (scRNA-Seq)
4.3. Bulk RNA Sequencing (Bulk RNA-Seq)
4.4. Cell Ranger Data Processing
4.5. Seurat Pre-Processing and Quality Control
4.6. STACAS Integration
4.7. UMAP Generation
4.8. Identification of Differentially Expressed Genes
4.9. Gene Set Enrichment Analysis
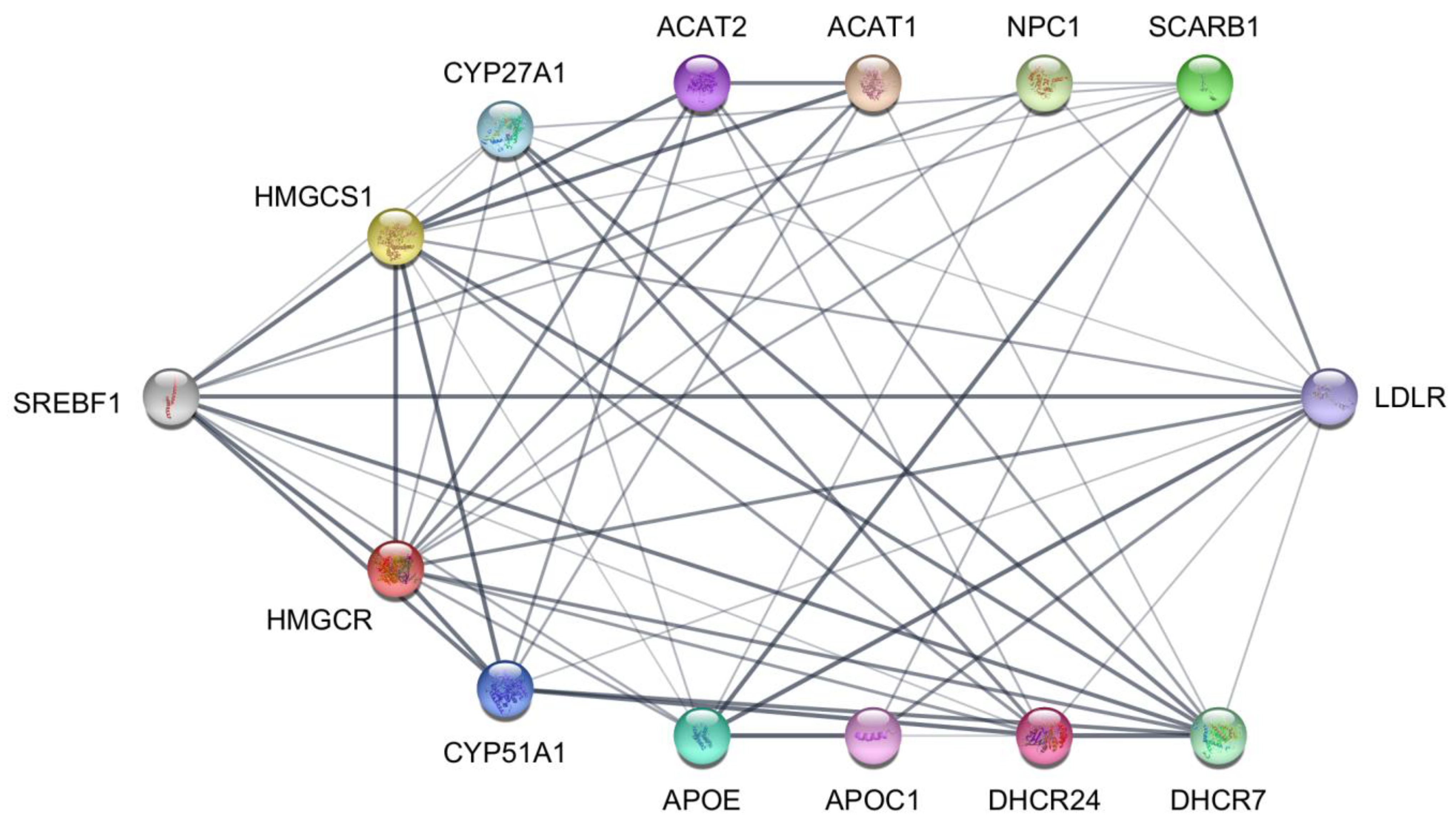
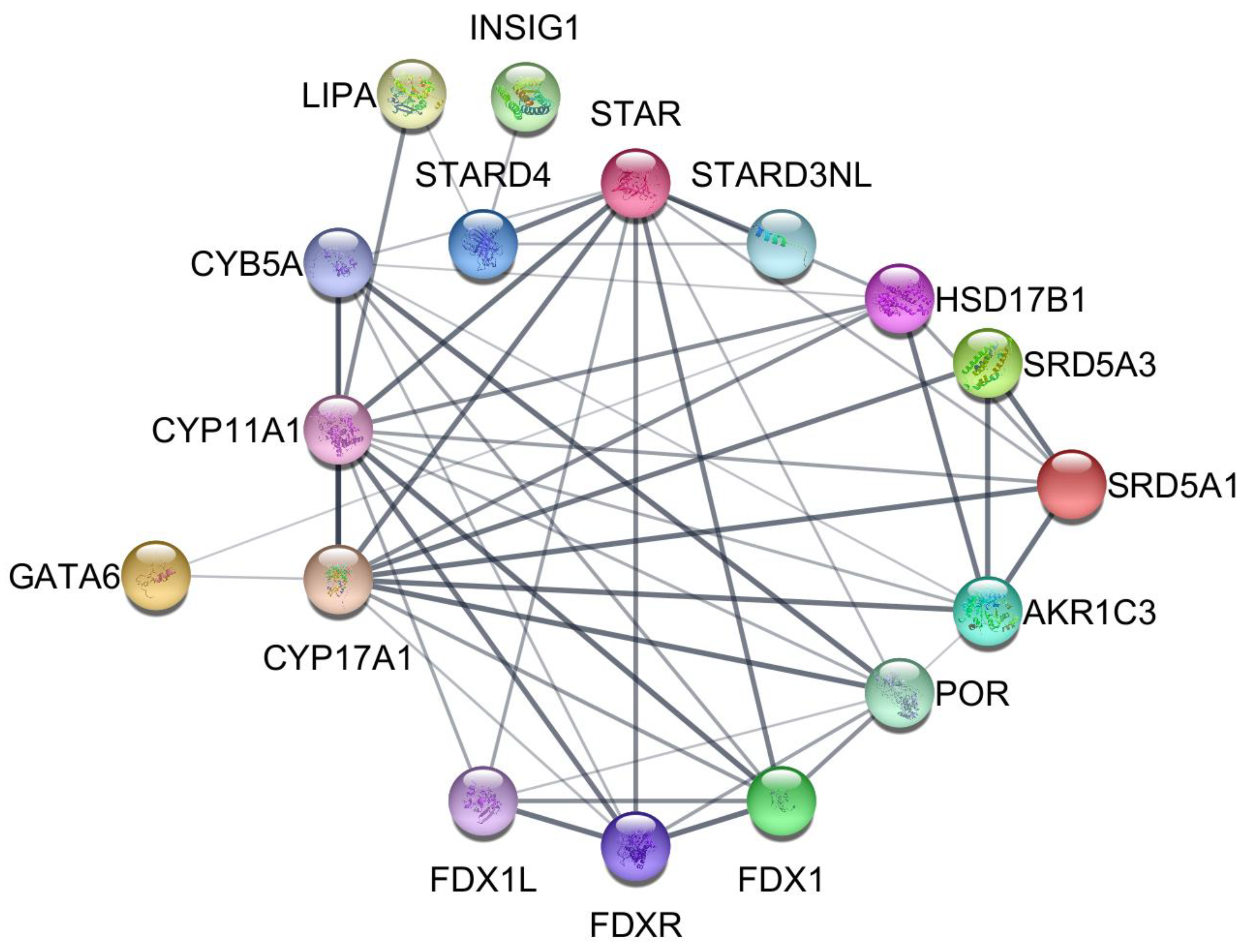
4.10. STRING Protein–Protein Interaction Database and Network Visualization
4.11. Bulk RNA Sequencing Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Azziz, R.; Dumesic, D.A.; Goodarzi, M.O. Polycystic Ovary Syndrome: An Ancient Disorder? Fertil. Steril. 2011, 95, 1544–1548. [Google Scholar] [CrossRef]
- Balen, A.; Homburg, R.; Franks, S. Defining Polycystic Ovary Syndrome. BMJ 2009, 338, 426. [Google Scholar] [CrossRef] [PubMed]
- Franks, S.; Webber, L.J.; Goh, M.; Valentine, A.; White, D.M.; Conway, G.S.; Wiltshire, S.; McCarthy, M.I. Ovarian Morphology Is a Marker of Heritable Biochemical Traits in Sisters with Polycystic Ovaries. J. Clin. Endocrinol. Metab. 2008, 93, 3396–3402. [Google Scholar] [CrossRef]
- Franks, S.; Stark, J.; Hardy, K. Follicle Dynamics and Anovulation in Polycystic Ovary Syndrome. Hum. Reprod. Update 2008, 14, 367–378. [Google Scholar] [CrossRef]
- McAllister, J.M.; Legro, R.S.; Modi, B.P.; Strauss, J.F. Functional Genomics of PCOS: From GWAS to Molecular Mechanisms. Trends Endocrinol. Metab. 2015, 26, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Dapas, M.; Dunaif, A. Deconstructing a Syndrome: Genomic Insights into PCOS Causal Mechanisms and Classification. Endocr. Rev. 2022, 43, 927–965. [Google Scholar] [CrossRef]
- McAllister, J.M.; Modi, B.; Miller, B.A.; Biegler, J.; Bruggeman, R.; Legro, R.S.; Strauss, J.F. Overexpression of a DENND1A Isoform Produces a Polycystic Ovary Syndrome Theca Phenotype. Proc. Natl. Acad. Sci. USA 2014, 111. [Google Scholar] [CrossRef] [PubMed]
- Azziz, R. PCOS in 2015: New Insights into the Genetics of Polycystic Ovary Syndrome. Nat. Rev. Endocrinol. 2016, 12, 74. [Google Scholar] [CrossRef]
- Azziz, R. Introduction: Determinants of Polycystic Ovary Syndrome. Fertil. Steril. 2016, 106, 4–5. [Google Scholar] [CrossRef]
- Legro, R.S.; Arslanian, S.A.; Ehrmann, D.A.; Hoeger, K.M.; Murad, M.H.; Pasquali, R.; Welt, C.K. Diagnosis and Treatment of Polycystic Ovary Syndrome: An Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2013, 98, 4565–4592. [Google Scholar] [CrossRef]
- McAllister, J.M.; Kerin, J.F.P.; Trant, J.M.; Estabrook, R.W.; Mason, J.I.; Waterman, M.R.; Simpson, E.R. Regulation of Cholesterol Side-Chain Cleavage and 17 Alpha-Hydroxylase/Lyase Activities in Proliferating Human Theca Interna Cells in Long Term Monolayer Culture. Endocrinology 1989, 125, 1959–1966. [Google Scholar] [CrossRef]
- Wickenheisser, J.K.; Nelson-DeGrave, V.L.; McAllister, J.M. Human Ovarian Theca Cells in Culture. Trends Endocrinol. Metab. 2006, 17, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Nelson, V.L.; Legro, R.S.; Strauss, J.F., 3rd; McAllister, J.M. Augmented Androgen Production Is a Stable Steroidogenic Phenotype of Propagated Theca Cells from Polycystic Ovaries. Mol. Endocrinol. 1999, 13, 946–957. [Google Scholar] [CrossRef]
- McAllister, J.M. Functional, Long-Term Human Theca and Granulosa Cell Cultures from Polycystic Ovaries. Endocrine 1995, 3, 143–149. [Google Scholar] [CrossRef]
- Comim, F.V.; Teerds, K.; Hardy, K.; Franks, S. Increased Protein Expression of LHCG Receptor and 17α-Hydroxylase/17-20-Lyase in Human Polycystic Ovaries. Hum. Reprod. 2013, 28, 3086–3092. [Google Scholar] [CrossRef] [PubMed]
- Gilling-Smith, C.; Willis, D.S.; Beard, R.W.; Franks, S. Hypersecretion of Androstenedione by Isolated Thecal Cells from Polycystic Ovaries. J. Clin. Endocrinol. Metab. 1994, 79, 1158–1165. [Google Scholar] [CrossRef]
- Gilling-Smith, C.; Story, H.; Rogers, V.; Franks, S. Evidence for a Primary Abnormality of Thecal Cell Steroidogenesis in the Polycystic Ovary Syndrome. Clin. Endocrinol. 1997, 47, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Mcallister, J.M.; Han, A.X.; Modi, B.P.; Teves, M.E.; Mavodza, G.R.; Anderson, Z.L.; Shen, T.; Christenson, L.K.; Archer, K.J.; Strauss, J.F. MiRNA Profiling Reveals MiRNA-130b-3p Mediates DENND1A Variant 2 Expression and Androgen Biosynthesis. Endocrinology 2019, 160, 1964–1981. [Google Scholar] [CrossRef]
- Alan Harris, R.; Archer, K.J.; Goodarzi, M.O.; York, T.P.; Rogers, J.; Dunaif, A.; McAllister, J.M.; Strauss, J.F. Loci on Chromosome 12q13.2 Encompassing ERBB3, PA2G4 and RAB5B Are Associated with Polycystic Ovary Syndrome. Gene 2023, 852, 147062. [Google Scholar] [CrossRef] [PubMed]
- Waterbury, J.S.; Teves, M.E.; Gaynor, A.; Han, A.X.; Mavodza, G.; Newell, J.; Strauss, J.F.; McAllister, J.M. The PCOS GWAS Candidate Gene ZNF217 Influences Theca Cell Expression of DENND1A.V2, CYP17A1, and Androgen Production. J. Endocr. Soc. 2022, 6, bvac078. [Google Scholar] [CrossRef]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef]
- Andreatta, M.; Carmona, S.J. STACAS: Sub-Type Anchor Correction for Alignment in Seurat to Integrate Single-Cell RNA-Seq Data. Bioinformatics 2021, 37, 882–884. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Cillo, A. Singleseqgset 2019. Available online: https://github.com/arc85/singleseqgset (accessed on 19 June 2023).
- Auchus, R.J. Steroid 17-Hydroxylase and 17,20-Lyase Deficiencies, Genetic and Pharmacologic. J. Steroid Biochem. Mol. Biol. 2017, 165, 71–78. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, S.; Yan, Z.; Niu, Y.; Du, W.; Liu, B.; Han, B.; Liu, X.; Zhao, S.; Song, H.; et al. Phenotypic Heterogeneity and Fertility Potential of Patients with 17-Hydroxylase/17,20-Lyase Deficiency. J. Clin. Endocrinol. Metab. 2022, 107, E2610–E2618. [Google Scholar] [CrossRef]
- Miller, W.L.; Tee, M.K. The Post-Translational Regulation of 17,20 Lyase Activity. Mol. Cell. Endocrinol. 2015, 408, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Watari, H.; Blanchette-Mackie, E.J.; Dwyer, N.K.; Sun, G.; Glick, J.M.; Patel, S.; Neufeld, E.B.; Pentchev, P.G.; Strauss, J.F. NPC1-Containing Compartment of Human Granulosa-Lutein Cells: A Role in the Intracellular Trafficking of Cholesterol Supporting Steroidogenesis. Exp. Cell Res. 2000, 255, 56–66. [Google Scholar] [CrossRef]
- Miller, W.L. Steroid Hormone Synthesis in Mitochondria. Mol. Cell. Endocrinol. 2013, 379, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.R.; Nelson, V.L.; Ho, C.; Jansen, E.; Wang, C.Y.; Urbanek, M.; McAllister, J.M.; Mosselman, S.; Strauss, J.F. The Molecular Phenotype of Polycystic Ovary Syndrome (PCOS) Theca Cells and New Candidate PCOS Genes Defined by Microarray Analysis. J. Biol. Chem. 2003, 278, 26380–26390. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, H.; Patel, J.; Jain, N.K.; Joshi, R. The Role of Polymorphism in Various Potential Genes on Polycystic Ovary Syndrome Susceptibility and Pathogenesis. J. Ovarian Res. 2021, 14, 125. [Google Scholar] [CrossRef]
- Pericleous, M.; Kelly, C.; Wang, T.; Livingstone, C.; Ala, A. Wolman’s Disease and Cholesteryl Ester Storage Disorder: The Phenotypic Spectrum of Lysosomal Acid Lipase Deficiency. Lancet Gastroenterol. Hepatol. 2017, 2, 670–679. [Google Scholar] [CrossRef]
- Miller, W.L. Disorders in the Initial Steps of Steroid Hormone Synthesis. J. Steroid Biochem. Mol. Biol. 2017, 165, 18–37. [Google Scholar] [CrossRef] [PubMed]
- Christenson, L.K.; Osborne, T.F.; Mcallister, J.M.; Strauss, J.F. Conditional Response of the Human Steroidogenic Acute Regulatory Protein Gene Promoter to Sterol Regulatory Element Binding Protein-1a. Endocrinology 2001, 142, 28–36. [Google Scholar] [CrossRef]
- Wood, J.R.; Ho, C.K.M.; Nelson-Degrave, V.L.; McAllister, J.M.; Strauss, J.F. The Molecular Signature of Polycystic Ovary Syndrome (PCOS) Theca Cells Defined by Gene Expression Profiling. J. Reprod. Immunol. 2004, 63, 51–60. [Google Scholar] [CrossRef]
- Ho, C.K.M.; Wood, J.R.; Stewart, D.R.; Ewens, K.; Ankener, W.; Wickenheisser, J.; Nelson-Degrave, V.; Zhang, Z.; Legro, R.S.; Dunaif, A.; et al. Increased Transcription and Increased Messenger Ribonucleic Acid (MRNA) Stability Contribute to Increased GATA6 MRNA Abundance in Polycystic Ovary Syndrome Theca Cells. J. Clin. Endocrinol. Metab. 2005, 90, 6596–6602. [Google Scholar] [CrossRef]
- Chen, J.; Huang, C.; Zhang, T.; Gong, W.; Deng, X.; Liu, H.; Liu, J.; Guo, Y. The Effects of Statins on Hyperandrogenism in Women with Polycystic Ovary Syndrome: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Reprod. Biol. Endocrinol. 2021, 19, 189. [Google Scholar] [CrossRef]
- Flück, C.E.; Miller, W.L. GATA-4 and GATA-6 Modulate Tissue-Specific Transcription of the Human Gene for P450c17 by Direct Interaction with Sp1. Mol. Endocrinol. 2004, 18, 1144–1157. [Google Scholar] [CrossRef]
- Sewer, M.B.; Waterman, M.R. ACTH Modulation of Transcription Factors Responsible for Steroid Hydroxylase Gene Expression in the Adrenal Cortex. Microsc. Res. Tech. 2003, 61, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Wickenheisser, J.K.; Biegler, J.M.; Nelson-DeGrave, V.L.; Legro, R.S.; Strauss, J.F.; McAllister, J.M. Cholesterol Side-Chain Cleavage Gene Expression in Theca Cells: Augmented Transcriptional Regulation and MRNA Stability in Polycystic Ovary Syndrome. PLoS ONE 2012, 7, e48963. [Google Scholar] [CrossRef] [PubMed]
- Wickenheisser, J.K.; Nelson-Degrave, V.L.; McAllister, J.M. Dysregulation of Cytochrome P450 17alpha-Hydroxylase Messenger Ribonucleic Acid Stability in Theca Cells Isolated from Women with Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2005, 90, 1720–1727. [Google Scholar] [CrossRef]
- Arzalluz-Luqueángeles, Á; Conesa, A. Single-Cell RNAseq for the Study of Isoforms-How Is That Possible? Genome Biol. 2018, 19, 110. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, B.; Lin, L.L.; Zhao, S. Evaluation and Comparison of Computational Tools for RNA-Seq Isoform Quantification. BMC Genom. 2017, 18, 583. [Google Scholar] [CrossRef] [PubMed]
- Tee, M.K.; Speek, M.; Legeza, B.; Modi, B.; Teves, M.E.; McAllister, J.M.; Strauss, J.F.; Miller, W.L. Alternative Splicing of DENND1A, a PCOS Candidate Gene, Generates Variant 2. Mol. Cell. Endocrinol. 2016, 434, 25–35. [Google Scholar] [CrossRef]
- Kulkarni, R.; Teves, M.E.; Han, A.X.; McAllister, J.M.; Strauss, J.F., 3rd. Colocalization of Polycystic Ovary Syndrome Candidate Gene Products in Theca Cells Suggests Novel Signaling Pathways. J. Endocr. Soc. 2019, 3, 2204–2223. [Google Scholar] [CrossRef] [PubMed]
- Nelson, V.L.; Qin, K.N.; Rosenfield, R.L.; Wood, J.R.; Penning, T.M.; Legro, R.S.; Strauss, J.F.; McAllister, J.M. The Biochemical Basis for Increased Testosterone Production in Theca Cells Propagated from Patients with Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2001, 86, 5925–5933. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harris, R.A.; McAllister, J.M.; Strauss, J.F., III. Single-Cell RNA-Seq Identifies Pathways and Genes Contributing to the Hyperandrogenemia Associated with Polycystic Ovary Syndrome. Int. J. Mol. Sci. 2023, 24, 10611. https://doi.org/10.3390/ijms241310611
Harris RA, McAllister JM, Strauss JF III. Single-Cell RNA-Seq Identifies Pathways and Genes Contributing to the Hyperandrogenemia Associated with Polycystic Ovary Syndrome. International Journal of Molecular Sciences. 2023; 24(13):10611. https://doi.org/10.3390/ijms241310611
Chicago/Turabian StyleHarris, R. Alan, Jan M. McAllister, and Jerome F. Strauss, III. 2023. "Single-Cell RNA-Seq Identifies Pathways and Genes Contributing to the Hyperandrogenemia Associated with Polycystic Ovary Syndrome" International Journal of Molecular Sciences 24, no. 13: 10611. https://doi.org/10.3390/ijms241310611
APA StyleHarris, R. A., McAllister, J. M., & Strauss, J. F., III. (2023). Single-Cell RNA-Seq Identifies Pathways and Genes Contributing to the Hyperandrogenemia Associated with Polycystic Ovary Syndrome. International Journal of Molecular Sciences, 24(13), 10611. https://doi.org/10.3390/ijms241310611





