Silver Ions Inhibit Bacterial Movement and Stall Flagellar Motor
Abstract
1. Introduction
2. Results and Discussions
2.1. Lower Motility of Bacteria Caused by Ag+ Ions
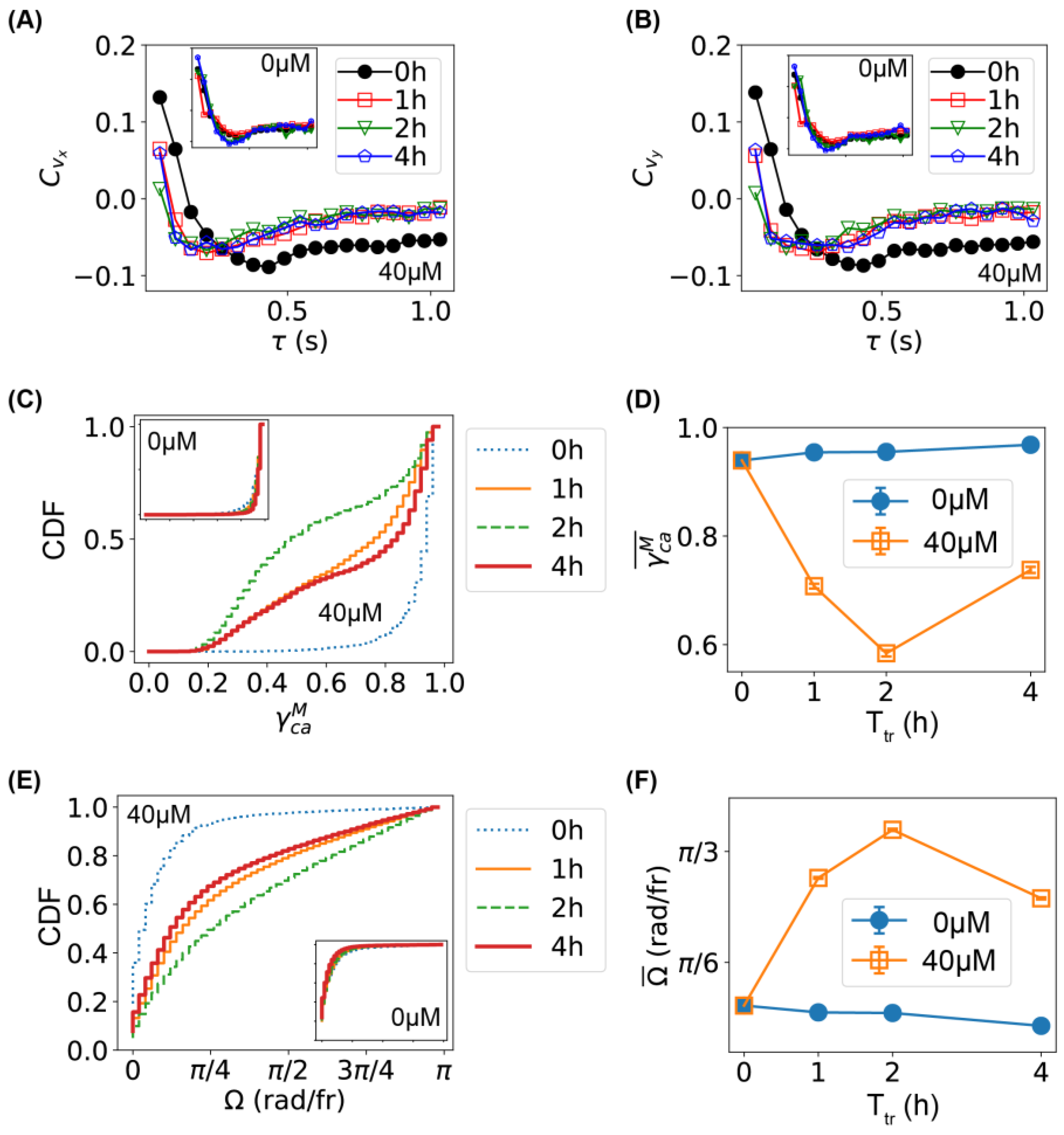
2.2. Comparison of Bacterial Movement before and after Ag+ Treatment
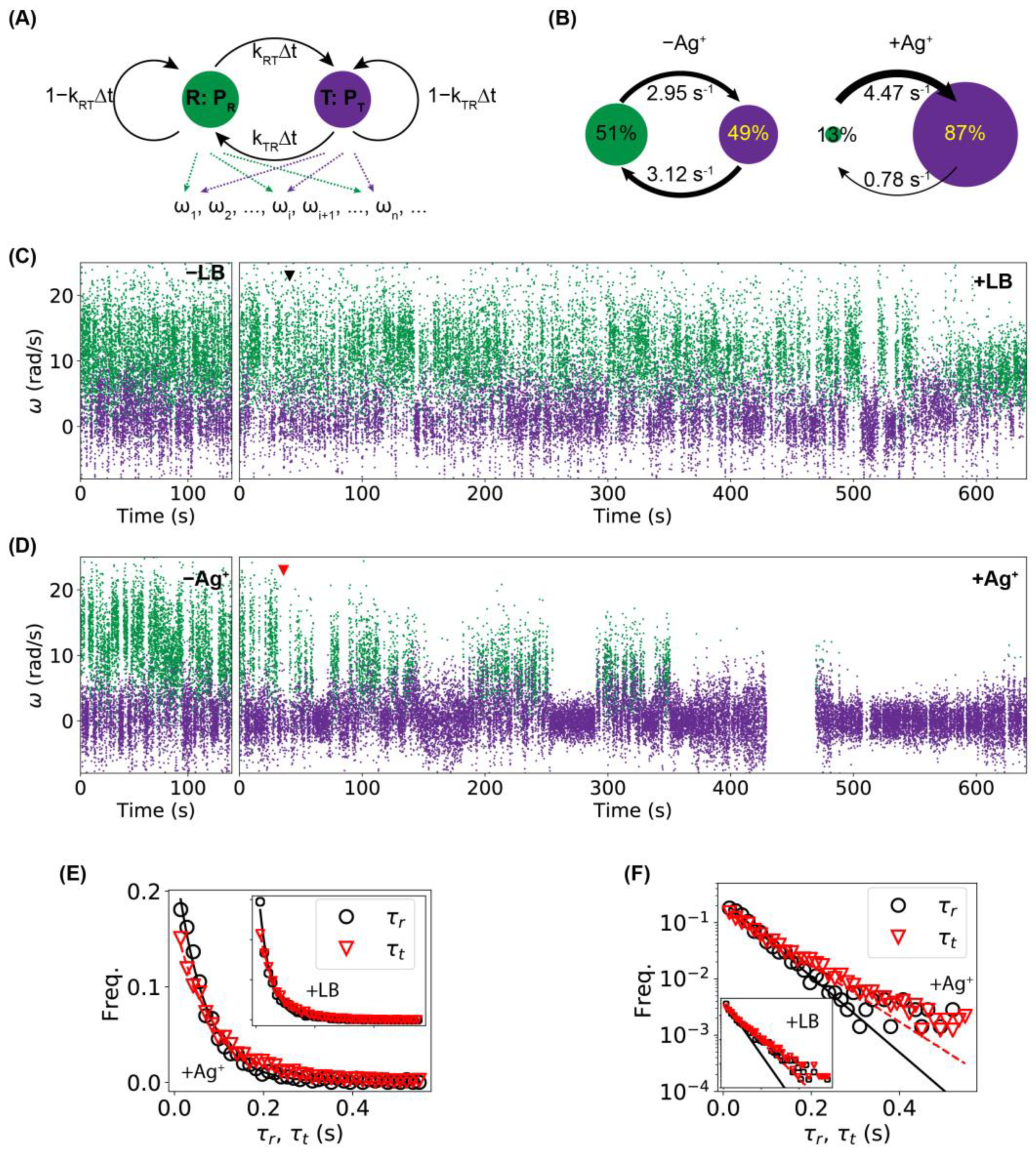
2.3. Higher Frequency of Tumbling/Pausing Caused by Ag+ Ions in Tethering Assays
2.4. Higher Frequency of Tumbling/Pausing Caused by Ag+ Ions in Untethered Rotation Assays
2.5. Stalling of Flagellar Motors Caused by Ag+ Ions
3. Materials and Methods
3.1. Bacterial Strain and Growth
3.2. Phase-Contrast Microscopy
3.3. Swimming Assay
3.4. Tethering Assay
3.5. Untethered Rotation Assay
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United State; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019. [Google Scholar]
- Allen, H.K.; Trachsel, J.; Looft, T.; Casey, T.A. Finding Alternatives to Antibiotics. Ann. N. Y. Acad. Sci. 2014, 1323, 91–100. [Google Scholar] [CrossRef] [PubMed]
- François, B.; Jafri, H.S.; Bonten, M. Alternatives to Antibiotics. Intensive Care Med. 2016, 42, 2034–2036. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.A.; Imamura, R.; Brown, G.A.; Krishnamurthi, V.R.; Niyonshuti, I.I.; Marcelle, T.; Mathurin, L.E.; Chen, J.; Wang, Y. An Experiment-Based Model Quantifying Antimicrobial Activity of Silver Nanoparticles on Escherichia coli. RSC Adv. 2017, 7, 56173–56182. [Google Scholar] [CrossRef]
- Durán, N.; Durán, M.; de Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver Nanoparticles: A New View on Mechanistic Aspects on Antimicrobial Activity. Nanomedicine 2016, 12, 789–799. [Google Scholar] [CrossRef]
- Rai, M.; Yadav, A.; Gade, A. Silver Nanoparticles as a New Generation of Antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef]
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A Mechanistic Study of the Antibacterial Effect of Silver Ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668. [Google Scholar] [CrossRef]
- Radzig, M.A.; Nadtochenko, V.A.; Koksharova, O.A.; Kiwi, J.; Lipasova, V.A.; Khmel, I.A. Antibacterial Effects of Silver Nanoparticles on Gram-Negative Bacteria: Influence on the Growth and Biofilms Formation, Mechanisms of Action. Colloids Surf. B Biointerfaces 2013, 102, 300–306. [Google Scholar] [CrossRef]
- Marambio-Jones, C.; Hoek, E.M.V. A Review of the Antibacterial Effects of Silver Nanomaterials and Potential Implications for Human Health and the Environment. J. Nanopart. Res. 2010, 12, 1531–1551. [Google Scholar] [CrossRef]
- Ivask, A.; Elbadawy, A.; Kaweeteerawat, C.; Boren, D.; Fischer, H.; Ji, Z.; Chang, C.H.; Liu, R.; Tolaymat, T.; Telesca, D.; et al. Toxicity Mechanisms in Escherichia coli Vary for Silver Nanoparticles and Differ from Ionic Silver. ACS Nano 2014, 8, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Sadoon, A.A.; Khadka, P.; Freeland, J.; Gundampati, R.K.; Manso, R.H.; Ruiz, M.; Krishnamurthi, V.R.; Thallapuranam, S.K.; Chen, J.; Wang, Y. Silver Ions Caused Faster Diffusive Dynamics of Histone-like Nucleoid-Structuring Proteins in Live Bacteria. Appl. Environ. Microbiol. 2020, 86, e02479-19. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthi, V.R.; Chen, J.; Wang, Y. Silver Ions Cause Oscillation of Bacterial Length of Escherichia coli. Sci. Rep. 2019, 9, 11745. [Google Scholar] [CrossRef]
- Josenhans, C.; Suerbaum, S. The Role of Motility as a Virulence Factor in Bacteria. Int. J. Med. Microbiol. 2002, 291, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Sowa, Y.; Berry, R.M. Bacterial Flagellar Motor. Q. Rev. Biophys. 2008, 41, 103–132. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhao, Y.; Zhuang, X.-Y.; Lo, W.-C.; Baker, M.A.B.; Lo, C.-J.; Bai, F. Frequent Pauses in Escherichia coli Flagella Elongation Revealed by Single Cell Real-Time Fluorescence Imaging. Nat. Commun. 2018, 9, 1885. [Google Scholar] [CrossRef] [PubMed]
- Minamino, T.; Imada, K. The Bacterial Flagellar Motor and Its Structural Diversity. Trends Microbiol. 2015, 23, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.S.; Hazelbauer, G.L.; Falke, J.J. Signaling and Sensory Adaptation in Escherichia coli Chemoreceptors: 2015 Update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef]
- Silverman, M.; Simon, M. Flagellar Rotation and the Mechanism of Bacterial Motility. Nature 1974, 249, 73–74. [Google Scholar] [CrossRef]
- Lapidus, I.R.; Welch, M.; Eisenbach, M. Pausing of Flagellar Rotation Is a Component of Bacterial Motility and Chemotaxis. J. Bacteriol. 1988, 170, 3627–3632. [Google Scholar] [CrossRef]
- Sourjik, V.; Wingreen, N.S. Responding to Chemical Gradients: Bacterial Chemotaxis. Curr. Opin. Cell Biol. 2012, 24, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Gugala, N.; Lemire, J.; Chatfield-Reed, K.; Yan, Y.; Chua, G.; Turner, R.J. Using a Chemical Genetic Screen to Enhance Our Understanding of the Antibacterial Properties of Silver. Genes 2018, 9, 344. [Google Scholar] [CrossRef] [PubMed]
- Alqahtany, M.; Khadka, P.; Niyonshuti, I.; Krishnamurthi, V.R.; Sadoon, A.A.; Challapalli, S.D.; Chen, J.; Wang, Y. Nanoscale Reorganizations of Histone-like Nucleoid Structuring Proteins in Escherichia coli Are Caused by Silver Nanoparticles. Nanotechnology 2019, 30, 385101. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ayala, J.C.; Benitez, J.A.; Silva, A.J. RNA-Seq Analysis Identifies New Genes Regulated by the Histone-like Nucleoid Structuring Protein (H-NS) Affecting Vibrio Cholerae Virulence, Stress Response and Chemotaxis. PLoS ONE 2015, 10, e0118295. [Google Scholar] [CrossRef]
- Bertin, P.; Terao, E.; Lee, E.H.; Lejeune, P.; Colson, C.; Danchin, A.; Collatz, E. The H-NS Protein Is Involved in the Biogenesis of Flagella in Escherichia coli. J. Bacteriol. 1994, 176, 5537–5540. [Google Scholar] [CrossRef]
- Garuglieri, E.; Cattò, C.; Villa, F.; Zanchi, R.; Cappitelli, F. Effects of Sublethal Concentrations of Silver Nanoparticles on Escherichia coli and Bacillus subtilis under Aerobic and Anaerobic Conditions. Biointerphases 2016, 11, 04B308. [Google Scholar] [CrossRef]
- West, M.; Burdash, N.M.; Freimuth, F. Simplified Silver-Plating Stain for Flagella. J. Clin. Microbiol. 1977, 6, 414–419. [Google Scholar] [CrossRef]
- Ortega-Calvo, J.-J.; Molina, R.; Jimenez-Sanchez, C.; Dobson, P.J.; Thompson, I.P. Bacterial Tactic Response to Silver Nanoparticles. Environ. Microbiol. Rep. 2011, 3, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Choi, O.; Deng, K.K.; Kim, N.-J.; Ross, L.; Surampalli, R.Y.; Hu, Z. The Inhibitory Effects of Silver Nanoparticles, Silver Ions, and Silver Chloride Colloids on Microbial Growth. Water Res. 2008, 42, 3066–3074. [Google Scholar] [CrossRef]
- Theves, M.; Taktikos, J.; Zaburdaev, V.; Stark, H.; Beta, C. A Bacterial Swimmer with Two Alternating Speeds of Propagation. Biophys. J. 2013, 105, 1915–1924. [Google Scholar] [CrossRef] [PubMed]
- Theves, M.; Taktikos, J.; Zaburdaev, V.; Stark, H.; Beta, C. Random Walk Patterns of a Soil Bacterium in Open and Confined Environments. Europhys. Lett. 2015, 109, 28007. [Google Scholar] [CrossRef]
- Lauga, E.; DiLuzio, W.R.; Whitesides, G.M.; Stone, H.A. Swimming in Circles: Motion of Bacteria near Solid Boundaries. Biophys. J. 2006, 90, 400–412. [Google Scholar] [CrossRef]
- Pottash, A.E.; McKay, R.; Virgile, C.R.; Ueda, H.; Bentley, W.E. TumbleScore: Run and Tumble Analysis for Low Frame-Rate Motility Videos. BioTechniques 2017, 62, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Boulos, L.; Prévost, M.; Barbeau, B.; Coallier, J.; Desjardins, R. LIVE/DEAD BacLight: Application of a New Rapid Staining Method for Direct Enumeration of Viable and Total Bacteria in Drinking Water. J. Microbiol. Methods 1999, 37, 77–86. [Google Scholar] [CrossRef]
- Michalet, X. Mean Square Displacement Analysis of Single-Particle Trajectories with Localization Error: Brownian Motion in an Isotropic Medium. Phys. Rev. E Stat. Nonlin Soft Matter Phys. 2010, 82, 041914. [Google Scholar] [CrossRef]
- Metzler, R.; Jeon, J.-H.; Cherstvy, A.G.; Barkai, E. Anomalous Diffusion Models and Their Properties: Non-Stationarity, Non-Ergodicity, and Ageing at the Centenary of Single Particle Tracking. Phys. Chem. Chem. Phys. 2014, 16, 24128–24164. [Google Scholar] [CrossRef]
- Sadoon, A.A.; Wang, Y. Anomalous, Non-Gaussian, Viscoelastic, and Age-Dependent Dynamics of Histonelike Nucleoid-Structuring Proteins in Live Escherichia coli. Phys. Rev. E 2018, 98, 042411. [Google Scholar] [CrossRef]
- Metzler, R.; Klafter, J. The Random Walk’s Guide to Anomalous Diffusion: A Fractional Dynamics Approach. Phys. Rep. 2000, 339, 1–77. [Google Scholar] [CrossRef]
- Amsler, C.D. Use of Computer-Assisted Motion Analysis for Quantitative Measurements of Swimming Behavior in Peritrichously Flagellated Bacteria. Anal. Biochem. 1996, 235, 20–25. [Google Scholar] [CrossRef]
- Alon, U.; Camarena, L.; Surette, M.G.; Aguera y Arcas, B.; Liu, Y.; Leibler, S.; Stock, J.B. Response Regulator Output in Bacterial Chemotaxis. EMBO J. 1998, 17, 4238–4248. [Google Scholar] [CrossRef] [PubMed]
- Schniederberend, M.; Abdurachim, K.; Murray, T.S.; Kazmierczak, B.I. The GTPase Activity of FlhF Is Dispensable for Flagellar Localization, but Not Motility, in Pseudomonas Aeruginosa. J. Bacteriol. 2013, 195, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Schniederberend, M.; Williams, J.F.; Shine, E.; Shen, C.; Jain, R.; Emonet, T.; Kazmierczak, B.I. Modulation of Flagellar Rotation in Surface-Attached Bacteria: A Pathway for Rapid Surface-Sensing after Flagellar Attachment. PLoS Pathog. 2019, 15, e1008149. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y. Rotation Measurements of Tethered Cells. Methods Mol. Biol. 2017, 1593, 163–174. [Google Scholar] [CrossRef]
- Dominick, C.N.; Wu, X.-L. Rotating Bacteria on Solid Surfaces without Tethering. Biophys. J. 2018, 115, 588–594. [Google Scholar] [CrossRef]
- Wang, F.; Yuan, J.; Berg, H.C. Switching Dynamics of the Bacterial Flagellar Motor near Zero Load. Proc. Natl. Acad. Sci. USA 2014, 111, 15752–15755. [Google Scholar] [CrossRef]
- Fahrner, K.A.; Ryu, W.S.; Berg, H.C. Bacterial Flagellar Switching under Load. Nature 2003, 423, 938. [Google Scholar] [CrossRef]
- Yuan, J.; Berg, H.C. Resurrection of the Flagellar Rotary Motor near Zero Load. Proc. Natl. Acad. Sci. USA 2008, 105, 1182–1185. [Google Scholar] [CrossRef]
- Yuan, J.; Fahrner, K.A.; Berg, H.C. Switching of the Bacterial Flagellar Motor Near Zero Load. J. Mol. Biol. 2009, 390, 394–400. [Google Scholar] [CrossRef]
- Qian, C.; Wong, C.C.; Swarup, S.; Chiam, K.-H. Bacterial Tethering Analysis Reveals a “Run-Reverse-Turn” Mechanism for Pseudomonas Species Motility. Appl. Environ. Microbiol. 2013, 79, 4734–4743. [Google Scholar] [CrossRef] [PubMed]
- Manson, M.D. Dynamic Motors for Bacterial Flagella. Proc. Natl. Acad. Sci. USA 2010, 107, 11151–11152. [Google Scholar] [CrossRef] [PubMed]
- Guyon, S.; Rhen, M.; Römling, U. Rapid Preparation of Unsheathed Bacterial Flagella. Bio-Protoc. 2015, 5, e1425. [Google Scholar] [CrossRef]
- Korobkova, E.A.; Emonet, T.; Park, H.; Cluzel, P. Hidden Stochastic Nature of a Single Bacterial Motor. Phys. Rev. Lett. 2006, 96, 058105. [Google Scholar] [CrossRef]
- Lele, P.P.; Hosu, B.G.; Berg, H.C. Dynamics of Mechanosensing in the Bacterial Flagellar Motor. Proc. Natl. Acad. Sci. USA 2013, 110, 11839–11844. [Google Scholar] [CrossRef]
- Johnston, R.R.M.; Spiro, M. Diffusion Coefficients of the Silver Ion and the Disulfitosilver (I) Ion by the Rotating Disk Method. J. Phys. Chem. 1967, 71, 3784–3790. [Google Scholar] [CrossRef]
- Mazouchi, A.; Milstein, J.N. Fast Optimized Cluster Algorithm for Localizations (FOCAL): A Spatial Cluster Analysis for Super-Resolved Microscopy. Bioinformatics 2016, 32, 747–754. [Google Scholar] [CrossRef]
- Zhang, M.; Chang, H.; Zhang, Y.; Yu, J.; Wu, L.; Ji, W.; Chen, J.; Liu, B.; Lu, J.; Liu, Y.; et al. Rational Design of True Monomeric and Bright Photoactivatable Fluorescent Proteins. Nat. Methods 2012, 9, 727–729. [Google Scholar] [CrossRef]
- Tang, H.; Blair, D.F. Regulated Underexpression of the FliM Protein of Escherichia coli and Evidence for a Location in the Flagellar Motor Distinct from the MotA/MotB Torque Generators. J. Bacteriol. 1995, 177, 3485–3495. [Google Scholar] [CrossRef]
- Berg, H.C.; Brown, D.A. Chemotaxis in Escherichia coli Analysed by Three-Dimensional Tracking. Nature 1972, 239, 500–504. [Google Scholar] [CrossRef]
- Del Tito, B.J.; Ward, J.M.; Hodgson, J.; Gershater, C.J.; Edwards, H.; Wysocki, L.A.; Watson, F.A.; Sathe, G.; Kane, J.F. Effects of a Minor Isoleucyl TRNA on Heterologous Protein Translation in Escherichia coli. J. Bacteriol. 1995, 177, 7086–7091. [Google Scholar] [CrossRef]
- Edelstein, A.; Amodaj, N.; Hoover, K.; Vale, R.; Stuurman, N. Computer Control of Microscopes Using uManager. Curr. Protoc. Mol. Biol. 2010, 92, 14.20.1–14.20.17. [Google Scholar] [CrossRef] [PubMed]
- Edelstein, A.D.; Tsuchida, M.A.; Amodaj, N.; Pinkard, H.; Vale, R.D.; Stuurman, N. Advanced Methods of Microscope Control Using ΜManager Software. J. Biol. Methods 2014, 1, e10. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Crocker, J.C.; Grier, D.G. Methods of Digital Video Microscopy for Colloidal Studies. J. Colloid. Interface Sci. 1996, 179, 298–310. [Google Scholar] [CrossRef]
- Stracy, M.; Lesterlin, C.; Garza de Leon, F.; Uphoff, S.; Zawadzki, P.; Kapanidis, A.N. Live-Cell Superresolution Microscopy Reveals the Organization of RNA Polymerase in the Bacterial Nucleoid. Proc. Natl. Acad. Sci. USA 2015, 112, E4390–E4399. [Google Scholar] [CrossRef]
- Allan, D.B.; Caswell, T.; Keim, N.C.; van der Wel, C.M. Trackpy: Trackpy V0.4.1. Zenodo; CERN: Geneva, Switzerland, 2018. [Google Scholar] [CrossRef]
- Da Silva, S.; Grosjean, L.; Ternan, N.; Mailley, P.; Livache, T.; Cosnier, S. Biotinylated Polypyrrole Films: An Easy Electrochemical Approach for the Reagentless Immobilization of Bacteria on Electrode Surfaces. Bioelectrochemistry 2004, 63, 297–301. [Google Scholar] [CrossRef]
- Kim, J.-S.; Kim, Y.J.; Seo, S.; Seong, M.-J.; Lee, K. Functional Role of Bdm during Flagella Biogenesis in Escherichia coli. Curr. Microbiol. 2015, 70, 369–373. [Google Scholar] [CrossRef]
- Francis, N.R.; Sosinsky, G.E.; Thomas, D.; DeRosier, D.J. Isolation, Characterization and Structure of Bacterial Flagellar Motors Containing the Switch Complex. J. Mol. Biol. 1994, 235, 1261–1270. [Google Scholar] [CrossRef]
- van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. scikit-image contributors Scikit-Image: Image Processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef] [PubMed]
- Patteson, A.E.; Gopinath, A.; Goulian, M.; Arratia, P.E. Running and Tumbling with E. coli in Polymeric Solutions. Sci. Rep. 2015, 5, 15761. [Google Scholar] [CrossRef] [PubMed]
- Cheong, F.C.; Wong, C.C.; Gao, Y.; Nai, M.H.; Cui, Y.; Park, S.; Kenney, L.J.; Lim, C.T. Rapid, High-Throughput Tracking of Bacterial Motility in 3D via Phase-Contrast Holographic Video Microscopy. Biophys. J. 2015, 108, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Persson, F.; Lindén, M.; Unoson, C.; Elf, J. Extracting Intracellular Diffusive States and Transition Rates from Single-Molecule Tracking Data. Nat. Methods 2013, 10, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Berg, H.C.E. Coli in Motion; Springer: New York, NY, USA, 2004; ISBN 978-0-387-21638-6. [Google Scholar]





| Assay | Replicate | Sample Size | |
|---|---|---|---|
| Swimming Assay | 5 | 0 µM, 0 h: 5906 | 40 µM, 0 h: 1872 |
| 0 µM, 1 h: 6579 | 40 µM, 1 h: 2921 | ||
| 0 µM, 2 h: 588 | 40 µM, 2 h: 2108 | ||
| 0 µM, 4 h: 1502 | 40 µM, 4 h: 2028 | ||
| Tethering Assay | 10 | 0 µM: 10 | 40 µM: 15 |
| Untethered Rotation Assay | 3 | 18 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russell, B.; Rogers, A.; Yoder, R.; Kurilich, M.; Krishnamurthi, V.R.; Chen, J.; Wang, Y. Silver Ions Inhibit Bacterial Movement and Stall Flagellar Motor. Int. J. Mol. Sci. 2023, 24, 11704. https://doi.org/10.3390/ijms241411704
Russell B, Rogers A, Yoder R, Kurilich M, Krishnamurthi VR, Chen J, Wang Y. Silver Ions Inhibit Bacterial Movement and Stall Flagellar Motor. International Journal of Molecular Sciences. 2023; 24(14):11704. https://doi.org/10.3390/ijms241411704
Chicago/Turabian StyleRussell, Benjamin, Ariel Rogers, Ryan Yoder, Matthew Kurilich, Venkata Rao Krishnamurthi, Jingyi Chen, and Yong Wang. 2023. "Silver Ions Inhibit Bacterial Movement and Stall Flagellar Motor" International Journal of Molecular Sciences 24, no. 14: 11704. https://doi.org/10.3390/ijms241411704
APA StyleRussell, B., Rogers, A., Yoder, R., Kurilich, M., Krishnamurthi, V. R., Chen, J., & Wang, Y. (2023). Silver Ions Inhibit Bacterial Movement and Stall Flagellar Motor. International Journal of Molecular Sciences, 24(14), 11704. https://doi.org/10.3390/ijms241411704





