A Pilot Study to Detect Viable Salmonella spp. in Diarrheal Stool Using Viability Real-Time PCR as a Culture-Independent Diagnostic Tool in a Clinical Setting
Abstract
1. Introduction
2. Results
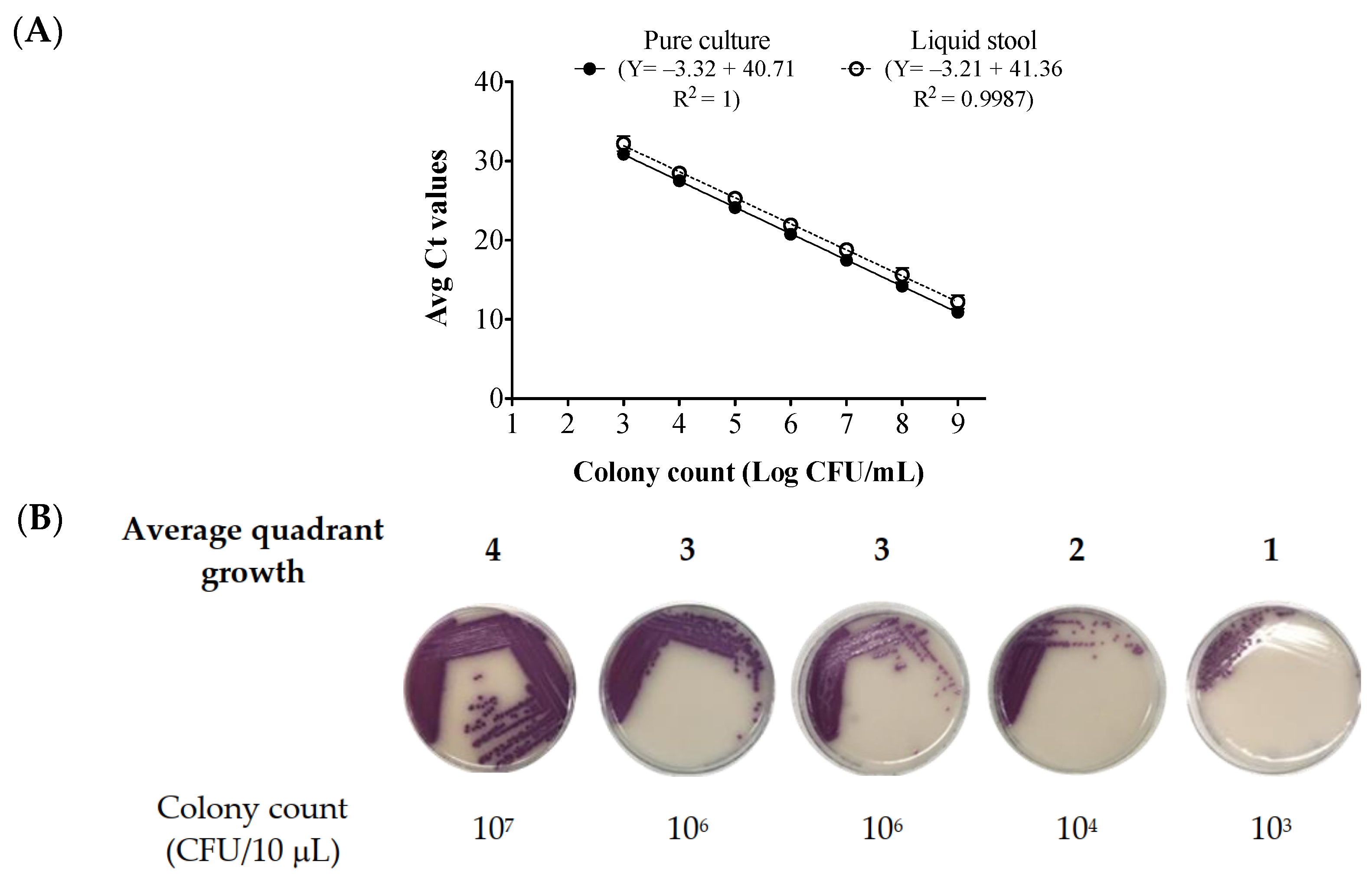
2.1. Standard Curve Generated from Bacteria Cells Spiked in Salmonella—Negative Stool Broth
2.2. Clinical Stool Analysis
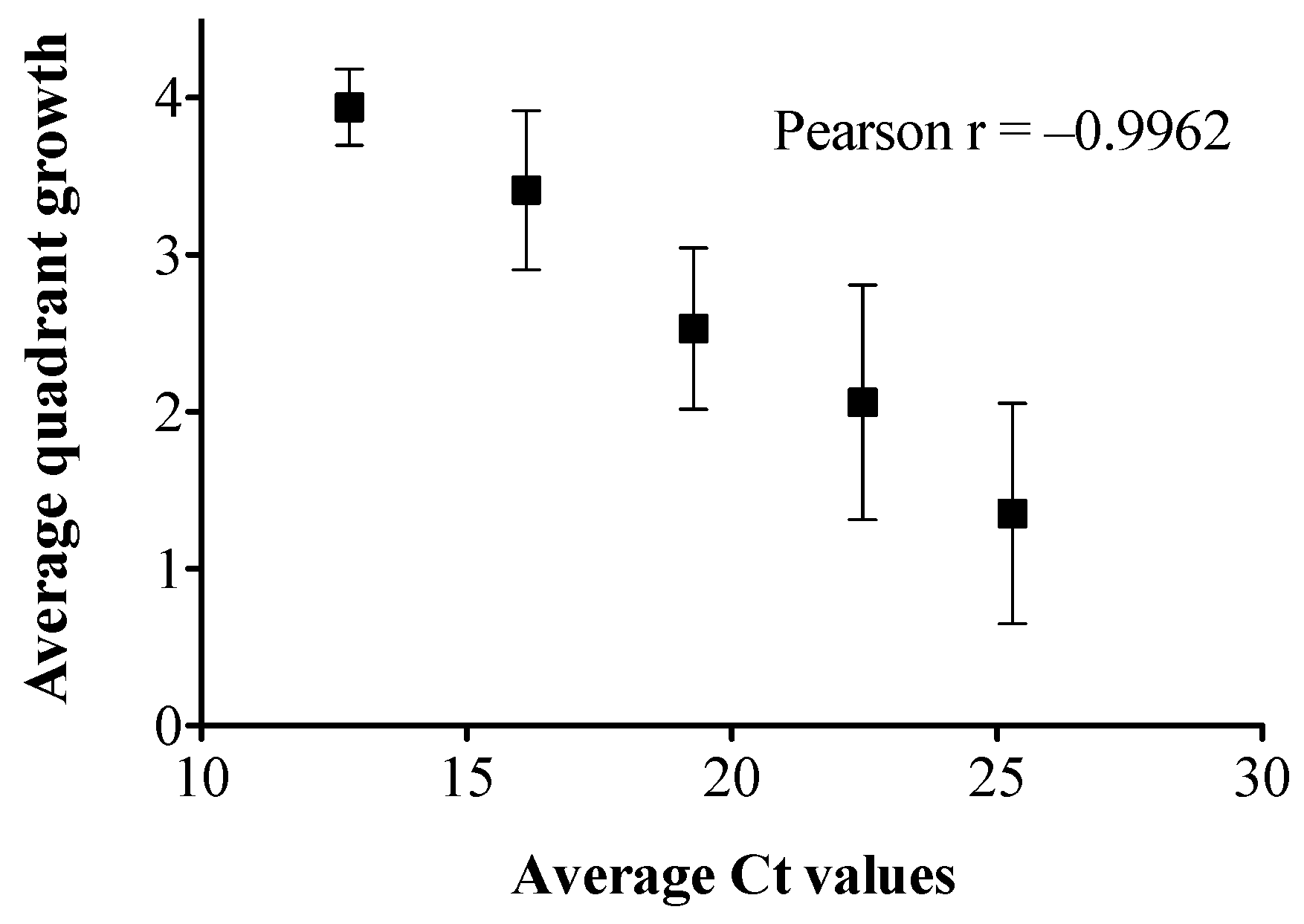
2.3. Colony Count Estimation in Enriched Clinical Stools Using the Stool Broth Standard Curve
3. Discussion
4. Materials and Methods
4.1. Standard Curve Construction
4.1.1. Bacterial Strains and Growth Conditions
4.1.2. Stool Spiking with S. Enteritidis
4.2. Nucleic Acid Extraction
4.3. qPCR and vPCR Assay Conditions
qPCR Assay
4.4. Clinical Stool Analysis
4.4.1. Clinical Stool Samples
4.4.2. PMAxx™ Preparation for vPCR Assay
4.4.3. qPCR and Viability Assessment via vPCR and Plate Growth
4.4.4. Enrichment of Stools with Low Bacterial Loads in Mannitol Selenite Broth
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomas, M.K.; Murray, R.; Flockhart, L.; Pintar, K.; Fazil, A.; Nesbitt, A.; Marshall, B.; Tataryn, J.; Pollari, F. Estimates of Foodborne Illness-Related Hospitalizations and Deaths in Canada for 30 Specified Pathogens and Unspecified Agents. Foodborne Pathog. Dis. 2015, 12, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Cronquist, A.B.; Mody, R.K.; Atkinson, R.; Besser, J.; D’Angelo, M.T.; Hurd, S.; Robinson, T.; Nicholson, C.; Mahon, B.E. Impacts of Culture-Independent Diagnostic Practices on Public Health Surveillance for Bacterial Enteric Pathogens. Clin. Infect. Dis. 2012, 54, S432–S439. [Google Scholar] [CrossRef] [PubMed]
- Berenger, B.; Chui, L.; Reimer, A.; Allen, V.; Alexander, D.; Domingo, M.-C.; Haldane, D.; Hoang, L.; Levett, P.; MacKeen, A.; et al. Canadian Public Health Laboratory Network Position Statement: Non-Culture Based Diagnostics for Gastroenteritis and Implications for Public Health Investigations. Can. Commun. Dis. Rep. 2017, 43, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Shea, S.; Kubota, K.A.; Maguire, H.; Gladbach, S.; Woron, A.; Atkinson-Dunn, R.; Couturier, M.R.; Miller, M.B. Clinical Microbiology Laboratories’ Adoption of Culture-Independent Diagnostic Tests Is a Threat to Foodborne-Disease Surveillance in the United States. J. Clin. Microbiol. 2017, 55, 10–15. [Google Scholar] [CrossRef]
- Ray, L.C.; Griffin, P.M.; Wymore, K.; Wilson, E.; Hurd, S.; Laclair, B.; Wozny, S.; Eikmeier, D.; Nicholson, C.; Burzlaff, K.; et al. Changing Diagnostic Testing Practices for Foodborne Pathogens, Foodborne Diseases Active Surveillance Network, 2012–2019. Open Forum Infect. Dis. 2022, 9, ofac344. [Google Scholar] [CrossRef]
- Imdad, A.; Retzer, F.; Thomas, L.S.; McMillian, M.; Garman, K.; Rebeiro, P.F.; Deppen, S.A.; Dunn, J.R.; Woron, A.M. Impact of Culture-Independent Diagnostic Testing on Recovery of Enteric Bacterial Infections. Clin. Infect. Dis. 2018, 66, 1892–1898. [Google Scholar] [CrossRef]
- Nocker, A.; Cheung, C.Y.; Camper, A.K. Comparison of Propidium Monoazide with Ethidium Monoazide for Differentiation of Live vs. Dead Bacteria by Selective Removal of DNA from Dead Cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef]
- Nogva, H.K.; Drømtorp, S.M.; Nissen, H.; Rudi, K. Ethidium Monoazide for DNA-Based Differentiation of Viable and Dead Bacteria by 5′-Nuclease PCR. Biotechniques 2003, 34, 804–813. [Google Scholar] [CrossRef]
- Nocker, A.; Camper, A.K. Selective Removal of DNA from Dead Cells of Mixed Bacterial Communities by Use of Ethidium Monoazide. Appl. Environ. Microbiol. 2006, 72, 1997–2004. [Google Scholar] [CrossRef]
- Nocker, A.; Sossa, K.E.; Camper, A.K. Molecular Monitoring of Disinfection Efficacy Using Propidium Monoazide in Combination with Quantitative PCR. J. Microbiol. Methods 2007, 70, 252–260. [Google Scholar] [CrossRef]
- Nocker, A.; Sossa-Fernandez, P.; Burr, M.D.; Camper, A.K. Use of Propidium Monoazide for Live/Dead Distinction in Microbial Ecology. Appl. Environ. Microbiol. 2007, 73, 5111–5117. [Google Scholar] [CrossRef]
- Fittipaldi, M.; Nocker, A.; Codony, F. Progress in Understanding Preferential Detection of Live Cells Using Viability Dyes in Combination with DNA Amplification. J. Microbiol. Methods 2012, 91, 276–289. [Google Scholar] [CrossRef]
- Nocker, A.; Camper, A.K. Novel Approaches toward Preferential Detection of Viable Cells Using Nucleic Acid Amplification Techniques. FEMS Microbiol. Lett. 2009, 291, 137–142. [Google Scholar] [CrossRef]
- Barbau-Piednoir, E.; Mahillon, J.; Pillyser, J.; Coucke, W.; Roosens, N.H.; Botteldoorn, N. Evaluation of Viability-QPCR Detection System on Viable and Dead Salmonella Serovar Enteritidis. J. Microbiol. Methods 2014, 103, 131–137. [Google Scholar] [CrossRef]
- Thilakarathna, S.H.; Stokowski, T.; Chui, L. An Improved Real-Time Viability PCR Assay to Detect Salmonella in a Culture-Independent Era. Int. J. Mol. Sci. 2022, 23, 14708. [Google Scholar] [CrossRef]
- Bae, S.; Wuertz, S. Discrimination of Viable and Dead Fecal Bacteroidales Bacteria by Quantitative PCR with Propidium Monoazide. Appl. Environ. Microbiol. 2009, 75, 2940–2944. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mustapha, A. Detection of Viable Escherichia Coli O157: H7 in Ground Beef by Propidium Monoazide Real-Time PCR. Int. J. Food Microbiol. 2014, 170, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Elizaquível, P.; Sánchez, G.; Aznar, R. Quantitative Detection of Viable Foodborne E. Coli O157:H7, Listeria Monocytogenes and Salmonella in Fresh-Cut Vegetables Combining Propidium Monoazide and Real-Time PCR. Food Control 2012, 25, 704–708. [Google Scholar] [CrossRef]
- Lee, A.S.; Lamanna, O.K.; Ishida, K.; Hill, E.; Nguyen, A.; Hsieh, M.H. A Novel Propidium Monoazide-Based PCR Assay Can Measure Viable Uropathogenic E. Coli In Vitro and In Vivo. Front. Cell. Infect. Microbiol. 2022, 12, 794323. [Google Scholar] [CrossRef]
- Desneux, J.; Chemaly, M.; Pourcher, A.M. Experimental Design for the Optimization of Propidium Monoazide Treatment to Quantify Viable and Non-Viable Bacteria in Piggery Effluents. BMC Microbiol. 2015, 15, 164. [Google Scholar] [CrossRef]
- Papanicolas, L.E.; Wang, Y.; Choo, J.M.; Gordon, D.L.; Wesselingh, S.L.; Rogers, G.B. Optimisation of a Propidium Monoazide Based Method to Determine the Viability of Microbes in Faecal Slurries for Transplantation. J. Microbiol. Methods 2019, 156, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Ishikawa, D.; Sasaki, T.; Lu, Y.J.; Kuwahara-Arai, K.; Kamei, M.; Shibuya, T.; Osada, T.; Hiramatsu, K.; Nagahara, A. Faecal Freezing Preservation Period Influences Colonization Ability for Faecal Microbiota Transplantation. J. Appl. Microbiol. 2019, 126, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hui, L.; Biao, D.; Xiaochang, H.; Hongyu, C.; Ru, Q.; Yichen, H.; Luyun, Z.; Zhenni, G.; Yuehua, Y.; et al. Systematic Evaluation of the Viable Microbiome in the Human Oral and Gut Samples with Spike-in Gram +/− Bacteria. Msystems 2023, 8, e0073822. [Google Scholar] [CrossRef] [PubMed]
- Loublier, C.; Taminiau, B.; Heinen, J.; Lecoq, L.; Amory, H.; Daube, G.; Cesarini, C. Evaluation of Bacterial Composition and Viability of Equine Feces after Processing for Transplantation. Microorganisms 2023, 11, 231. [Google Scholar] [CrossRef]
- Lewis, S.J.; Heaton, K.W. Stool Form Scale as a Useful Guide to Intestinal Transit Time. Scand. J. Gastroenterol. 1997, 32, 920–924. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s Microbes: Tools for Distinguishing the Living from the Dead in Microbial Ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Trevethan, R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef]
- Anderson, N.W.; Buchan, B.W.; Ledeboer, N.A. Comparison of the BD MAX Enteric Bacterial Panel to Routine Culture Methods for Detection of Campylobacter, Enterohemorrhagic Escherichia Coli (O157), Salmonella, and Shigella Isolates in Preserved Stool Specimens. J. Clin. Microbiol. 2014, 52, 1222–1224. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Oethinger, M.; Tuohy, M.J.; Hall, G.S.; Bauer, T.W. Improving Clinical Significance of PCR: Use of Propidium Monoazide to Distinguish Viable from Dead Staphylococcus Aureus and Staphylococcus Epidermidis. J. Orthop. Res. 2009, 27, 1243–1247. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.D.; Freydière, A.M. The Application of Chromogenic Media in Clinical Microbiology. J. Appl. Microbiol. 2007, 103, 2046–2055. [Google Scholar] [CrossRef] [PubMed]
- May, F.J.; Stafford, R.J.; Carroll, H.; Robson, J.M.; Vohra, R.; Nimmo, G.R.; Bates, J.; Kirk, M.D.; Fearnley, E.J.; Polkinghorne, B.G. The Effects of Culture Independent Diagnostic Testing on the Diagnosis and Reporting of Enteric Bacterial Pathogens in Queensland, 2010 to 2014. Commun. Dis. Intell. Q Rep. 2017, 41, E223–E230. [Google Scholar] [PubMed]
- Kellner, T.; Parsons, B.; Chui, L.; Berenger, B.M.; Xie, J.; Burnham, C.A.D.; Tarr, P.I.; Lee, B.E.; Nettel-Aguirre, A.; Szelewicki, J.; et al. Comparative Evaluation of Enteric Bacterial Culture and a Molecular Multiplex Syndromic Panel in Children with Acute Gastroenteritis. J. Clin. Microbiol. 2019, 57, e00205-19. [Google Scholar] [CrossRef] [PubMed]
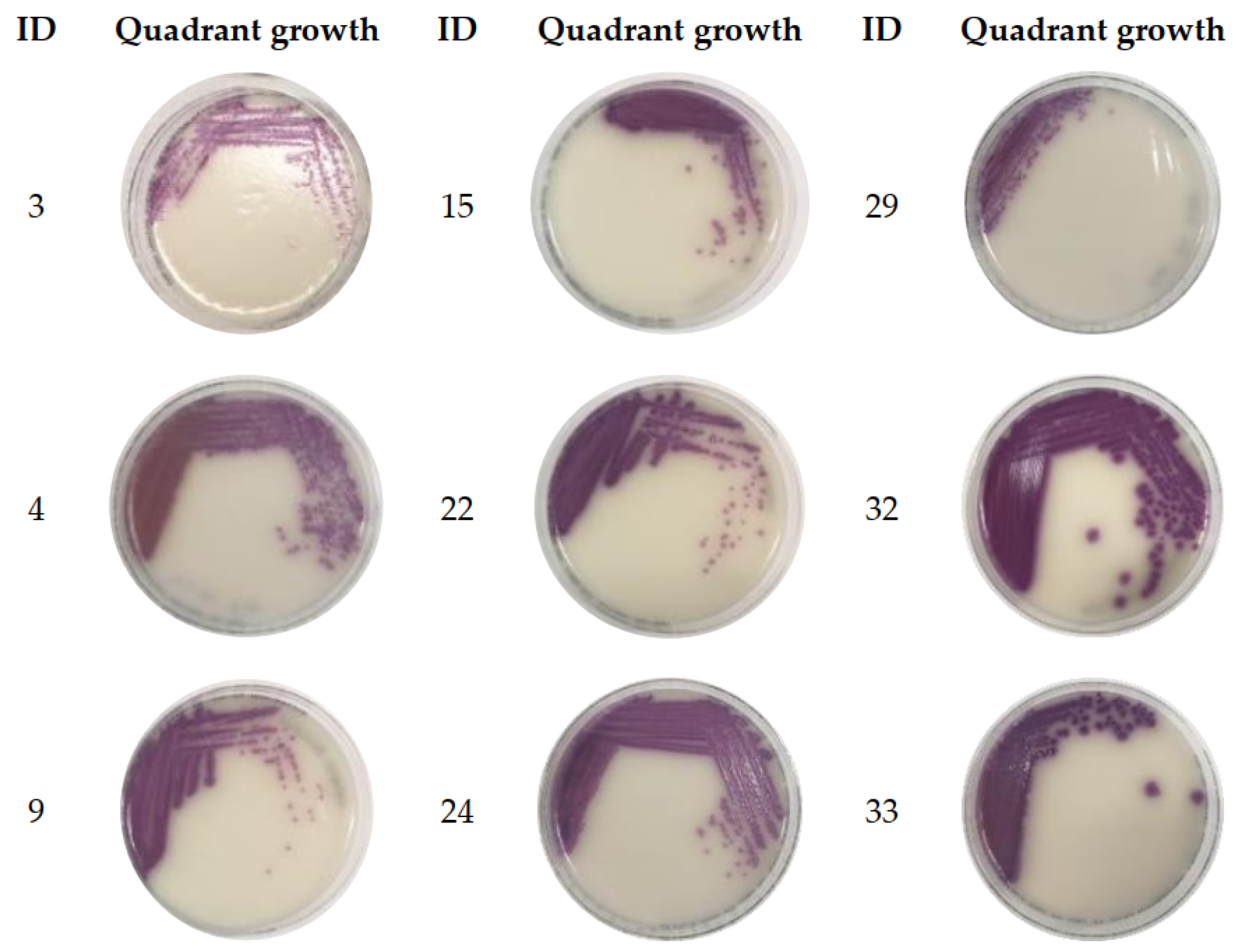
| ID | Pre-Enrichment Data | Post-Enrichment Data | |||
|---|---|---|---|---|---|
| qPCR (Ct Value) | vPCR (Ct Value) | Stool Culture | qPCR (Ct Value) | Quadrant Growth | |
| 3 | 34.48 | 36.34 | POS | 13.76 | 3 |
| 4 | 29.71 | 32.67 | NEG # | 16.20 | 3 |
| 9 | 30.53 | 33.08 | POS | 12.12 | 3 |
| 15 | 30.43 | 34.69 | POS | 12.16 | 3 |
| 22 | 31.36 | 33.73 | POS | 13.01 | 3 |
| 24 | 29.88 | 31.92 | POS | 12.15 | 4 |
| 29 | 27.39 | 30.70 | POS | 12.34 | 1 |
| 32 | 31.82 | 31.88 | POS | 12.53 | 4 |
| 33 | UD | 32.25 | POS | 13.20 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thilakarathna, S.H.; Chui, L. A Pilot Study to Detect Viable Salmonella spp. in Diarrheal Stool Using Viability Real-Time PCR as a Culture-Independent Diagnostic Tool in a Clinical Setting. Int. J. Mol. Sci. 2023, 24, 9979. https://doi.org/10.3390/ijms24129979
Thilakarathna SH, Chui L. A Pilot Study to Detect Viable Salmonella spp. in Diarrheal Stool Using Viability Real-Time PCR as a Culture-Independent Diagnostic Tool in a Clinical Setting. International Journal of Molecular Sciences. 2023; 24(12):9979. https://doi.org/10.3390/ijms24129979
Chicago/Turabian StyleThilakarathna, Surangi H., and Linda Chui. 2023. "A Pilot Study to Detect Viable Salmonella spp. in Diarrheal Stool Using Viability Real-Time PCR as a Culture-Independent Diagnostic Tool in a Clinical Setting" International Journal of Molecular Sciences 24, no. 12: 9979. https://doi.org/10.3390/ijms24129979
APA StyleThilakarathna, S. H., & Chui, L. (2023). A Pilot Study to Detect Viable Salmonella spp. in Diarrheal Stool Using Viability Real-Time PCR as a Culture-Independent Diagnostic Tool in a Clinical Setting. International Journal of Molecular Sciences, 24(12), 9979. https://doi.org/10.3390/ijms24129979






