A Novel Bioactive Peptide, T14, Selectively Activates mTORC1 Signalling: Therapeutic Implications for Neurodegeneration and Other Rapamycin-Sensitive Applications
Abstract
1. Introduction
2. Results
2.1. T14 and p-mTOR s2448 Correlate in Midbrain Samples from Alzheimer’s Disease Patients
2.2. T30 Increases the Expression of p-mTOR s2448 in Differentiated PC12 Cells and Ex Vivo Rat Brain Slices Containing Substantia Nigra
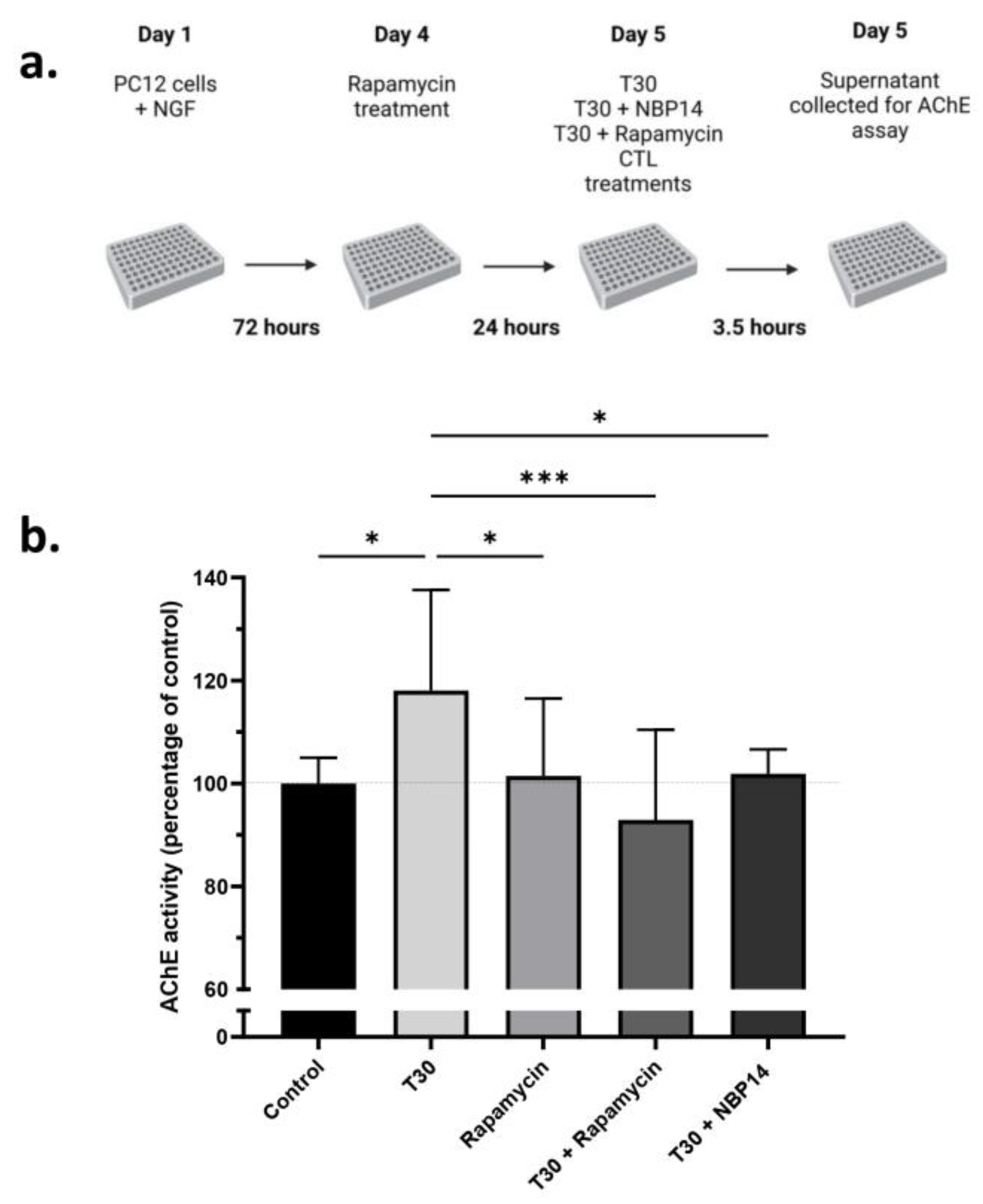
2.3. Rapamycin Attenuates the T30 Induced Increase in AChE Release by Differentiated PC12 Cells
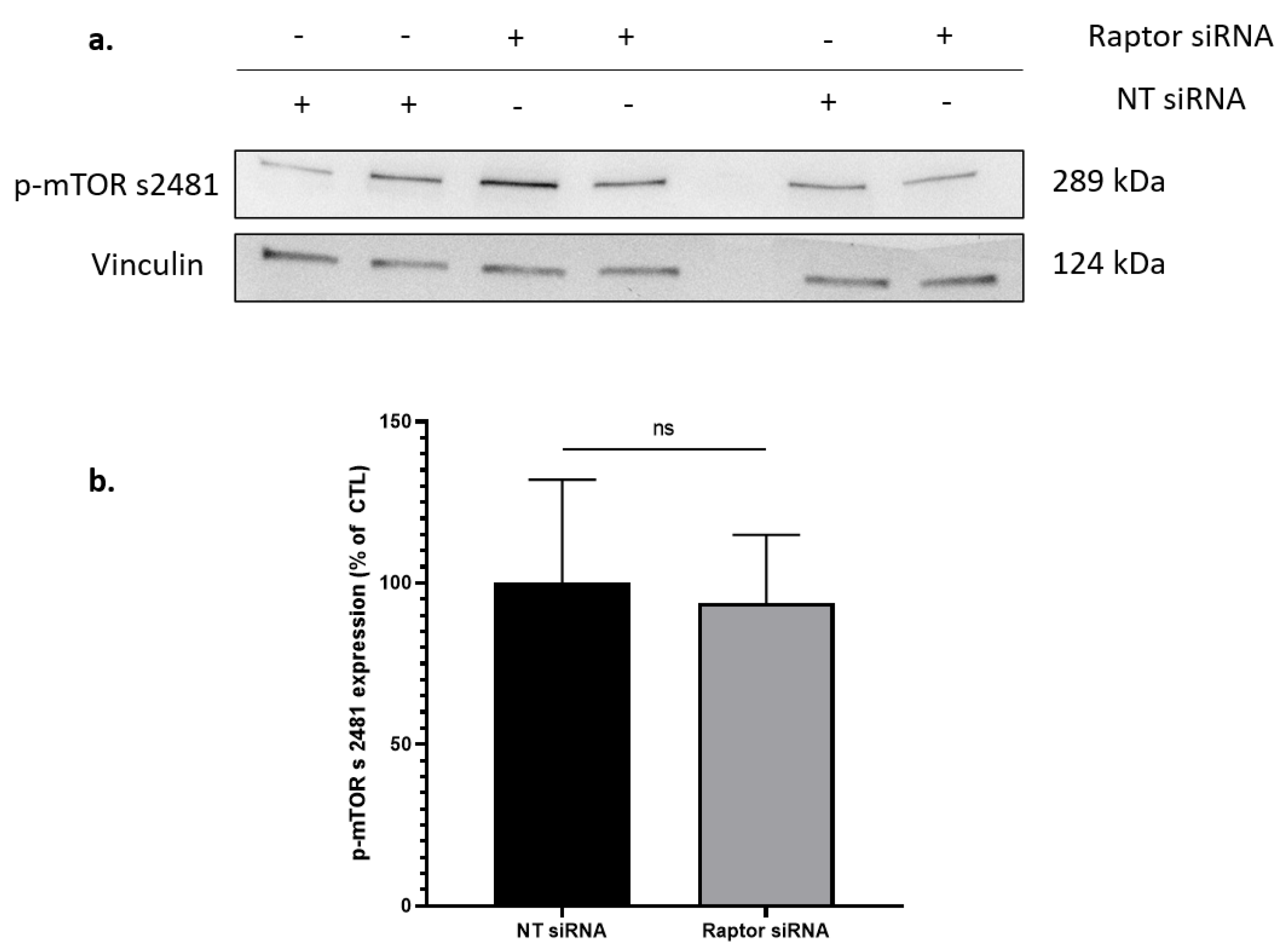
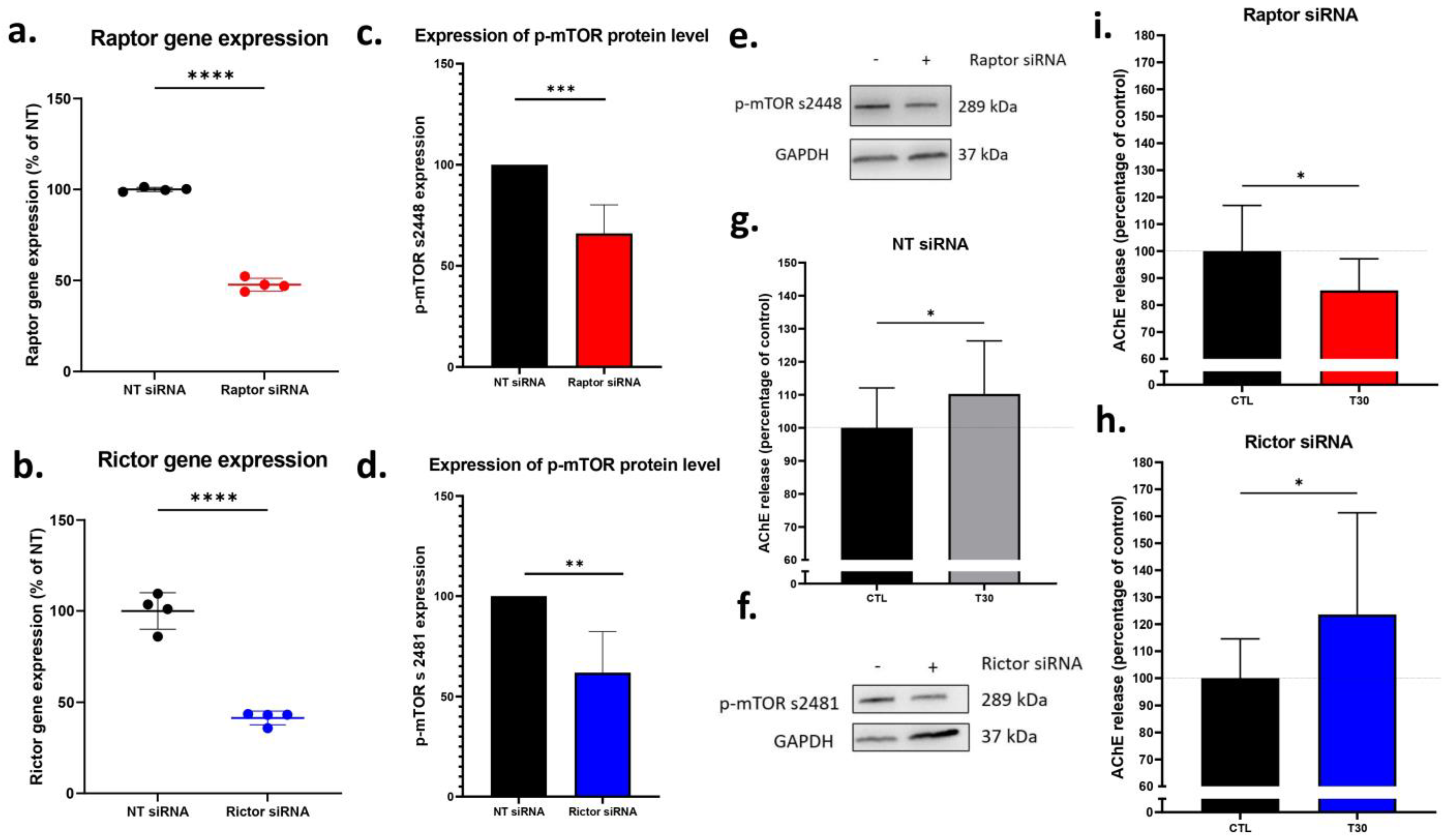
2.4. Silencing Raptor, but Not Rictor Blocked T30 Induced Increase in AChE Release by Undifferentiated PC12 Cells
2.5. Rapamycin Reduces the Number of Viable Undifferentiated PC12 Cells
3. Discussion
3.1. In-Vitro, Ex Vivo and Post-Mortem Preparations
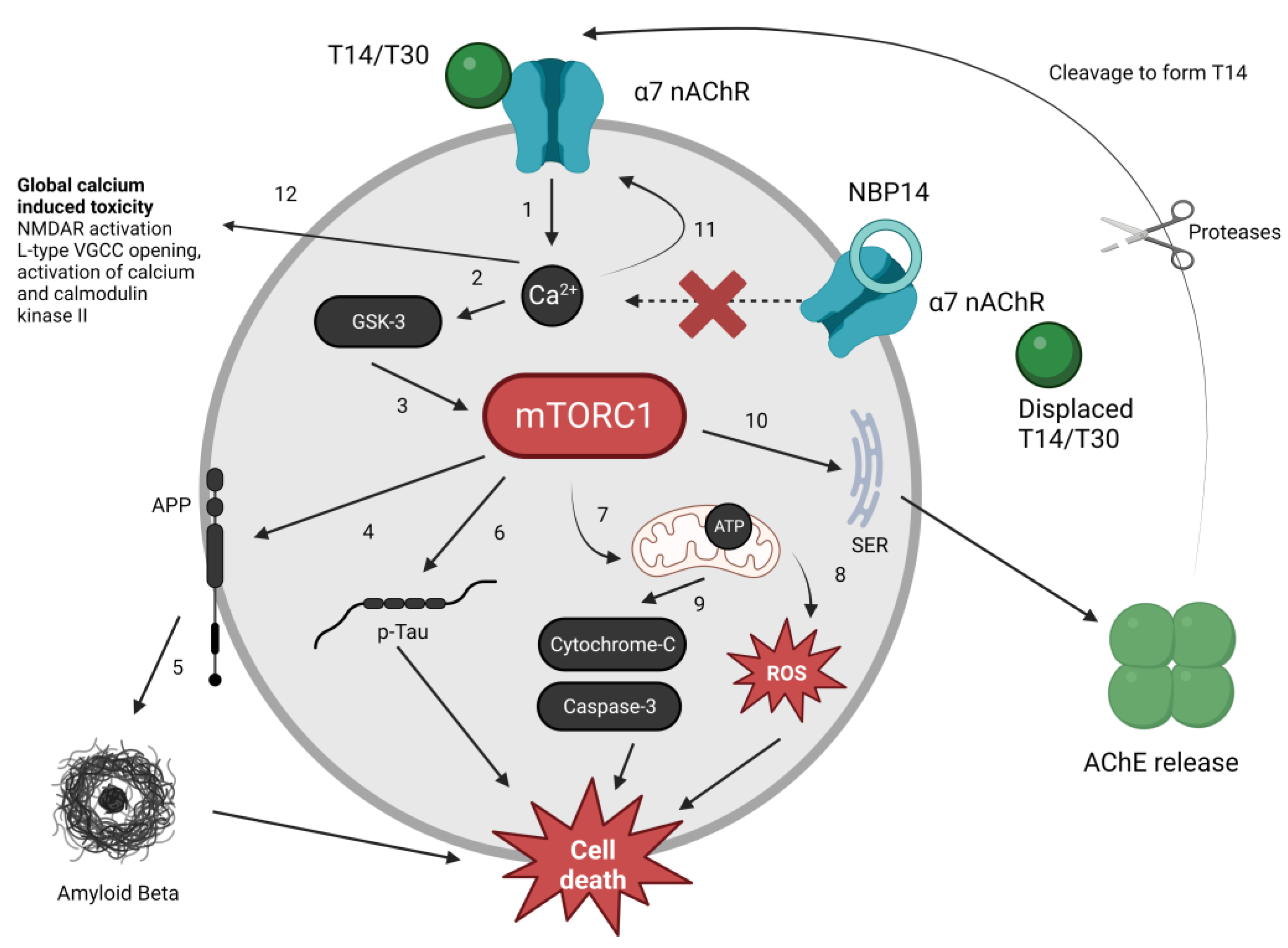
3.2. The mTORC1 Pathway and the T14 Signalling System
4. Materials and Methods
4.1. PC12 Cell Culture and Reagents
4.2. Ex Vivo Brain Slices
4.3. Cell Viability Assay
4.4. AChE Release Assay
4.5. siRNA Transfection
4.6. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.7. Human Clinical Samples
4.8. Western Blotting
4.9. Statistical Analysis
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


References
- Greenfield, S.; Vaux, D.J. Parkinson’s disease, Alzheimer’s disease and motor neurone disease: Identifying a common mechanism. Neuroscience 2002, 113, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Day, T.; Greenfield, S.A. Bioactivity of a peptide derived from acetylcholinesterase in hippocampal organotypic cultures. Exp. Brain Res. 2004, 155, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, S.A.; Day, T.; Mann, E.O.; Bermudez, I. A novel peptide modulates alpha7 nicotinic receptor responses: Implications for a possible trophic-toxic mechanism within the brain. J. Neurochem. 2004, 90, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Day, T.; Greenfield, S.A. A peptide derived from acetylcholinesterase induces neuronal cell death: Characterisation of possible mechanisms. Exp. Brain Res. 2003, 153, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Bond, C.E.; Zimmermann, M.; Greenfield, S.A. Upregulation of α7 Nicotinic Receptors by Acetylcholinesterase C-Terminal Peptides. PLoS ONE 2009, 4, e4846. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ratés, S.; Lewis, M.; Worrall, R.; Greenfield, S. Additive Toxicity of β-Amyloid by a Novel Bioactive Peptide In Vitro: Possible Implications for Alzheimer’s Disease. PLoS ONE 2013, 8, e54864. [Google Scholar] [CrossRef]
- Garcia-Ratés, S.; Morrill, P.; Tu, H.; Pottiez, G.; Badin, A.-S.; Tormo-Garcia, C.; Heffner, C.; Coen, C.W.; Greenfield, S.A. (I) Pharmacological profiling of a novel modulator of the α7 nicotinic receptor: Blockade of a toxic acetylcholinesterase-derived peptide increased in Alzheimer brains. Neuropharmacology 2016, 105, 487–499. [Google Scholar] [CrossRef]
- Greenfield, S.A.; Cole, G.M.; Coen, C.W.; Frautschy, S.; Singh, R.P.; Mekkittikul, M.; Garcia-Ratés, S.; Morrill, P.; Hollings, O.; Passmore, M.; et al. A novel process driving Alzheimer’s disease validated in a mouse model: Therapeutic potential. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2022, 8, e12274. [Google Scholar] [CrossRef]
- Hasan, S.; Ahmed, M.; Garcia-Ratés, S.; Greenfield, S. Antagonising a novel toxin “T14” in Alzheimer’s disease: Comparison of receptor blocker versus antibody effects in vitro. Biomed. Pharmacother. 2023, 158, 114120. [Google Scholar] [CrossRef]
- Greenfield, S.A.; Ferrati, G.; Coen, C.W.; Vadisiute, A.; Molnár, Z.; Garcia-Rates, S.; Frautschy, S.; Cole, G.M. Characterization of a Bioactive Peptide T14 in the Human and Rodent Substantia Nigra: Implications for Neurodegenerative Disease. Int. J. Mol. Sci. 2022, 23, 13119. [Google Scholar] [CrossRef]
- Brai, E.; Simon, F.; Cogoni, A.; Greenfield, S.A. Modulatory Effects of a Novel Cyclized Peptide in Reducing the Expression of Markers Linked to Alzheimer’s Disease. Front. Neurosci. 2018, 12, 362. [Google Scholar] [CrossRef] [PubMed]
- Onganer, P.U.; Djamgoz, M.B.A.; Whyte, K.; Greenfield, S.A. An acetylcholinesterase-derived peptide inhibits endocytic membrane activity in a human metastatic breast cancer cell line. Biochim. Biophys. Acta BBA—Gen. Subj. 2006, 1760, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Pepper, C.; Tu, H.; Morrill, P.; Garcia-Rates, S.; Fegan, C.; Greenfield, S. Tumor cell migration is inhibited by a novel therapeutic strategy antagonizing the alpha-7 receptor. Oncotarget 2017, 8, 11414–11424. [Google Scholar] [CrossRef]
- Garcia-Ratés, S.; Greenfield, S. Cancer and neurodegeneration: Two sides, same coin? Oncotarget 2017, 8, 22307–22308. [Google Scholar] [CrossRef]
- Tan, V.P.; Miyamoto, S. Nutrient-sensing mTORC1, Integration of metabolic and autophagic signals. J. Mol. Cell. Cardiol. 2016, 95, 31–41. [Google Scholar] [CrossRef]
- Tokunaga, C.; Yoshino, K.; Yonezawa, K. mTOR integrates amino acid- and energy-sensing pathways. Biochem. Biophys. Res. Commun. 2004, 313, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Ben-Sahra, I.; Manning, B.D. mTORC1 signaling and the metabolic control of cell growth. Curr. Opin. Cell. Biol. 2017, 45, 72–82. [Google Scholar] [CrossRef]
- Wang, X.; Proud, C.G. The mTOR Pathway in the Control of Protein Synthesis. Physiology 2006, 21, 362–369. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Yuan, H.-X.; Guan, K.-L. Structural insights of mTOR complex 1. Cell Res. 2016, 26, 267–268. [Google Scholar] [CrossRef]
- Szwed, A.; Kim, E.; Jacinto, E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 2021, 101, 1371–1426. [Google Scholar] [CrossRef] [PubMed]
- Tramutola, A.; Triplett, J.C.; Di Domenico, F.; Niedowicz, D.M.; Murphy, M.P.; Coccia, R.; Perluigi, M.; Butterfield, D.A. Alteration of mTOR signaling occurs early in the progression of Alzheimer disease (AD): Analysis of brain from subjects with pre-clinical AD, amnestic mild cognitive impairment and late-stage AD. J. Neurochem. 2015, 133, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Majd, S.; Power, J.; Majd, Z. Alzheimer’s Disease and Cancer: When Two Monsters Cannot Be Together. Front. Neurosci. 2019, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz López, K.G.; Toledo Guzmán, M.E.; Sánchez, E.O.; García Carrancá, A. mTORC1 as a Regulator of Mitochondrial Functions and a Therapeutic Target in Cancer. Front. Oncol. 2019, 9, 1373. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-X.; Ji, X.; Mao, X.; Xie, L.; Jia, J.; Galvan, V.; Greenberg, D.A.; Jin, K. Differential Activation of mTOR Complex 1 Signaling in Human Brain with Mild to Severe Alzheimer’s Disease. J. Alzheimers Dis. 2013, 38, 437–444. [Google Scholar] [CrossRef]
- Graur, A.; Kabbani, N. The Human Acetylcholinesterase c-Terminal T30 Peptide Activates Neural Growth through an Alpha 7 Nicotinic Acetylcholine Receptor mTOR Pathway. bioRxiv 2023. Available online: http://biorxiv.org/content/early/2023/04/07/2023.04.07.536081.abstract (accessed on 25 April 2023).
- Li, J.; Kim, S.G.; Blenis, J. Rapamycin: One Drug, Many Effects. Cell Metab. 2014, 19, 373–379. [Google Scholar] [CrossRef]
- Fletcher, L.; Evans, T.M.; Watts, L.T.; Jimenez, D.F.; Digicaylioglu, M. Rapamycin Treatment Improves Neuron Viability in an In Vitro Model of Stroke. PLoS ONE 2013, 8, e68281. [Google Scholar] [CrossRef]
- Copp, J.; Manning, G.; Hunter, T. TORC-Specific Phosphorylation of Mammalian Target of Rapamycin (mTOR): Phospho-Ser2481 Is a Marker for Intact mTOR Signaling Complex 2. Cancer Res. 2009, 69, 1821–1827. [Google Scholar] [CrossRef]
- Verhave, J.; Boucher, A.; Dandavino, R.; Collette, S.; Senécal, L.; Hebert, M.-J.; Girardin, C.; Cardinal, H. The incidence, management, and evolution of rapamycin-related side effects in kidney transplant recipients. Clin. Transplant. 2014, 28, 616–622. [Google Scholar] [CrossRef]
- Soefje, S.A.; Karnad, A.; Brenner, A.J. Common toxicities of mammalian target of rapamycin inhibitors. Target. Oncol. 2011, 6, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Carosi, J.M.; Sargeant, T.J. Rapamycin and Alzheimer disease: A double-edged sword? Autophagy 2019, 15, 1460–1462. [Google Scholar] [CrossRef] [PubMed]
- Dumas, S.N.; Lamming, D.W. Next Generation Strategies for Geroprotection via mTORC1 Inhibition. J. Gerontol. Ser. A 2020, 75, 14–23. [Google Scholar] [CrossRef]
- Adebayo Michael, A.O.; Ko, S.; Tao, J.; Moghe, A.; Yang, H.; Xu, M.; Russell, J.O.; Pradhan-Sundd, T.; Liu, S.; Singh, S.; et al. Inhibiting Glutamine-Dependent mTORC1 Activation Ameliorates Liver Cancers Driven by β-Catenin Mutations. Cell Metab. 2019, 29, 1135–1150.e6. [Google Scholar] [CrossRef] [PubMed]
- Rosner, M.; Siegel, N.; Valli, A.; Fuchs, C.; Hengstschläger, M. mTOR phosphorylated at S2448 binds to raptor and rictor. Amino Acids 2010, 38, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Sengupta, S.; Sheen, J.H.; Hsu, P.P.; Bagley, A.F.; Markhard, A.L.; Sabatini, D.M. Prolonged Rapamycin Treatment Inhibits mTORC2 Assembly, A.k.t./.P.K.B. Mol. Cell 2006, 22, 159–168. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a Novel Binding Partner of mTOR, Defines a Rapamycin-Insensitive and Raptor-Independent Pathway that Regulates the Cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef]
- Westerink, R.H.S.; Ewing, A.G. The PC12 cell as model for neurosecretion. Acta Physiol. 2007, 192, 273–285. [Google Scholar] [CrossRef]
- Teng, K.; Angelastro, J.; Cunningham, M.; Greene, L. Cultured PC12 Cells A Model for Neuronal Function, Differentiation, and Survival. In Cell Biology; Elsevier: Amsterdam, The Netherlands, 2006; pp. 171–176. [Google Scholar]
- Youdim, M.B.H. PC12 cells as a window for the differentiation of neural crest into adrenergic nerve ending and adrenal medulla. In Recent Advances in Neuropharmacology; Springer: Vienna, Austria, 1991; pp. 61–67. [Google Scholar]
- Melega, W.P.; Howard, B.D. Choline and acetylcholine metabolism in PC12 secretory cells. Biochemistry 1981, 20, 4477–4483. [Google Scholar] [CrossRef]
- Wiatrak, B.; Kubis-Kubiak, A.; Piwowar, A.; Barg, E. PC12 Cell Line: Cell Types, Coating of Culture Vessels, Differentiation and Other Culture Conditions. Cells 2020, 9, 958. [Google Scholar] [CrossRef]
- Izumiyama, N.; Asami, E.; Itoh, Y.; Ohtsubo, K. Alzheimer’s neurofibrillary tangles and paired helical filaments in the pheochromocytoma cells of the adrenal medulla ?Electron microscopic and immunoelectron microscopic observations. Acta Neuropathol. 1990, 81, 213–216. [Google Scholar] [CrossRef]
- Parker, E.M.; Monopoli, A.; Ongini, E.; Lozza, G.; Babij, C.M. Rapamycin, but not FK506 and GPI-1046, increases neurite outgrowth in PC12 cells by inhibiting cell cycle progression. Neuropharmacology 2000, 39, 1913–1919. [Google Scholar] [CrossRef] [PubMed]
- Ferrati, G.; Brai, E.; Stuart, S.; Marino, C.; Greenfield, S. A Multidisciplinary Approach Reveals an Age-Dependent Expression of a Novel Bioactive Peptide, Already Involved in Neurodegeneration, in the Postnatal Rat Forebrain. Brain Sci. 2018, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, M.; Nakamura, A.; Ebashi, M.; Hirokawa, K.; Takahashi, R.; Uchihara, T. Brainstem tau pathology in Alzheimer’s disease is characterized by increase of three repeat tau and independent of amyloid β. Acta Neuropathol. Commun. 2018, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Woolf, N.J. Global and serial neurons form a hierarchically arranged interface proposed to underlie memory and cognition. Neuroscience 1996, 74, 625–651. [Google Scholar] [CrossRef]
- Garcia-Ratés, S.; Greenfield, S. When a trophic process turns toxic: Alzheimer’s disease as an aberrant recapitulation of a developmental mechanism. Int. J. Biochem. Cell Biol. 2022, 149, 106260. [Google Scholar] [CrossRef]
- Kosciuczuk, E.M.; Saleiro, D.; Platanias, L.C. Dual targeting of eIF4E by blocking MNK and mTOR pathways in leukemia. Cytokine 2017, 89, 116–121. [Google Scholar] [CrossRef]
- Majeed, S.T.; Batool, A.; Majeed, R.; Bhat, N.N.; Zargar, M.A.; Andrabi, K.I. mTORC1 induces eukaryotic translation initiation factor 4E interaction with TOS-S6 kinase 1 and its activation. Cell Cycle 2021, 20, 839–854. [Google Scholar] [CrossRef]
- Deleyto-Seldas, N.; Efeyan, A. The mTOR–Autophagy Axis and the Control of Metabolism. Front. Cell Dev. Biol. 2021, 9, 655731. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Galvan, V. Rapamycin and Alzheimer’s disease: Time for a clinical trial? Sci. Transl. Med. 2019, 11, eaar4289. [Google Scholar] [CrossRef]
- Paplomata, E.; Zelnak, A.; O’Regan, R. Everolimus: Side effect profile and management of toxicities in breast cancer. Breast Cancer Res. Treat. 2013, 140, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Reho, J.J.; Guo, D.-F.; Morgan, D.A.; Rahmouni, K. mTORC1 (Mechanistic Target of Rapamycin Complex 1) Signaling in Endothelial and Smooth Muscle Cells Is Required for Vascular Function. Hypertension 2021, 77, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Hahn, D.; Hodson, E.M.; Hamiwka, L.A.; Lee, V.W.; Chapman, J.R.; Craig, J.C.; Webster, A.C. Target of rapamycin inhibitors (TOR-I; sirolimus and everolimus) for primary immunosuppression in kidney transplant recipients. Cochrane Database Syst. Rev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Selvarani, R.; Mohammed, S.; Richardson, A. Effect of rapamycin on aging and age-related diseases—Past and future. Geroscience 2021, 43, 1135–1158. [Google Scholar] [CrossRef]
- Evangelisti, C.; Chiarini, F.; Paganelli, F.; Marmiroli, S.; Martelli, A.M. Crosstalks of GSK3 signaling with the mTOR network and effects on targeted therapy of cancer. Biochim. Biophys. Acta BBA—Mol. Cell Res. 2020, 1867, 118635. [Google Scholar] [CrossRef]
- Cai, Z.; Zhou, Y.; Xiao, M.; Yan, L.-J.; He, W. Activation of mTOR: A culprit of Alzheimer’s disease? Neuropsychiatr. Dis. Treat. 2015, 11, 1015–1030. [Google Scholar] [CrossRef]
- Tang, Z.; Bereczki, E.; Zhang, H.; Wang, S.; Li, C.; Ji, X.; Branca, R.; Lehtiö, J.; Guan, Z.; Filipcik, P.; et al. Mammalian Target of Rapamycin (mTor) Mediates Tau Protein Dyshomeostasis. J. Biol. Chem. 2013, 288, 15556–15570. [Google Scholar] [CrossRef]
- Jean, L.; Thomas, B.; Tahiri-Alaoui, A.; Shaw, M.; Vaux, D.J. Heterologous Amyloid Seeding: Revisiting the Role of Acetylcholinesterase in Alzheimer’s Disease. PLoS ONE 2007, 2, e652. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates, 6th ed.; Academic Press: San Diego, CA, USA, 2006. [Google Scholar]



| Gene | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) | Product Size (bp) |
|---|---|---|---|
| Raptor | ACGGTGAATGGAGAGGTCTG | CAGCATTGGAGCAGTCGTAG | 124 |
| Rictor | TGCTTCCTTGTTTCCGAGTT | GCTACCACCTCTGGGTTCTG | 138 |
| GAPDH | GGGCTCTCTGCTCCTCCCTGT | CAGGCGTCCGATACGGCCAAA | 119 |
| Beta-actin | CCACACCCGCCACCAGTTCG | TACAGCCCGGGGAGCATCGT | 112 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranglani, S.; Ashton, A.; Mahfooz, K.; Komorowska, J.; Graur, A.; Kabbani, N.; Garcia-Rates, S.; Greenfield, S. A Novel Bioactive Peptide, T14, Selectively Activates mTORC1 Signalling: Therapeutic Implications for Neurodegeneration and Other Rapamycin-Sensitive Applications. Int. J. Mol. Sci. 2023, 24, 9961. https://doi.org/10.3390/ijms24129961
Ranglani S, Ashton A, Mahfooz K, Komorowska J, Graur A, Kabbani N, Garcia-Rates S, Greenfield S. A Novel Bioactive Peptide, T14, Selectively Activates mTORC1 Signalling: Therapeutic Implications for Neurodegeneration and Other Rapamycin-Sensitive Applications. International Journal of Molecular Sciences. 2023; 24(12):9961. https://doi.org/10.3390/ijms24129961
Chicago/Turabian StyleRanglani, Sanskar, Anna Ashton, Kashif Mahfooz, Joanna Komorowska, Alexandru Graur, Nadine Kabbani, Sara Garcia-Rates, and Susan Greenfield. 2023. "A Novel Bioactive Peptide, T14, Selectively Activates mTORC1 Signalling: Therapeutic Implications for Neurodegeneration and Other Rapamycin-Sensitive Applications" International Journal of Molecular Sciences 24, no. 12: 9961. https://doi.org/10.3390/ijms24129961
APA StyleRanglani, S., Ashton, A., Mahfooz, K., Komorowska, J., Graur, A., Kabbani, N., Garcia-Rates, S., & Greenfield, S. (2023). A Novel Bioactive Peptide, T14, Selectively Activates mTORC1 Signalling: Therapeutic Implications for Neurodegeneration and Other Rapamycin-Sensitive Applications. International Journal of Molecular Sciences, 24(12), 9961. https://doi.org/10.3390/ijms24129961






