Bioactivity Profiling and Quantification of Gastrodin in Gastrodia elata Cultivated in the Field versus Facility via Hyphenated High-Performance Thin-Layer Chromatography
Abstract
1. Introduction
2. Results
2.1. Origin, Harvest, and Post-Harvest Processing of Samples
2.2. Selection of the Extractant
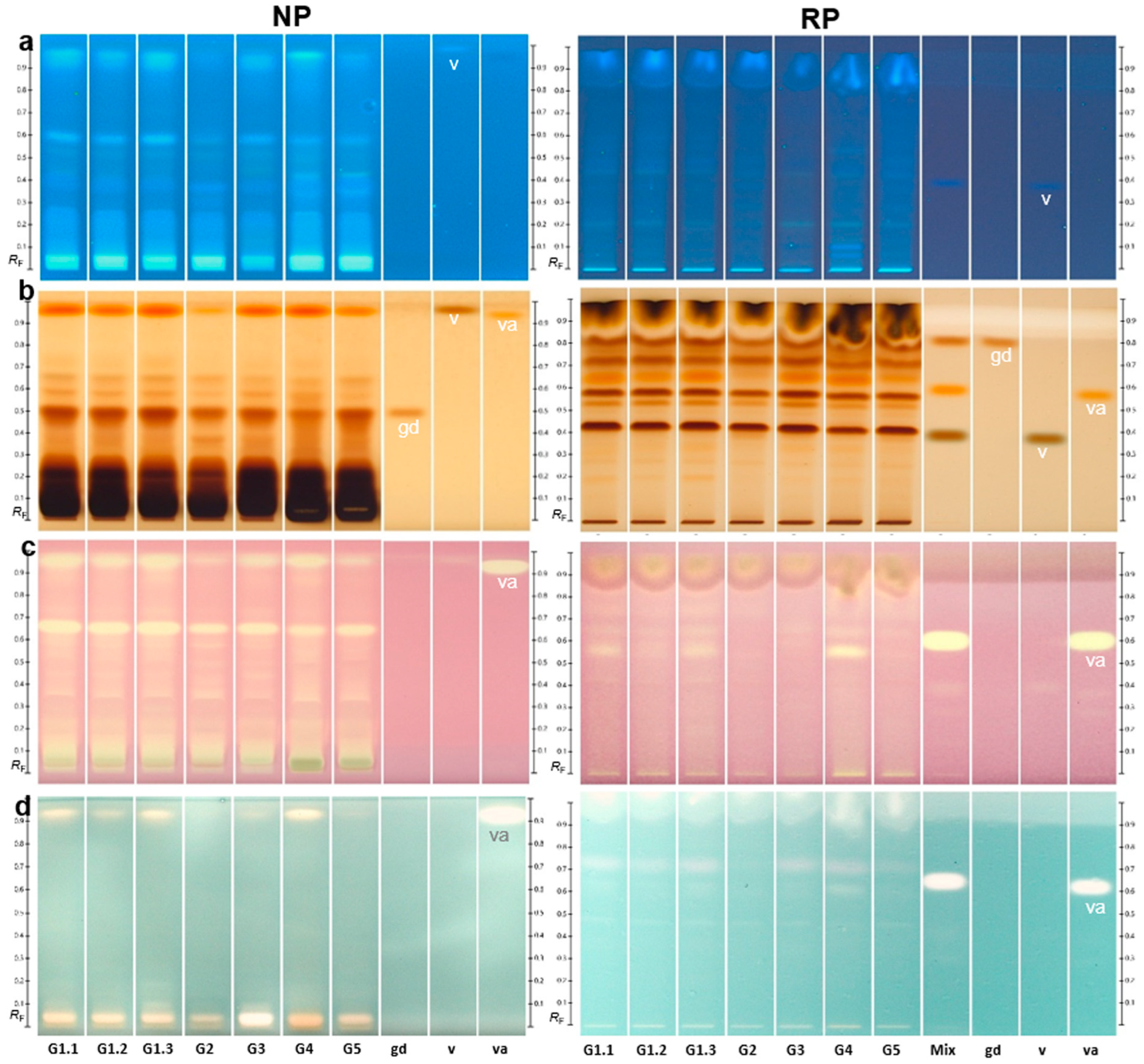
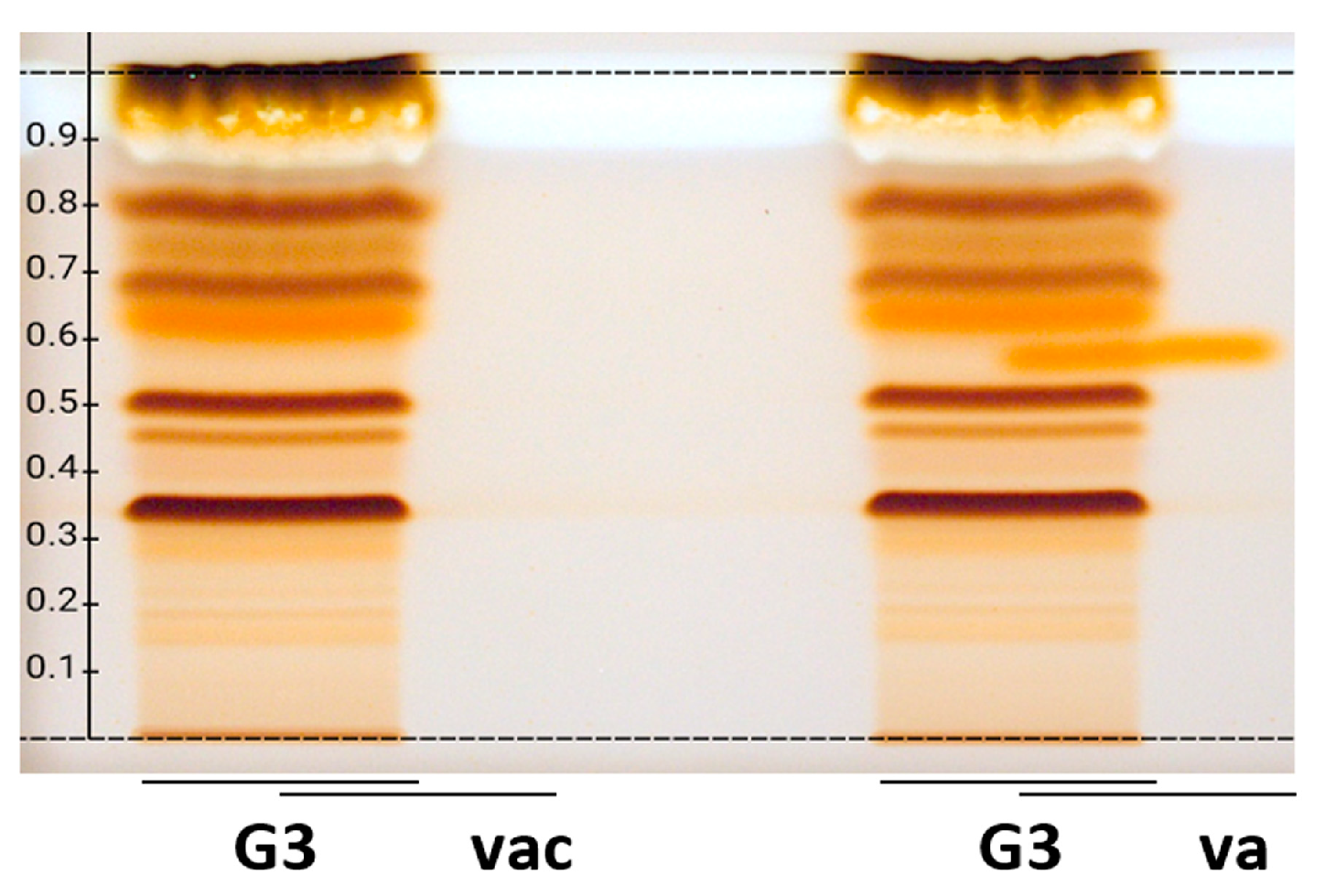
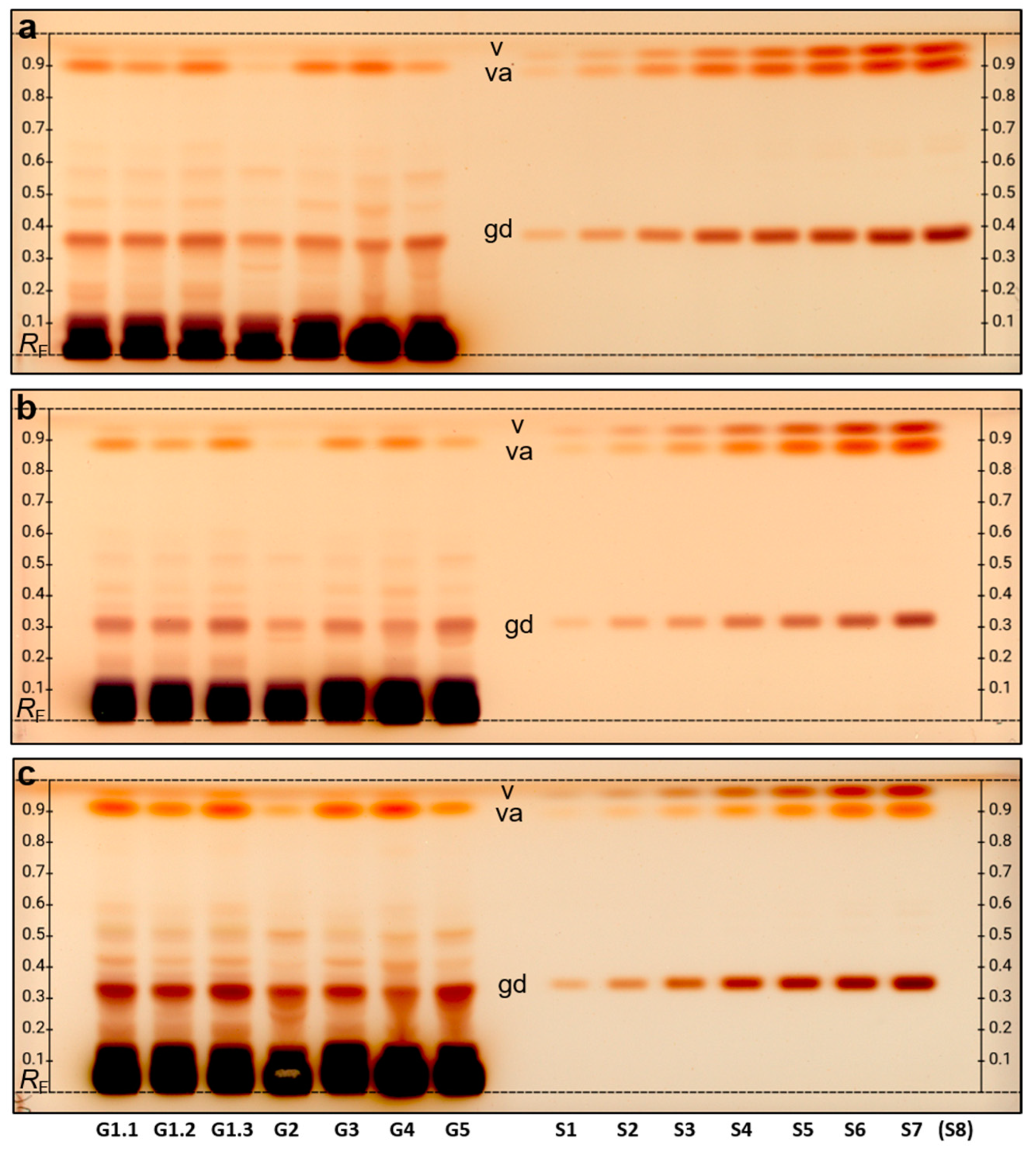
2.3. Comparative Chemical and Effect-Directed Profiling
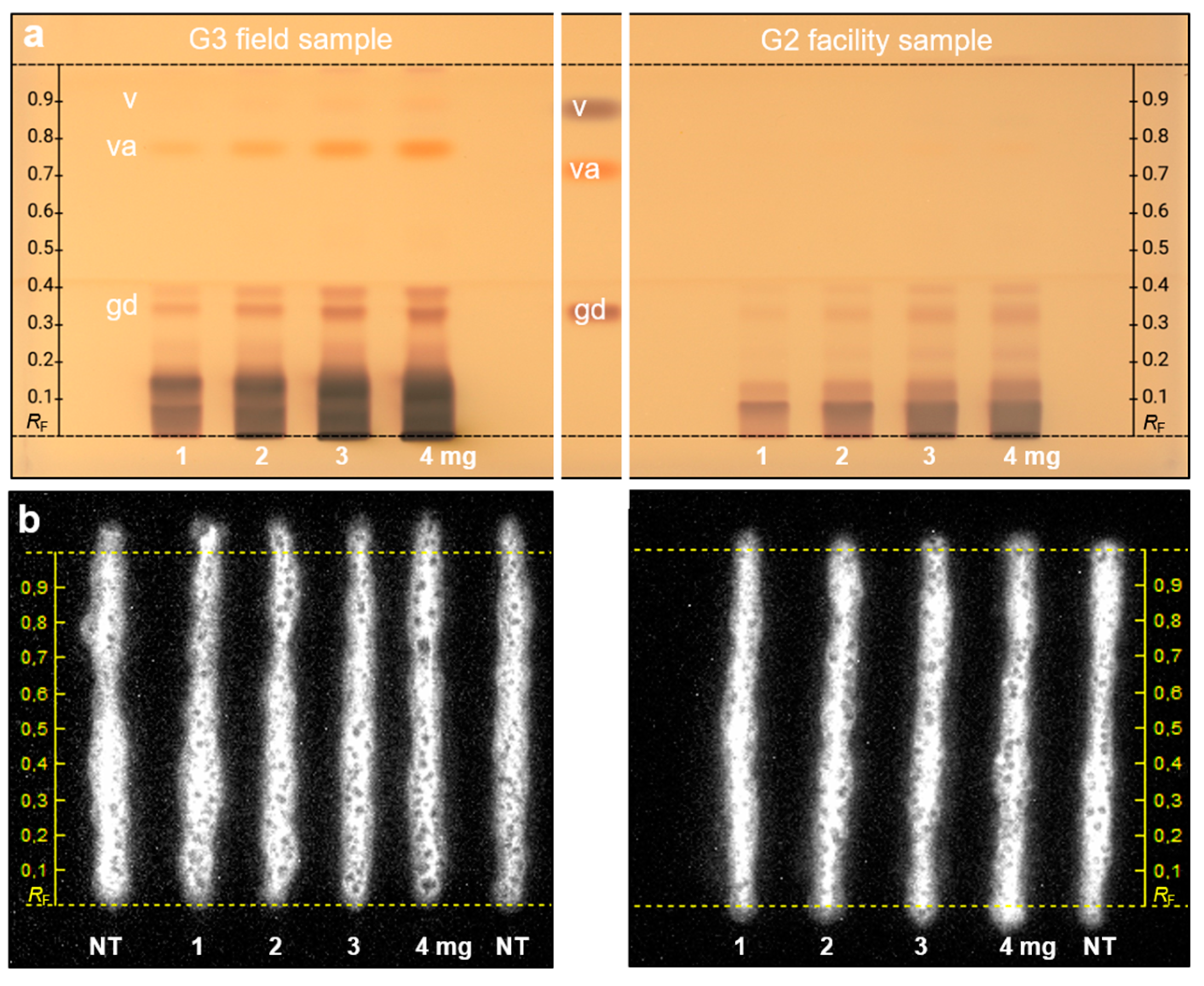
2.4. Quantification of the Marker Compound Gastrodin
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Cultivation, Harvest, and Post-Harvest Processing
4.3. Extraction and Standard Solutions
4.4. HPTLC–UV/Vis/FLD Method
4.5. Bioactivity Profiling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kew Royal Botanic Gardens. Gastrodia elata: Plants of the World Online. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:635522-1 (accessed on 1 May 2023).
- Zhan, H.-D.; Zhou, H.-Y.; Sui, Y.-P.; Du, X.-L.; Wang, W.-H.; Dai, L.; Sui, F.; Huo, H.-R.; Jiang, T.-L. The rhizome of Gastrodia elata Blume-An ethnopharmacological review. J. Ethnopharmacol. 2016, 189, 361–385. [Google Scholar] [CrossRef]
- An, H.; Kim, I.S.; Koppula, S.; Kim, B.W.; Park, P.J.; Lim, B.O.; Choi, W.S.; Lee, K.H.; Choi, D.K. Protective effects of Gastrodia elata Blume on MPP+-induced cytotoxicity in human dopaminergic SH-SY5Y cells. J. Ethnopharmacol. 2010, 130, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-Y.; Yuan, Y.-H.; Yan, J.-Q.; Wang, Y.-N.; Chu, S.-F.; Zhu, C.-G.; Guo, Q.-L.; Shi, J.-G.; Chen, N.-H. 20C, a bibenzyl compound isolated from Gastrodia elata, protects PC12 cells against rotenone-induced apoptosis via activation of the Nrf2/ARE/HO-1 signaling pathway. Acta Pharmacol. Sin. 2016, 37, 731–740. [Google Scholar] [CrossRef]
- Jung, T.-Y.; Suh, S.-I.; Lee, H.; Kim, I.-S.; Kim, H.-J.; Yoo, H.-S.; Lee, S.-R. Protective effects of several components of Gastrodia elata on lipid peroxidation in gerbil brain homogenates. Phytother. Res. 2007, 21, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-E.; Lin, C.-H.; Ho, E.-P.; Ke, Y.-C.; Petridi, S.; Elliott, C.J.; Sheen, L.-Y.; Chien, C.-T. Glial Nrf2 signaling mediates the neuroprotection exerted by Gastrodia elata Blume in Lrrk2-G2019S Parkinson’s disease. Elife 2021, 10, e73753. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wan, Q.; Huang, S.; Dai, H.; Wu, Y.; Zhou, J.; Luo, H.; Zhao, Y. Phenolic Constituents with Inhibitory Activities on Acetylcholinesterase from the Rhizomes of Gastrodia elata. Chem. Nat. Compd. 2015, 51, 158–160. [Google Scholar] [CrossRef]
- Seok, P.R.; Oh, S.J.; Choi, J.W.; Lim, C.R.; Choi, J.R.; Kim, J.H.; Shin, J.-H. The protective effects of Gastrodia elata Blume extracts on middle cerebral artery occlusion in rats. Food Sci. Biotechnol. 2019, 28, 857–864. [Google Scholar] [CrossRef]
- Liang, W.-Z.; Jan, C.-R.; Hsu, S.-S. Cytotoxic effects of gastrodin extracted from the rhizome of Gastrodia elata Blume in glioblastoma cells, but not in normal astrocytes, via the induction of oxidative stress-associated apoptosis that involved cell cycle arrest and p53 activation. Food Chem. Toxicol. 2017, 107, 280–292. [Google Scholar] [CrossRef]
- Huang, Z.-B.; Wu, Z.; Chen, F.-K.; Zou, L.-B. The protective effects of phenolic constituents from Gastrodia elata on the cytotoxicity induced by KCl and glutamate. Arch. Pharm. Res. 2006, 29, 963–968. [Google Scholar] [CrossRef]
- Lu, C.; Qu, S.; Zhong, Z.; Luo, H.; Lei, S.S.; Zhong, H.-J.; Su, H.; Wang, Y.; Chong, C.-M. The effects of bioactive components from the rhizome of Gastrodia elata blume (Tianma) on the characteristics of Parkinson’s disease. Front. Pharmacol. 2022, 13, 963327. [Google Scholar] [CrossRef]
- Lin, Y.-E.; Chou, S.-T.; Lin, S.-H.; Lu, K.-H.; Panyod, S.; Lai, Y.-S.; Ho, C.-T.; Sheen, L.-Y. Antidepressant-like effects of water extract of Gastrodia elata Blume on neurotrophic regulation in a chronic social defeat stress model. J. Ethnopharmacol. 2018, 215, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-J.; Sheen, L.-Y. Gastrodiae Rhizoma (天麻tiān má): A review of biological activity and antidepressant mechanisms. J. Tradit. Complement. Med. 2011, 1, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mori, A. Antioxidant and free radical scavenging activities of Gastrodia elata Bl. and Uncaria rhynchophylla (Miq.) Jacks. Neuropharmacology 1992, 31, 1287–1298. [Google Scholar] [CrossRef]
- Shi, A.; Xiang, J.; He, F.; Zhu, Y.; Zhu, G.; Lin, Y.; Zhou, N. The Phenolic Components of Gastrodia elata improve Prognosis in Rats after Cerebral Ischemia/Reperfusion by Enhancing the Endogenous Antioxidant Mechanisms. Oxid. Med. Cell. Longev. 2018, 2018, 7642158. [Google Scholar] [CrossRef]
- Song, E.; Chung, H.; Shim, E.; Jeong, J.-K.; Han, B.-K.; Choi, H.-J.; Hwang, J. Gastrodia elata Blume Extract Modulates Antioxidant Activity and Ultraviolet A-Irradiated Skin Aging in Human Dermal Fibroblast Cells. J. Med. Food 2016, 19, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Zheng, Q.; Feng, K.; Feng, X.; Zhong, W.; Liao, C.; Li, S.; Liu, Y.; Hu, W. Neuroprotection of Gastrodia elata polyphenols against H2O2-induced PC12 cell cytotoxicity by reducing oxidative stress. Front. Pharmacol. 2022, 13, 1050775. [Google Scholar] [CrossRef]
- Xian, J.W.; Choi, A.Y.-T.; Lau, C.B.-S.; Leung, W.N.; Ng, C.F.; Chan, C.W. Gastrodia and Uncaria (tianma gouteng) water extract exerts antioxidative and antiapoptotic effects against cerebral ischemia in vitro and in vivo. Chin. Med. 2016, 11, 27. [Google Scholar] [CrossRef]
- Yu, S.J.; Kim, J.R.; Lee, C.K.; Han, J.E.; Lee, J.H.; Kim, H.-S.; Hong, J.H.; Kang, S.G. Gastrodia elata blume and an active component, p-hydroxybenzyl alcohol reduce focal ischemic brain injury through antioxidant related gene expressions. Biol. Pharm. Bull. 2005, 28, 1016–1020. [Google Scholar] [CrossRef]
- Hwang, S.M.; Lee, Y.J.; Kang, D.G.; Lee, H.S. Anti-inflammatory effect of Gastrodia elata rhizome in human umbilical vein endothelial cells. Am. J. Chin. Med. 2009, 37, 395–406. [Google Scholar] [CrossRef]
- Ahn, E.-K.; Jeon, H.-J.; Lim, E.-J.; Jung, H.-J.; Park, E.-H. Anti-inflammatory and anti-angiogenic activities of Gastrodia elata Blume. J. Ethnopharmacol. 2007, 110, 476–482. [Google Scholar] [CrossRef]
- He, P.; Hu, Y.; Huang, C.; Wang, X.; Zhang, H.; Zhang, X.; Dai, H.; Wang, R.; Gao, Y. N-Butanol Extract of Gastrodia elata Suppresses Inflammatory Responses in Lipopolysaccharide-Stimulated Macrophages and Complete Freund’s Adjuvant- (CFA-) Induced Arthritis Rats via Inhibition of MAPK Signaling Pathway. Evid. Based Complement. Alternat. Med. 2020, 2020, 1658618. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jang, Y.W.; Kang, H.S.; Moon, H.; Sim, S.S.; Kim, C.J. Anti-inflammatory action of phenolic compounds from Gastrodia elata root. Arch. Pharm. Res. 2006, 29, 849–858. [Google Scholar] [CrossRef]
- UNEP-United Nations Environment Program. Appendix II-CITES-Convention on International Trade in Endangered Species of Wild Fauna and Flora. Available online: https://cites.org/sites/default/files/eng/app/2023/E-Appendices-2023-02-23.pdf (accessed on 1 May 2023).
- Sim, C.M.; Seong, B.J.; Kim, D.W.; Kim, Y.B.; Wi, S.G.; Kim, G.; Oh, H.; Kim, T.; Chung, B.Y.; Song, J.Y.; et al. Continuous cropping of endangered therapeutic plants via electron beam soil-treatment and neutron tomography. Sci. Rep. 2018, 8, 2136. [Google Scholar] [CrossRef] [PubMed]
- Long, L.l.p.; Luo, L.f.l. Effects of different years of natural recovery of Gastrodia elata on the community structure of bacteria and fungi in rhizosphere soil. Res. Sq. 2021, Preprint. [Google Scholar] [CrossRef]
- Krstić, Đ.D.; Ristivojević, P.M.; Gašić, U.M.; Lazović, M.; Fotirić Akšić, M.M.; Milivojević, J.; Morlock, G.E.; Milojković-Opsenica, D.M.; Trifković, J.Đ. Authenticity assessment of cultivated berries via phenolic profiles of seeds. Food Chem. 2023, 402, 134184. [Google Scholar] [CrossRef]
- Mügge, F.L.B.; Morlock, G.E. Planar bioluminescent cytotoxicity assay via genetically modified adherent human reporter cell lines, applied to authenticity screening of Saussurea costus root. J. Chromatogr. A 2022, 1683, 463522. [Google Scholar] [CrossRef] [PubMed]
- Chepngeno, J.; Imathiu, S.; Owino, W.O.; Morlock, G.E. Baobab pulp authenticity and quality control by multi-imaging high-performance thin-layer chromatography. Food Chem. 2022, 390, 133108. [Google Scholar] [CrossRef]
- Guzelmeric, E.; Ristivojević, P.; Trifković, J.; Dastan, T.; Yilmaz, O.; Cengiz, O.; Yesilada, E. Authentication of Turkish propolis through HPTLC fingerprints combined with multivariate analysis and palynological data and their comparative antioxidant activity. LWT 2018, 87, 23–32. [Google Scholar] [CrossRef]
- Frommenwiler, D.A.; Booker, A.; Vila, R.; Heinrich, M.; Reich, E.; Cañigueral, S. Comprehensive HPTLC fingerprinting as a tool for a simplified analysis of purity of ginkgo products. J. Ethnopharmacol. 2019, 243, 112084. [Google Scholar] [CrossRef]
- Morlock, G.E.; Heil, J. HI-HPTLC-UV/Vis/FLD-HESI-HRMS and bioprofiling of steviol glycosides, steviol, and isosteviol in Stevia leaves and foods. Anal. Bioanal. Chem. 2020, 412, 6431–6448. [Google Scholar] [CrossRef]
- Schreiner, T.; Morlock, G.E. Non-target bioanalytical eight-dimensional hyphenation including bioassay, heart-cut trapping, online desalting, orthogonal separations and mass spectrometry. J. Chromatogr. A 2021, 1647, 462154. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, T.; Ronzheimer, A.; Friz, M.; Morlock, G.E. Multiplex planar bioassay with reduced diffusion on normal phase, identifying androgens, verified antiandrogens and synergists in botanicals via 12D hyphenation. Food Chem. 2022, 395, 133610. [Google Scholar] [CrossRef] [PubMed]
- Darwish, R.S.; Shawky, E.; Hammoda, H.M.; Harraz, F.M. Peroxidase inhibitory and antioxidant constituents from Juniperus L. species guided by HPTLC-bioautography and molecular docking studies. Nat. Prod. Res. 2021, 35, 4653–4657. [Google Scholar] [CrossRef] [PubMed]
- Darwish, R.S.; Hammoda, H.M.; Ghareeb, D.A.; Abdelhamid, A.S.A.; Harraz, F.M.; Shawky, E. Seasonal dynamics of the phenolic constituents of the cones and leaves of oriental Thuja (Platycladus orientalis L.) reveal their anti-inflammatory biomarkers. RSC Adv. 2021, 11, 24624–24635. [Google Scholar] [CrossRef]
- Morlock, G.E.; Sabir, G. Comparison of two orthogonal liquid chromatographic methods for quantitation of sugars in food. J. Liq. Chromatogr. Relat. Technol. 2011, 34, 902–919. [Google Scholar] [CrossRef]
- Krüger, S.; Winheim, L.; Morlock, G.E. Planar chromatographic screening and quantification of coumarin in food, confirmed by mass spectrometry. Food Chem. 2018, 239, 1182–1191. [Google Scholar] [CrossRef]
- Krüger, S.; Bergin, A.; Morlock, G.E. Effect-directed analysis of ginger (Zingiber officinale) and its food products, and quantification of bioactive compounds via high-performance thin-layer chromatography and mass spectrometry. Food Chem. 2018, 243, 258–268. [Google Scholar] [CrossRef]
- Morlock, G.E.; Busso, M.; Tomeba, S.; Sighicelli, A. Effect-directed profiling of 32 vanilla products, characterization of multi-potent compounds and quantification of vanillin and ethylvanillin. J. Chromatogr. A 2021, 1652, 462377. [Google Scholar] [CrossRef]
- Abdel Salam, N.A.; Ghazy, N.M.; Shawky, E.; Sallam, S.M.; Shenouda, M.L. Validated HPTLC Method for Dihydrokaempferol-4′-O-glucopyranoside Quantitative Determination in Alcea Species. J. Chromatogr. Sci. 2018, 56, 518–523. [Google Scholar] [CrossRef]
- Alqarni, M.H.; Alam, P.; Foudah, A.I.; Muharram, M.M.; Shakeel, F. Combining Normal/Reversed-Phase HPTLC with Univariate Calibration for the Piperine Quantification with Traditional and Ultrasound-Assisted Extracts of Various Food Spices of Piper nigrum L. under Green Analytical Chemistry Viewpoint. Molecules 2021, 26, 732. [Google Scholar] [CrossRef]
- Morlock, G.; Vega-Herrera, M. Two new derivatization reagents for planar chromatographic quantification of sucralose in dietetic products. J. Planar Chromatogr.–Mod. TLC 2007, 20, 411–417. [Google Scholar] [CrossRef]
- Saçıcı, E.; Yesilada, E. Development of new and validated HPTLC methods for the qualitative and quantitative analysis of hyperforin, hypericin and hyperoside contents in Hypericum species. Phytochem. Anal. 2022, 33, 355–364. [Google Scholar] [CrossRef]
- Morlock, G.E. Planar chromatographic super-hyphenations for rapid dereplication. Phytochem. Rev. 2022. [Google Scholar] [CrossRef]
- Schreiner, T.; Sauter, D.; Friz, M.; Heil, J.; Morlock, G.E. Is Our Natural Food Our Homeostasis? Array of a Thousand Effect-Directed Profiles of 68 Herbs and Spices. Front. Pharmacol. 2021, 12, 755941. [Google Scholar] [CrossRef]
- Wagner, H.; Bauer, R.; Melchart, D.; Xiao, P.-G.; Staudinger, A. Rhizoma Gastrodiae Tianma. In Chromatographic Fingerprint Analysis of Herbal Medicines: Thin-Layer and High Performance Liquid Chromatography of Chinese Drugs; Wagner, H., Bauer, R., Melchart, D., Xiao, P.-G., Staudinger, A., Eds.; Springer: Vienna, Austria, 2011; pp. 255–262. ISBN 978-3-7091-0763-8. [Google Scholar]
- Ong, E.S.; Heng, M.Y.; Tan, S.N.; Hong Yong, J.W.; Koh, H.; Teo, C.C.; Hew, C.S. Determination of gastrodin and vanillyl alcohol in Gastrodia elata Blume by pressurized liquid extraction at room temperature. J. Sep. Sci. 2007, 30, 2130–2137. [Google Scholar] [CrossRef]
- Klingelhöfer, I.; Pham Ngoc, L.; van der Burg, B.; Morlock, G.E. A bioimaging system combining human cultured reporter cells and planar chromatography to identify novel bioactive molecules. Anal. Chim. Acta 2021, 1183, 338956. [Google Scholar] [CrossRef]
- Yang, X.D.; Zhu, J.; Yang, R.; Liu, J.P.; Li, L.; Zhang, H.B. Phenolic constituents from the rhizomes of Gastrodia elata. Nat. Prod. Res. 2007, 21, 180–186. [Google Scholar] [CrossRef]
- Wald, J.P.; Morlock, G.E. Quantification of steviol glycosides in food products, Stevia leaves and formulations by planar chromatography, including proof of absence for steviol and isosteviol. J. Chromatogr. A 2017, 1506, 109–119. [Google Scholar] [CrossRef]
- Oberlerchner, J.T.; Böhmdorfer, S.; Rosenau, T.; Potthast, A. A matrix-resistant HPTLC method to quantify monosaccharides in wood-based lignocellulose biorefinery streams. Holzforschung 2018, 72, 645–652. [Google Scholar] [CrossRef]
- Klingelhöfer, I.; Morlock, G.E. Challenges in quantitative high-performance thin-layer chromatography—Part 2: Influence of the application mode on the result. JPC-J. Planar Chromatogr.-Mod. TLC 2017, 30, 411–417. [Google Scholar] [CrossRef]
- Morlock, G.E.; Koch, J.; Schwack, W. Miniaturized open-source 2LabsToGo screening of lactose-free dairy products and saccharide-containing foods. J. Chromatogr. A 2023, 1688, 463720. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.-D.; Fan, Y.-X.; Jin, C.-C.; Wang, F.; Xin, G.-Z.; Li, P.; Li, H.-J. Specific targeted quantification combined with non-targeted metabolite profiling for quality evaluation of Gastrodia elata tubers from different geographical origins and cultivars. J. Chromatogr. A 2016, 1450, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.-K.; Zhang, J.-Q.; Guo, L.-P.; Yang, Y.; Xiao, C.-H.; Yuan, Q.-S.; Wang, X.; Zhou, T. Thoughts and suggestions on ecological cultivation of Gastrodia elata. China J. Chin. Mater. Med. 2022, 47, 2277–2280. [Google Scholar] [CrossRef]
- Wu, Z.; Gao, R.; Li, H.; Liao, X.; Tang, X.; Wang, X.; Su, Z. How steaming and drying processes affect the active compounds and antioxidant types of Gastrodia elata Bl. f. glauca S. chow. Food Res. Int. 2022, 157, 111277. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wang, L.; Liu, X.; Cheng, M.; Xiao, H. Chemical fingerprint and metabolic profile analysis of ethyl acetate fraction of Gastrodia elata by ultra performance liquid chromatography/quadrupole-time of flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1011, 233–239. [Google Scholar] [CrossRef]
- Teo, C.-C.; Tan, S.-N.; Yong, J.-W.; Hew, C.-S.; Ong, E.-S. Evaluation of the extraction efficiency of thermally labile bioactive compounds in Gastrodia elata Blume by pressurized hot water extraction and microwave-assisted extraction. J. Chromatogr. A. 2008, 1182, 34–40. [Google Scholar] [CrossRef]
- Kim, I.S.; Choi, D.-K.; Jung, H.J. Neuroprotective Effects of Vanillyl Alcohol in Gastrodia elata Blume Through Suppression of Oxidative Stress and Anti-Apoptotic Activity in Toxin-Induced Dopaminergic MN9D Cells. Molecules 2011, 16, 5349–5361. [Google Scholar] [CrossRef]
- Chen, J.; Huang, Y.; Hu, X.; Bian, X.; Nian, S. Gastrodin prevents homocysteine-induced human umbilical vein endothelial cells injury via PI3K/Akt/eNOS and Nrf2/ARE pathway. J. Cell. Mol. Med. 2021, 25, 345–357. [Google Scholar] [CrossRef]
- Fan, S.; Tian, W.; Wang, Q.; Shangguan, C.; Liu, X.; Zhang, X.; Yue, L.; Chen, C. Evaluation of the changes in active substances and their effects on intestinal microflora during simulated digestion of Gastrodia elata. LWT 2022, 169, 113924. [Google Scholar] [CrossRef]
- Chen, S.; Hao, X.; Yu, L.; Zhang, P.; Cao, W.; Chen, H.; Zhu, D. Gastrodin causes vasodilation by activating KATP channels in vascular smooth muscles via PKA-dependent signaling pathway. J. Recept. Signal Transduct. Res. 2017, 37, 543–549. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, Y.; Yao, C.; He, Q.; Chen, F.; Yu, H.; Lu, G.; Jiang, N.; Liu, X. The effects of fresh Gastrodia elata Blume on the cognitive deficits induced by chronic restraint stress. Front. Pharmacol. 2022, 13, 890330. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, J.; Peng, M.; Meng, H.; Ma, H.; Cai, P.; Xu, Y.; Zhao, Q.; Si, G. A Review on Central Nervous System Effects of Gastrodin. Front. Pharmacol. 2018, 9, 24. [Google Scholar] [CrossRef]
- Lu, J.; Ma, X.; Gao, W.-C.; Zhang, X.; Fu, Y.; Liu, Q.; Tian, L.; Qin, X.-D.; Yang, W.; Zheng, H.-Y.; et al. Gastrodin Exerts Cardioprotective Action via Inhibition of Insulin-Like Growth Factor Type 2/Insulin-Like Growth Factor Type 2 Receptor Expression in Cardiac Hypertrophy. ACS Omega 2021, 6, 16763–16774. [Google Scholar] [CrossRef]
- Park, S.; Kim, D.S.; Kang, S. Gastrodia elata Blume water extracts improve insulin resistance by decreasing body fat in diet-induced obese rats: Vanillin and 4-hydroxybenzaldehyde are the bioactive candidates. Eur. J. Nutr. 2011, 50, 107–118. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.-C.; Qin, L.-Y.; He, H.-Y.; Yu, X.-L.; Yang, M.-Z.; Zhang, H.-B. Dynamics of fungal communities during Gastrodia elata growth. BMC Microbiol. 2019, 19, 158. [Google Scholar] [CrossRef]
- Yu, E.; Gao, Y.; Li, Y.; Zang, P.; Zhao, Y.; He, Z. An exploration of mechanism of high quality and yield of Gastrodia elata Bl. f. glauca by the isolation, identification and evaluation of Armillaria. BMC Plant Biol. 2022, 22, 621. [Google Scholar] [CrossRef]
- Tang, C.; Wu, B.; Wu, J.; Zhang, Z.; Yu, B. Novel Strategies Using Total Gastrodin and Gastrodigenin, or Total Gastrodigenin for Quality Control of Gastrodia elata. Molecules 2018, 23, 270. [Google Scholar] [CrossRef]
- Morlock, G.E.; Heil, J.; Inarejos-Garcia, A.M.; Maeder, J. Effect-Directed Profiling of Powdered Tea Extracts for Catechins, Theaflavins, Flavonols and Caffeine. Antioxid 2021, 10, 117. [Google Scholar] [CrossRef]
- Sherma, J. Review of the Determination of the Antioxidant Activity of Foods, Food Ingredients, and Dietary Supplements by Thin Layer Chromatography-Direct Bioautography, Spectrometry, and the Dot-Blot Procedure. J. AOAC Int. 2018, 101, 1285–1294. [Google Scholar] [CrossRef]
| Method Ruggedness: Mean Contents (n = 3–5 days/plates; ±Standard Deviation) | ||
|---|---|---|
| G. elata Samples | Gastrodin (µg/100 mg ± sd) | Preliminary Assigned Parishin E (µg/100 mg ± sd) Equivalently Calculated to Vanillyl Alcohol |
| G1.1 | 209 ± 22 | 203 ± 19 |
| G1.2 | 182 ± 16 | 129 ± 19 |
| G1.3 | 218 ± 28 | 218 ± 14 |
| G2 | 102 ± 27 | 24 ± 3 |
| G3 | 146 ± 20 | 186 ± 10 |
| G4 | 93 ± 11 | 235 ± 16 |
| G5 | 198 ± 22 | 92 ± 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mügge, F.L.B.; Sim, C.M.; Honermeier, B.; Morlock, G.E. Bioactivity Profiling and Quantification of Gastrodin in Gastrodia elata Cultivated in the Field versus Facility via Hyphenated High-Performance Thin-Layer Chromatography. Int. J. Mol. Sci. 2023, 24, 9936. https://doi.org/10.3390/ijms24129936
Mügge FLB, Sim CM, Honermeier B, Morlock GE. Bioactivity Profiling and Quantification of Gastrodin in Gastrodia elata Cultivated in the Field versus Facility via Hyphenated High-Performance Thin-Layer Chromatography. International Journal of Molecular Sciences. 2023; 24(12):9936. https://doi.org/10.3390/ijms24129936
Chicago/Turabian StyleMügge, Fernanda L. B., Cheul Muu Sim, Bernd Honermeier, and Gertrud E. Morlock. 2023. "Bioactivity Profiling and Quantification of Gastrodin in Gastrodia elata Cultivated in the Field versus Facility via Hyphenated High-Performance Thin-Layer Chromatography" International Journal of Molecular Sciences 24, no. 12: 9936. https://doi.org/10.3390/ijms24129936
APA StyleMügge, F. L. B., Sim, C. M., Honermeier, B., & Morlock, G. E. (2023). Bioactivity Profiling and Quantification of Gastrodin in Gastrodia elata Cultivated in the Field versus Facility via Hyphenated High-Performance Thin-Layer Chromatography. International Journal of Molecular Sciences, 24(12), 9936. https://doi.org/10.3390/ijms24129936







