Abstract
The construction of a genetic circuit requires the substitution and redesign of different promoters and terminators. The assembly efficiency of exogenous pathways will also decrease significantly when the number of regulatory elements and genes is increased. We speculated that a novel bifunctional element with promoter and terminator functions could be created via the fusion of a termination signal with a promoter sequence. In this study, the elements from a Saccharomyces cerevisiae promoter and terminator were employed to design a synthetic bifunctional element. The promoter strength of the synthetic element is apparently regulated through a spacer sequence and an upstream activating sequence (UAS) with a ~5-fold increase, and the terminator strength could be finely regulated by the efficiency element, with a ~5-fold increase. Furthermore, the use of a TATA box-like sequence resulted in the adequate execution of both functions of the TATA box and the efficiency element. By regulating the TATA box-like sequence, UAS, and spacer sequence, the strengths of the promoter-like and terminator-like bifunctional elements were optimally fine-tuned with ~8-fold and ~7-fold increases, respectively. The application of bifunctional elements in the lycopene biosynthetic pathway showed an improved pathway assembly efficiency and higher lycopene yield. The designed bifunctional elements effectively simplified pathway construction and can serve as a useful toolbox for yeast synthetic biology.
1. Introduction
A key concept in synthetic biology is the development of standardized multifunctional components [1]. Specifically, structurally different elements have the potential to develop new properties in new combinatorial structures, which will result in the creation of novel genetic elements [2]. Many natural gene elements, such as promoters and terminators, are essential transcriptional regulators of gene expression and pathway engineering that exhibit excellent structural properties and whose functions are apparent [3,4,5,6]. With the improvement in natural gene elements, synthetic genetic elements have the advantage of a streamlined structure for easy regulation and a controlled synthesis length, with minimal sequence homology and better performance. Generally, random or rational construction methods have been widely used to develop novel synthetic gene elements, which have been successfully applied to the expression of metabolic engineering-related enzymes in recent decades. As the most basic functional element in metabolic engineering, promoters are the most effective tool for regulating metabolic pathways at the transcriptional level. By analyzing the structure of eukaryotic promoters, Juven et al. [7] described the design of a super core promoter (SCP) to enhance gene expression, which contained a TATA box, initiator (Inr), motif ten element (MTE), and downstream promoter element (DPE). This promoter directs high levels of transcription by RNA polymerase II in eukaryotic cells. The copy number of the UAS was found to be another important factor. The intensity of the promoter could be increased 400 times by changing the copy number of the UAS [8]. Sen et al. [9] constructed a broad-spectrum promoter by integrating the core promoter regions of E. coli, S. cerevisiae, and B. subtilis, which demonstrated normal function in a range of different hosts.
Transcription terminators are another important element for regulating gene expression in synthetic gene networks. The terminator sequence in eukaryotic genes are located in the 3′ untranslated region, and its activity is directly related to mRNA stability, and thus the regulation of gene expression [10]. Guo et al. [5,11] found that a yeast terminator usually consists of three elements: an efficiency element, a positioning element, and a poly(A) site. In addition, linker 1 (situated between the efficiency and positioning elements) and linker 2 (between the positioning element and the poly(A) site) can finely regulate the intensity of the terminator [12]. Considering the transcription process, it begins at the promoter and terminates at a particular RNA sequence encoded by the DNA terminator, thereby affecting a particular pause in RNA polymerase (RNAP) with ultimate breakdown of the transcriptional complex to release the RNA product. Kang et.al [13] reported that the population of RNA polymerase that bound to the intrinsic terminator usually releases RNA transcription products without departing from the DNA template, and the remaining RNA polymerase molecules diffuse one-dimensionally in both directions on DNA, even reinitiating transcription at nearby promoters.
The assembly of regulatory elements and genes is a basic procedure. However, in yeast, standard parts, such as promoters and terminators, are repeatedly used for genetic circuit construction, which may lead to the rearrangement of repetitive sequences in multigene assemblies [14,15,16,17,18]. This problem is a major obstacle when assembling complex synthetic circuits or pathways. Moreover, as the number of assembled fragments increases, the efficiency of DNA ligation decreases [2,19,20]. DNA assembly involving multiple fragments is the most time-consuming, and the cost of quality control will also increase. Therefore, the need for highly robust and efficient multifunctional regulatory elements has become an important direction for synthetic biology research.
In this study, we thus aimed to establish a regulatory element capable of simultaneously regulating the expression of two genes in yeast. That is, an element in a gene circuit not only performs the function of transcriptional termination of upstream genes but also possesses the ability to recruit RNA polymerase II to induce transcription by acting on the promoters of downstream genes. The terminator–promoter bifunctional element simplifies the construction process in gene circuits and pathway engineering, serving as a useful toolbox for the development of yeast metabolic engineering.
2. Results and Discussion
2.1. Design of Synthetic Bifunctional Element
The basic sequences for the promoter and terminator were used to build the synthetic bifunctional element with terminator and promoter functions. First, sequences that may help the basal transcription for the core promoter were determined. In S. cerevisiae, TATATAA is recognized as being the optimal binding site sequence for recognition by the TATA-box binding protein (TBP) and is conserved [21]. The finest conserved sequence of the TFIIB recognition element (BRE) is GGACGCC, which is adjacent to the TATA box and synergistic with the TATA box [22,23]. The initiator (Inr), which contains the transcription start site, mainly recruits the cofactors and assembles a preinitiation complex (PIC) consisting of Pol II and general transcription factors (GTFs), such as TBP, TFIID, and TFIIB [24]. The conservative DPE sequence is GACGTGC, which can serve as a binding site for transcription factor TFIID by cooperating with the Inr element when a TATA box is lacking [25]. The conserved MTE sequence, with a strong synergistic effect between the TATA box and DPE, is GAGCCGAGCA [26]. Based on these elements, a core promoter, called DCP, was assembled by connecting the BRE, TATA box, Inr, MTE, and DPE via linker sequences. Additionally, an upstream activating sequence (UAS) that contains yeast transcription factor binding sites was designed and added upstream of the core promoter DCP (Figure 1A,B) [4]. The UAS can help the core promoter to recruit transcription factors and RNA polymerase II for transcription initiation, thereby enhancing gene expression or achieving specific regulation [6,27,28]. The combination of the UAS and DCP formed a synthetic promoter named DP (Figure 1C).
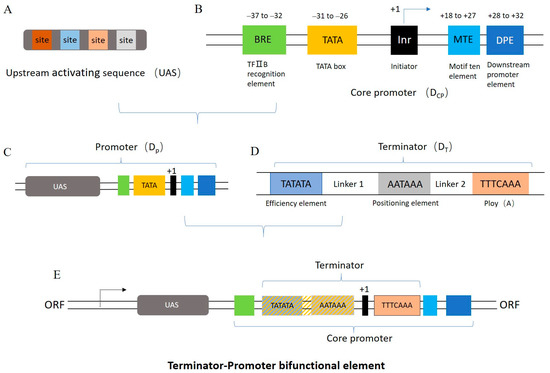
Figure 1.
Design of functional elements. (A) Structural sketch of upstream activating sequences. The transcription factor binding sites (TFBSs) contain Rap1, Gcr1, Mig1, and Gcr2. Color code does not refer specifically to a site. (B) Schematic design of yeast core promoter DCP. (C) Complete promoter element DP was formed by combining UAS and DCP. (D) Terminator element DT composed of efficiency element 5′-TATATA-3′, positioning element 5′-AATAAA-3′, and Poly(A). (E) Schematic diagram of the terminator–promoter bifunctional element Bif-b1, where terminator sequences are fused into promoter sequences. That is, orange stripes were added to the blue and gray rectangles to indicate that these elements have now acquired the functionality of a TATA box. This is one of several fusion methods, which we discuss in detail in Section 2.4.
Previous studies have shown that a terminator implementing transcriptional termination requires at least three basic elements: an efficiency element, a positioning element, and a poly(A) site [11]. The sequence TATATA, as an efficiency element, is considered to be the strongest termination signal [29]. The positioning element is regarded as essential for proper 3′ end formation, as well as cleavage and polyadenylation, and the optimal sequence is AATAAA [30,31]. The poly(A) site plays a role in mRNA transcription and translation, and the optimal sequence is TTTCAAA [11,32]. The sequential ligation of TATATA, linker 1, AATAAA, linker 2, and TTTCAAA produced a synthetic terminator (DT) (Figure 1D). After determining the sequences of the promoter and terminator, their different elements were aligned and fused to produce a synthetic element with promoter and terminator functions, and the overall construct is referred to as the terminator–promoter bifunctional element (Figure 1E, Table S1).
2.2. Regulation of Promoter Activity by Spacer Sequence and UAS
The synthetic promoter consisting of a core promoter region and UAS was designed to guide the expression of green fluorescent protein in plasmid pRS-Exp1 with CYC1t as a terminator to indirectly characterize its activity in terms of fluorescence intensity (Figure S1A). Conserved regions of the core promoter are generally not specifically regulated; therefore, the mutation of spacer sequences is a common method of engineering for regulating gene expression [1,33]. By altering the spacer sequences, even the binding of a promoter to specific activators to increase promoter activity by several orders of magnitude can be achieved [34].
The transcriptional function of the core promoter element DCP1, consisting of the BRE element (located at −37 to −32 bp of the transcription start site), TATA box (−31 to −26 bp), Inr (+1), MTE (+18 to +27 bp), and DPE (+28 to +32 bp), was first tested. In order to simplify the sequence composition, a short DCP1 was also designed by shortening the spacer sequence between the DCP1 elements to 5 bp. The super core promoter (SCP) and the core promoter regions of the natural promoters CYC1 and TYS1 were used as positive controls to verify the basic transcriptional ability of the core promoter element [7]. The control indicates the relative fluorescence values yielded by the transformation of the nonmodified pRS-Exp1 in S. cerevisiae. The results show that the core promoter designed in this study successfully promotes expression of the fluorescent protein. Compared with the native core promoter TYS1, the expression of DCP1 reaches 88%. However, the fluorescence intensity of short DCP1 decreased by 67%, indicating that the length of the spacer sequence appears to affect the intensity of the core promoter element (Figure 2A).
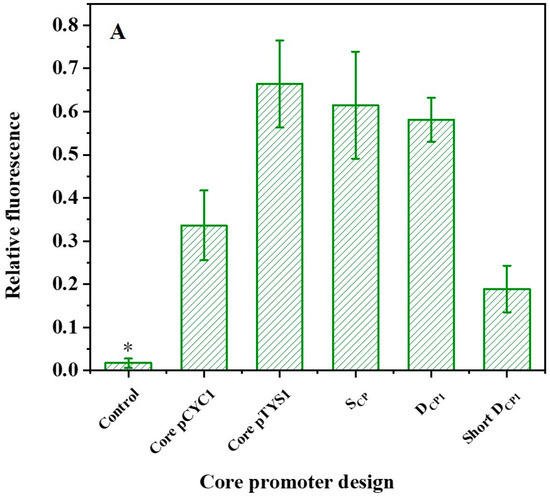
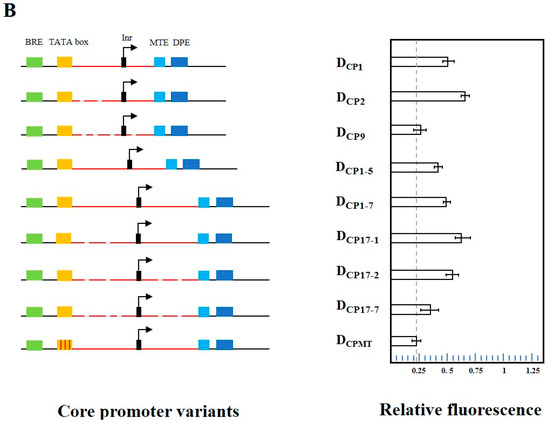
Figure 2.
Regulation of promoter activity. (A) The activities of designed core promoters. * The fluorescence levels are not statistically significantly different from the blank control. (B) Effect of non-conserved regions and TATA box on core promoter activity. The structure of the core promoter variants in the left panel corresponds to the activity presented in the right panel. Red line indicates a non-conserved region. The different lengths represent variations in the length of the spacer sequence, and the breakpoints represent mutations. Compared to DCP1, the non-conserved regions of DCP2 and DCP9 have the same spacer sequence length but different mutations, DCP1-5 and DCP1-7 both have different spacer sequence lengths; and DCP17-1, DCP17-2, and DCP17-3 have different spacer sequence lengths and different mutations. The TATA box sequence of the mutant DCPMT is mutated to GAGAGA. The left area of the gray line indicates relative fluorescence values below 0.25, defined as non-expression, i.e., no statistical difference relative to the negative control. Data represent the mean fluorescence intensity with error bars (standard error of the mean; SEM) from three independent experiments.
Furthermore, we designed five spacer sequences with different mutation regions to test the effect of non-conserved sequences on promoter activity. As shown in Figure 2B, mutations in non-conserved regions can achieve about 0.5~1.3-fold intensity. The negative control lacking the promoter region in the expression cassette was used as a threshold to assess whether the synthesized core promoter is active. In this assay, synthetic core promoters with relative fluorescence values below 0.25 are considered nonfunctional. Moreover, studies have shown that mutations in the TATA box region also affect the promoter strength [35,36]. In parallel, our results show that the mutant DCPMT lacks transcriptional activity. Apparently, the TATA box is an important promoter element.
Transcription factors Rap1, Gcr1, Gcr2, and Mig1 are thought to most effectively influence RNA polymerase II transcription initiation [37,38]. To increase the efficiency of RNA polymerase II binding to the promoter, the UAS region encompassing the binding sites for Rap1, Gcr1, Mig1, and Gcr2 was designed. Their corresponding binding sites were sequentially linked by 20 bp random sequences, which were designed using the R2oDNA Designer website to ensure that no yeast proteins or transcription factor binding sites were included [39]. The assembly of UAS1 to the 5′ end of the core promoter element DCP1 formed a complete promoter P-DCP1. This design resulted in an 86% increase in the fluorescent protein expression compared with DCP1, and its intensity reached 64% of the native promoter PTYS1 (Figure 3). Similarly, the UAS1 also enhanced the activities of the promoters P-DCP2 and P-DCP17-1, which were increased by 107% and 136% compared with DCP2 and DCP17-1, respectively. However, it is worth mentioning that when UAS1 is added to the 5′ end of DCPMT (P-DCPMT), it failed to increase the expression. This may because the mutations in the TATA box resulted in it no longer being recognized by RNA polymerase II (Figure 3). To verify the reproducibility of this result, we constructed another validation vector, pRS-Exp5 with blue fluorescent protein mTagBFP2 as a reporter (Figure S1E), and observed similar results.
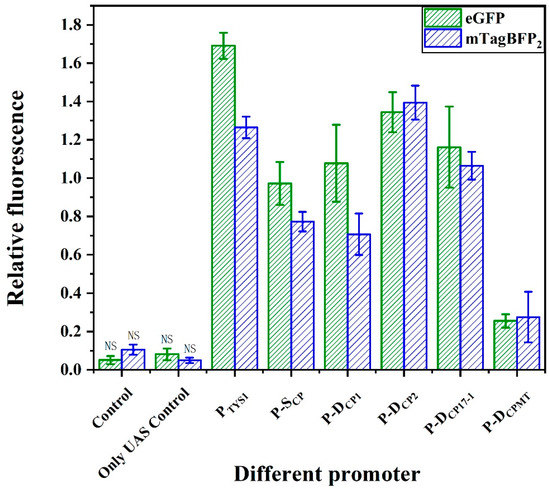
Figure 3.
Activity of synthetic promoters consisting of UAS and different core promoters. P-DCPn: the UAS assembled with DCPn to form different promoters. Among them, P-DCP2 showed the highest activity. Mean fluorescence intensity ± SEM values from triplicate experiments are shown. NS: Not Significant. The fluorescence levels are not statistically significantly different from the blank control.
Considering the modularity verified in the system, and allowing for sufficient spacing between the UAS and RNA polymerase II binding sequences, the determined core promoter–UAS spacer was maintained in all subsequent core promoter substitutions. That is, the core promoter spacing was set to 20 bp upstream of the BRE element. In another negative control, there was a complete loss of function when the core promoter was removed (only in the UAS control), which confirms that the UAS-only sequence is unable to initiate transcription.
2.3. Regulation of mRNA and Fluorescent Reporter Protein Levels by the Synthetic Terminator Elements
The vectors pRS-Exp2 with eGFP and pRS-Exp4 with mTagBFP2 were constructed (Figure S1B,D) to verify the regulation of synthetic terminator elements on mRNA and fluorescent protein levels. The natural strong promoter PTYS1 was used to control the transcriptional initiation of fluorescent proteins [40,41,42]. The terminator can significantly regulate gene expression in bacterial and fungal systems and human cells [43,44,45]. The selection of the terminators for heterologous expression can result in desirable or maximum protein yields in yeast. The minimal synthetic sequence required to achieve the terminator function has been demonstrated in previous studies [11]. In particular, the efficiency element is located upstream of the terminator sequence and is an important element affecting its activity [11,29]. Thus, terminators DT1, DT2, and DT3 were constructed with the efficiency elements TATATA, TACCTA, and TATGTC, respectively. Furthermore, because the linker 1 and linker 2 sequences also have a notable effect on its activity, the linker sequences TCCCTTTTCT and GAAAGGGC for the strongest terminator were used [12]. The DT1 with efficiency element TATATA demonstrated the same relative fluorescence levels as the natural terminator CYC1 (Figure 4A). Further, the relative mRNA of the fluorescent proteins controlled by different terminators designed in this paper was calculated based on the mRNA of the reference gene TUB1 (as shown in Figure 4B). The transcription levels and fluorescence values of fluorescent proteins showed similar trends. Therefore, the synthetic terminators in this study could regulate the fluorescent protein levels. When the efficiency element TATATA was replaced with TACCTA or TATGTC, the relative fluorescence decreased by 25% (DT2) or 35% (DT3) relative to that of DT1. Moreover, if the efficiency element TATATA is mutated to GAGAGA (DTMU), the relative fluorescence decreases significantly to only 21% that of DT1. Similarly, the vector pRS-Exp4 with blue fluorescent protein mTagBFP2 was applied to verify the fluorescent reporter protein levels with similar results. In short, the transcription termination function of the synthetic terminators can be finely regulated by the efficiency element.
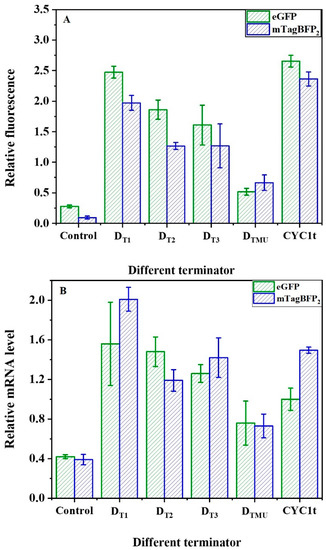
Figure 4.
Design of synthetic terminators (A) Effects of efficiency element on synthetic terminator (DTn) strength. Data represent the mean fluorescence intensity with error bars (standard error of the mean (SEM)) from three independent experiments. (B) The relative mRNA of terminators. Relative mRNA level was calculated based on the mRNA level of the reference gene TUB1. Error bars indicate standard deviation (SD). Data are means: SD (n = 3).
2.4. TATA Box-like Sequence Can Execute Both Functions of TATA Box and Efficiency Element
To construct a regulatory element capable of simultaneously regulating the expressions of two genes in yeast, we fused the strong promoter P-DCP2 and the strong terminator DT1. The direct assembly of DT1 and P-DCP2 generated new elements Bif-a1 (DT1-P-DCP2) or Bif-a2 (P-DCP2-DT1). For functional validation, pRS-Exp3 was also constructed (Figure S1C). In the vector pRS-Exp3, the natural terminator ADH1 and promoter FBA1 were selected as positive controls to regulate the transcription terminator of the upstream gene and the promoter of the downstream gene. The terminator-like strength and promoter-like strength were characterized by the relative fluorescence values of the blue fluorescent protein (mTagBFP2) and green fluorescent protein (eGFP), respectively. As shown in Figure 5, the terminator-like strength of the bifunctional element Bif-a1 was elevated by 11% compared to the natural terminator ADH1, and its promoter-like strength reached 90% of the promoter FBA1. However, the terminator-like and promoter-like strength of the bifunctional element Bif-a2 increased by only 9% and reached only 12% of the natural promoter FBA1. The results show that the assembly mode of the terminator–promoter achieved the expected functions (Figure S2). The combined mode of terminator–promoter caused the promoter insulation, failing to transcribe the downstream gene [46]. The combination mode of the terminator–promoter is no different from the conventional regulatory elements. Therefore, we attempted to insert the termination signal inside the promoter, not only to perform the transcriptional termination on upstream genes but also to regulate the expressions of downstream genes by recruiting RNA polymerase II.
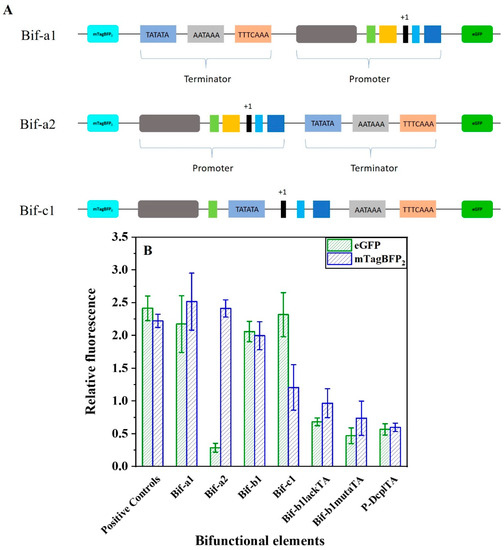
Figure 5.
Design and activity characterization of bifunctional elements. (A) A schematic of different designs for constructing the bifunctional elements. Their activities are characterized in the reporter plasmid pRS-Exp3, where mTagBFP was inserted upstream and eGFP downstream (B) Characterization of the activities of bifunctional elements. The relative FI of mTagBFP2 indirectly indicates the terminator-like strength of bifunctional element. For the eGFP, its relative FI indicates the promoter-like strength of bifunctional elements. Data represent the mean fluorescence intensity with error bars (standard error of the mean; SEM) from three independent experiments.
Considering the high homology of the TATA box with the efficiency element, we replaced the TATA box in Bif-a2 with the efficiency element to form the Bif-c1. (Figure 5A). The result of the activity verification showed that the terminator-like strength was significantly reduced. We speculated that the spacing of the efficiency element and the positioning element would have a significant effect on the terminator strength. Thus, the terminator DT1 was used to replace the TATA box to construct a novel regulatory element, Bif-b1 (Figure 1E). As shown in Figure 5B, the Bif-b1 successfully regulated the expressions of fluorescent proteins in S. cerevisiae. In short, the efficiency element and the positioning element combine to perform the function of a TATA box in a bifunctional element. To further confirm the essential TATA sequence in the bifunctional element, we constructed Bif-b1lackTA through the lack of a TATA box and Bif-b1mutaTA through a mutant of the TATA box (Figure S2). Compared to Bif-b1, the activities of the promoter-like and terminator-like Bif-b1lackTA were decreased by 67% and 52%, and those of Bif-b1mutaTA were decreased by 77% and 63%, respectively. These results support the hypothesis that a TATA box-like efficiency element is capable of recruiting RNA polymerase II and initiating the mRNA transcription.
We used a de novo design approach to synthesize promoter and terminator sequences in this study. The design was based on the structural analysis of eukaryotic transcription initiation and transcriptional termination to find the required motif. Synthetic promoters and terminators were first tested experimentally in the yeast. However, problems were caused by the multiple gene assemblies in the construction of engineered strains of Saccharomyces cerevisiae, mainly including sequence duplication rearrangement, time-consuming assembly, and inefficiency. In terms of genetic elements, the regulation of gene expression in yeast is altered only through promoters or terminators. One element with the two functions of terminator and promoter has not yet been studied. Therefore, we are interested in developing novel regulatory elements. We based the fusion on the sequences of a synthetic promoter and a synthetic terminator. As shown in Figure 5, the design approach of Bif-b1 (Figure 1E) for developing a sequence of elements with two functions was ultimately successful.
2.5. Fine-Tuning the Activities of the Bifunctional Element
To meet the requirements of finely regulating a genetic circuit or biosynthetic pathway, the activity of the terminator–promoter bifunctional element was further adjusted based on numerous considerations. Considering the significant effect of the efficiency element and TATA sequence on the terminator and promoter activity, and the peculiarity of the bifunctional element sharing a common TATA sequence [47,48], we first regulated the activity of the bifunctional element based on repetition and the type of TATA. Based on Bif-b1, a small bifunctional element library was constructed. Figure 6 shows that the terminator-like strength (mTagBFP2 expression) reaches the highest level when TATA is tripled. The promoter-like strength (eGFP expression) is the highest at 1*TA–3*TA. However, if there are more than three TATA repeats, the strength of the bifunctional elements decreases significantly. When the TATA sequence was mutated to other different types, the strength of the bifunctional elements decreases with the decreasing homology of the TATA type with TATTTA. By changing the length and type of TATA, the terminator-like strength of the bifunctional elements was modulated to as high as ~3-fold, and its promoter-like strength was modulated to as high as ~4-fold.
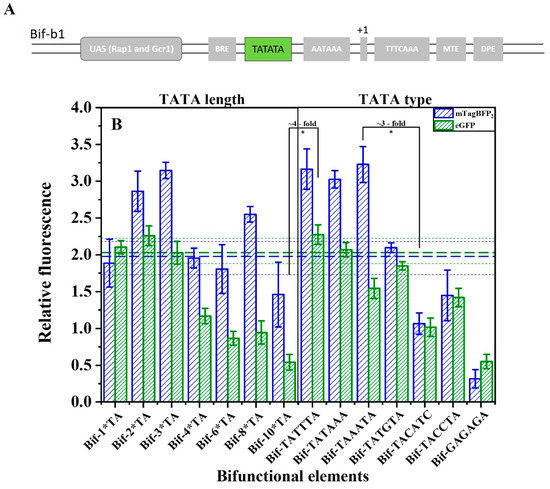
Figure 6.
(A) Schematic representation of TATA sequence to regulate the activity of bifunctional elements. (B) The effect of the number of TATA repeats and type on the activity of the bifunctional element. n*TA indicates the number of “TATA” repeats. Horizontal green bold dashed line indicates the activity of promoter-like of the Bif-b1, and the green dashed line represents the standard error. Horizontal blue bold dashed line indicates the terminator-like activity of the Bif-b1, and the blue dashed line represents the standard error. Data represent the mean fluorescence intensity with error bars (standard error of the mean; SEM) from three independent experiments. Asterisks * indicate a significant difference in the strength of elements in the library (* p ≤ 0.05).
Because a tandem copy of the upstream active sequence (UAS) can act as a synthetic transcriptional amplifier to increase promoter activity [8], a second attempt was made to finely regulate transcriptional initiation of the bifunctional elements. The different combinations of the transcription factor binding sites (TFBSs: Rap1, Gcr1, Mig1, and Gcr2) shown in Table S2 were linked to Bif-b1, generating another bifunctional element library (Table S3). As shown in Figure 7, the promoter-like strength of Bif-UAS5 and Bif-UAS9 reached 11% and 25% that of Bif-b1. The terminator-like strength of Bif-UAS7 and Bif-UAS10 reached 41% and 29% that of Bif-b1. The modification of the UAS achieved the highest increases of ~5-fold and ~3-fold for the promoter-like and terminator-like strength of bifunctional elements, respectively.
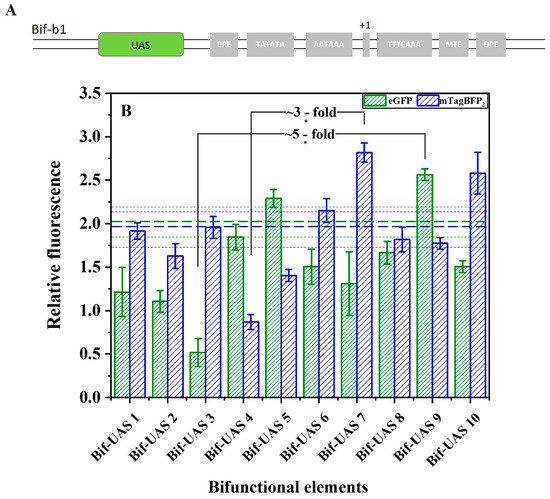
Figure 7.
Effect of UAS on the activity of the bifunctional element. (A) Schematic representation of UAS to regulate the activity of bifunctional elements. (B) UAS1–UAS10 indicate different combinations of Rap1, Gcr1, Mig1, and Gcr2 in the bifunctional element. Horizontal green bold dashed line indicates the activity of promoter-like of the Bif-b1, and the green dashed line represents the standard error. Horizontal blue bold dashed line indicates the terminator-like activity of the Bif-b1, and the blue dashed line represents the standard error. Data represent the mean fluorescence intensity with error bars (standard error of the mean; SEM) from three independent experiments. Asterisks * indicate a significant difference in the strength of elements in the library (* p ≤ 0.05).
In addition, to suppress undesirable homologous recombination events that are present in genetic circuits in yeast, another library was constructed by inserting different spacer sequences between the individual motifs of the bifunctional element (Figure 8A). The relative fluorescence values show that the spacer sequences designed by the R2oDNA designer could increase the terminator-like strength by ~7-fold and promoter-like strength by ~6-fold (Figure 8B). These results show that the tuning of bifunctional element strength can be achieved by varying the spacer sequences. Specifically, compared with Bif-b1, the promoter-like strength of U4 was increased by 31%, and the terminator-like strength of U6 was increased by 59%. Repeated sequences may be prone to recombination; thus, spacer sequences could be advantageous to the construction of genetic circuits and the fine regulation of pathways [17]. In short, through the abovementioned construction and regulation, the design of flexible bifunctional elements can also be achieved. The promoter-like strength of bifunctional elements was increased by 8-fold, and its terminator-like strength was increased by 7-fold.
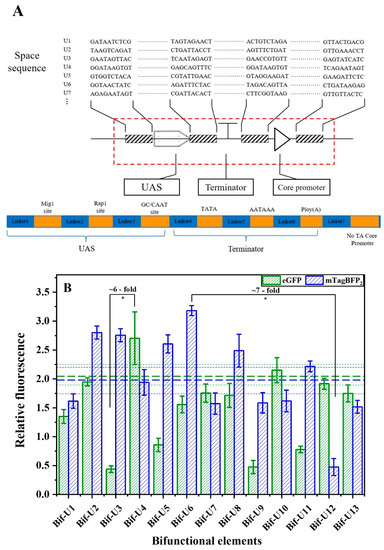
Figure 8.
Effect of spacer sequence on the activity of bifunctional element. (A) Schematic diagram of different spacer sequences inserted into bifunctional element. Blue box shows the spacer sequence designed by R2oDNA designer. (B) Activity of bifunctional elements containing different spacer sequences. Control indicates transformation of pRS-Exp3 plasmid, without any alteration, into yeast in vivo. Horizontal green bold dashed line indicates the promoter-like activity of the Bif-b1, and the green dashed line represents the standard error. Horizontal blue bold dashed line indicates the activity of terminator-like of the Bif-b1, and the blue dashed line represents the standard error. Data represent the mean fluorescence intensity with error bars (standard error of the mean; SEM) from three independent experiments. Asterisks * indicate a significant difference in the strength of elements in the library (* p ≤ 0.05).
2.6. Application of the Bifunctional Element in Pathway Engineering
As a proof of concept, the lycopene synthesis pathway based on the synthetic bifunctional elements was constructed in S. cerevisiae. Because yeast contains the MVA pathway to produce the intermediate product geranylgeranyl diphosphate (GGPP) for lycopene synthesis, exogenous phytoene synthase Crt B and phytoene desaturase Crt I need to be introduced to S. cerevisiae [49]. To verify the function of bifunctional elements in pathway engineering, plasmid-based strains were constructed by transforming gene expression cassettes pRS—Bif-2*TA—Crt B—Bif-UAS4—Crt I—Bif-U7, pRS—Bif-TATTTA—Crt B—Bif-UAS9—Crt I—Bif-U6, and pRS—Bif-3*TA—Crt B—Bif-UAS5—Crt I—Bif-U2 into S. cerevisiae dubbed as Sl-1, Sl-2 and Sl-3, respectively (Figure 9A). Strain Tl-1 was a control strain constructed by transforming the expression cassette pRS—CYC1p—Crt B—ADH1—TYS1—Crt I—ADAL. According to the results of HPLC analysis, lycopene was synthesized by all engineered strains, and the titer of lycopene increased by 85% in strain Sl-2 compared with Tl-1. The quantitative polymerase chain reaction (qPCR) showed that the transcription of Crt I and Crt B were significantly higher in strains Sl-2 and Sl-3 than in strain Tl-1 (Figure 9B). These results indicated that the synthetic bifunctional elements designed in this study possess the ability to recruit RNA polymerase II to act on the transcriptional initiation of the target gene, and the elements can also achieve transcriptional termination. It is further demonstrated that bifunctional elements could be a promising tool for application in pathway engineering for fine regulation. In addition, the use of bifunctional elements reduces the number of required assembly fragments compared with traditional assembly methods, resulting in an increase in assembly efficiency (Figure S3).
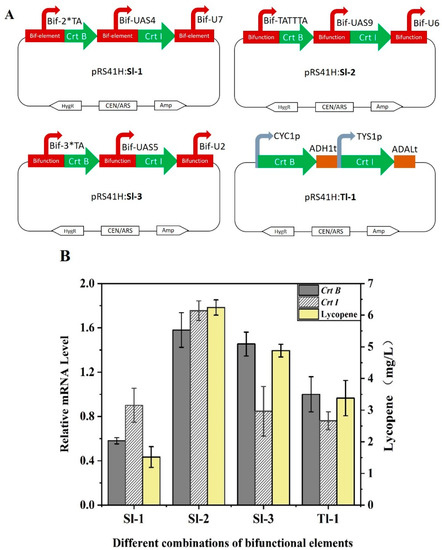
Figure 9.
Application of bifunctional elements in lycopene synthesis pathway. (A) The detailed genotype of engineered strains. Strains Sl-1, Sl-2 and Sl-3 were constructed using bifunctional elements. Strain Tl-1 was obtained using the native promoters and terminators. (B) Lycopene production and relative mRNA with engineered yeasts. Relative mRNA levels were calculated based on the mRNA level of the reference gene TUB1. Error bars indicate ± SD. Data are means ± SD (n = 3).
3. Methods
3.1. Strain, Culture Medium, and Materials
S. cerevisiae CEN.PK2-1C (MATa ura3-52 leu2-3, 112 trp1-289 his3DMAL2-8cSUC2) was used as the host strain. Yeast transformants in this study were selected on YPD plates (YPD plate, containing yeast extract (1%), peptone (2%), glucose (2%), agar (2%), and 500 mg/L hygromycin B). Yeast strains were cultured in YPD liquid medium (containing yeast extract (1%), peptone (2%), glucose (2%), and with corresponding antibiotics) at 30 °C and 220 rpm.
E. coli DH5a (Tiangen, Beijing, China) was used for clone and routine transformations after routine propagation in lysogeny broth (LB) medium (yeast extract (0.5%), tryptone (1%), NaCl (1%), and 100 μg/mL ampicillin).
Oligonucleotides were synthesized by GENEWIZ (Suzhou, China). DNA polymerase Phanta Max was obtained from Vazyme (Nanjing, China). Restriction enzymes and T7 DNA ligase were purchased from New England Biolabs (NEB, Ipswich, MA, USA). The kits for plasmid extraction were from TIANGEN (Beijing, China). The kits for DNA purification were from Vazyme (Nanjing, China). The chemical reagents were all of analytical grade. A spectrophotometer (UV-5100, Shanghai Metash Instrument Co., Ltd., Shanghai, China) was used to determine the cell concentration by measuring the optical density at 600 nm (OD600).
3.2. Plasmid Construction
Two fluorescence genes, eGFP and mTagBFP2 were synthesized by GENEWIZ after codon optimization (Table S4). The genes of phytoene synthase (Crt B, accession no.KC954270.1), phytoene desaturase (Crt I, accession no. AY177424.1) were cloned from the W1 strain constructed by Lixia et al. [41] Yeast promoters (CYC1 and TYS1), terminators (ADH1 and ADAL) were PCR-amplified from the S. cerevisiae CEN.PK2-1C genome.
The plasmid used as a backbone in the construction of expression vectors in this study was pRS41H (Beijing Biosea Biotechnology Co., Ltd., Beijing, China). Specificallmand); the ligated products were then inserted into the pRS41H plasmid linearized by NotI and SacI to form the scaffold plasmids pRS-Exp1 (Supplementary Figure S1). Five scaffold plasmids pRS-Exp1–5 were constructed according to a similar strategy. All tested promoters were cloned using primers containing sites for restriction by BamHI/NotI and then mixed with the linearized pRS-Exp1 for characterization of the promoter activity. Similarly, all tested terminators and bifunctional elements were cloned using primers containing restriction sites for BamHI/NotI. Using T7 ligase, they were each ligated into linearized scaffold plasmids pRS-Exp2 and pRS-Exp3.
The construction of lycopene producing strain Sl-1 was taken as an example. First, the gene expression plasmid pRS—Bif-2*TA—Crt B—Bif-UAS4—Crt I—Bif-U7 was constructed via Gibson assembly [13]. Plasmid-based strain Sl-1 was constructed by transforming gene expression plasmid into S. cerevisiae. Specifically, the primer pairs for adjacent fragments were designed with flanking overlap homology sequences, as shown in Table S5. Bif-2*TA-F/Bif-2*TA-R and Bif-U7-F/Bif-U7-R, were used to amplify upstream homologous arms, Bif-2*TA, and downstream homologous arms, Bif-U7, from the plasmid pRS41H, respectively. The target gene Crt B and Crt I were amplified using primer pairs, Crt B-F/Crt B-R and Crt I-F/Crt I-R, respectively. Bif-UAS4 was amplified using the primer pair, Bif-UAS4−F/Bif-UAS4-R. Strains Sl-2, Sl-3, and Tl-1 were constructed similarly to Sl-1. The gene expression plasmid pRS—Bif-TATTTA—Crt B—Bif-UAS9—Crt I—Bif-U6, pRS—Bif-3*TA—Crt B—Bif-UAS5—Crt I—Bif-U2, and pRS—CYC1p—Crt B—ADH1—TYS1—Crt I—ADAL were ligated by a similar strategy and were transformed into S. cerevisiae, respectively. Sequences were verified by Sanger sequencing (Jiangsu Saisofi Biotechnology Co., Ltd., Wuxi, China).
3.3. Transformation of Yeast
Yeast was cultured overnight in YPD medium and then transferred into YPD medium (50 mL) with a concentration ratio of 10% and continually cultivated for 4–6 h. The yeast broth was collected in 1.5 mL centrifuge tubes, and the medium was discarded after centrifugation at 1844× g for 1 min. The collected cells were washed with sterile water (2 × 1 mL) and then gently resuspended in 0.1 M LiAc (1 mL). After centrifugation at 844× g for 5 min, the supernatant (900 mL) was discarded, and the remaining cells were used as competent cells. Then, the components, including the DNA fragments (600 ng), linearized plasmid (400 ng), 10% denatured DNA from salmon sperm (40 µL; denatured at 100 °C for 5 min), poly(ethylene glycol) (PEG3350; 480 µL, 50% w/v), and 1 M LiAc (72 µL), were added, according to the indicated sequence, to the competent cells, and fully mixed using a high-speed spiral mixer for about 1 min. After incubation for 30 min at 30 °C, DMSO (72 µL) was added to the above system. The cells were immediately heat-shocked at 42 °C for 15 min, and the supernatant was discarded after centrifugation at 1180× g for 1 min. Finally, 5 mM CaCl2 (200 µL) was used to resuspend the yeast cells. After centrifugation, the yeast cells were re-suspended in double-distilled H2O (100 µL) and coated on the selective medium for incubation.
3.4. Determination of Element Strength
The element strength is indirectly indicated by the fluorescence intensity (FI) of eGFP and mTagBFP2. Yeast cells containing the genes of eGFP and mTagBFP2 were cultured in YPD medium in shaking flasks at 30 °C for 12 h and then collected by centrifugation, washed with 1× PBS (pH = 7.4), and diluted to 1 × 106 cell/mL for fluorescence analysis (Beckman Coulter, Kraemer Boulevard Brea, CA, USA). The green fluorescent protein eGFP (λex 488 mm, λem 507 nm) and the blue fluorescent protein mTagBFP2 (λex 399 mm, λem 454 nm) were detected using a FITC-A detector and PB450-A detector, respectively. Finally, the data were processed using CytExpert software (version 2.3.1.22, Beckman Coulter, Inc., Brea, CA, USA). The FI values for at least 10,000 cells in each sample were measured. The FI of eGFP and mTagBFP2 was obtained by normalization relative to that of the FI of S. cerevisiae CEN.PK2-1C with the plasmids TYS1p-eGFP-CYC1t and TYS1p-mTagBFP2-CYC1t, respectively. All experiments were repeated 3 times. The standard error was estimated from the square root of the estimated error variance of the quantity.
3.5. qPCR
Fresh samples of the cell culture were used to determine the relative expression ratio of genes via qPCR. The total RNA in the mid-log-phase yeast cells extracted was quantified by measurement of the absorbance at 260 nm with a NanoDrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA). Removing DNA Contamination from RNA Samples by treatment with DNase I (NEB, Ipswich, MA, USA). cDNA was transcribed from 500 ng total RNA using a high-capacity cDNA reverse transcription kit (PrimeScript RT Master Mix, TaKaRa, San Jose, CA, USA). qPCR was performed with a SYBR Premix Ex Taq II (TaKaRa) system and LightCycler® 96 (Roche). TUB1 was used as the reference gene. The primers were as follows: 5′-CCAAC TGGTT TCAAG ATCGG TA-3′ and 5′-TCCAC AGTGG CCAAT TGTGA-3′ for qPCR analysis, 5′-ACGCT GGTTC TCAAA TGCAC-3′ and 5′-AAGTG TCGTC CAA TT GAGAG-3′ for the Crt B gene, and 5′-GGTAA GGACG GTTTC GACAG-3′ and 5′-CGATG AATTG CAATC TCAAG-3′ for the Crt I gene. The PCR conditions consisted of 40 cycles at 95 °C for 5 s, 60 °C for 20 s, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. The transcription levels of the function genes in recombinant S. cerevisiae were calculated based on the level of TUB1 mRNA.
3.6. Statistical Analysis
The fluorescence was recorded by flow cytometry. Data represent the mean fluorescence intensity with error bars (Standard Error of the Mean; SEM) from three independent experiments. Statistical analysis was performed using Origin 64 software (Origin Lab Corporation, Northampton, MA, USA). Independent univariate analysis of variance (ANOVA) was used for the comparison of multiple books. The difference of 0.05 was statistically significant.
3.7. Samples Preparation and Metabolite Analysis by HPLC
All engineered yeasts were grown at 30 °C and 220 rpm for 36 h as a seed solution. Then, they were inoculated into unbaffled 150 mL culture flasks containing 100 mL medium and cultured in the same conditions for four days. Lycopene was extracted using the modified hot HCl/acetone method. The acetone phase containing extracted lycopene was filtered for HPLC analysis. An HPLC system (LC-2030 plus) equipped with an Inertsil ODS-3 C18 column (4.6 × 250 mm, 5 mm) and a photodiode array (PDA) detector was used to measure lycopene. The detection wavelength was 470 nm. The mobile phase was acetonitrile–methanol–dichloromethane solution (21:21:8) with a flow rate of 1.0 mL/min. All experiments were repeated three times. The standard error was estimated from the square root of the estimated error variance of the quantity.
4. Conclusions
A class of terminator–promoter bifunctional elements was designed and constructed. Three libraries of bifunctional elements were established by adjusting the TATA sequence, transcription factor binding site, and spacer region. The results show that the promoter and terminator activities of the bifunctional elements could be finely regulated by adjusting the TATA sequence, transcription factor binding site, and spacer region. The application of bifunctional elements was shown to not only improve the efficiency of pathway construction, but also increase the yield of lycopene.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms24129870/s1. Ref. [50] is cited in the Supplementary Materials.
Author Contributions
G.Z., Z.L. and X.N. designed this study. Z.L. and X.N. carried out the experiment and analyzed the data. J.G. assisted in experiments. X.N., Z.L. and G.Z. discussed the results and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by National Natural Science Foundation of China (Grant No. 22178226, 21676167), and Research Project of Youth Science and Technology Innovation Leader of XPCC (Grant No. 2017CB007).
Institutional Review Board Statement
This article does not contain any studies involving human participants or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this publication (and the Supplementary Materials).
Conflicts of Interest
The authors declare no competing financial interest.
Abbreviations
| upstream activating sequence | UASs |
| super core promoter | SCP |
| initiator | Inr |
| motif ten element | MTE |
| downstream promoter element | DPE |
| enhanced green fluorescent protein | eGFP |
| blue fluorescent protein | mTagBFP2 |
| TATA box binding protein | TBP |
| Transcription Factor IIB | TFIIB |
| TFIIB recognition element | BRE |
| general transcription factors | GTFs |
| preinitiation complex | PIC |
| RNA polymerase | RNAP |
| polymerase II | Pol II |
| Designed core promoter (in this study) | DCP |
| Designed terminator (in this study) | DT |
| Designed promoter (in this study) | P-DCP |
| geranylgeranyl diphosphate | GGPP |
References
- Portela, R.M.C.; Vogl, T.; Kniely, C.; Fischer, J.E.; Oliveira, R.; Glieder, A. Synthetic Core Promoters as Universal Parts for Fine-Tuning Expression in Different Yeast Species. ACS Synth. Biol. 2017, 6, 471–484. [Google Scholar] [CrossRef]
- Ellis, T.; Adie, T.; Baldwin, G.S. DNA assembly for synthetic biology: From parts to pathways and beyond. Integr. Biol. 2011, 3, 109–118. [Google Scholar] [CrossRef]
- Hahn, S.; Young, E.T. Transcriptional Regulation in Saccharomyces cerevisiae: Transcription Factor Regulation and Function, Mechanisms of Initiation, and Roles of Activators and Coactivators. Genetics 2011, 189, 705–736. [Google Scholar] [CrossRef]
- Gertz, J.; Siggia, E.D.; Cohen, B.A. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 2009, 457, 215-U113. [Google Scholar] [CrossRef]
- Guo, Z.J.; Sherman, F. 3’-end-forming signals of yeast mRNA. Trends Biochem. Sci. 1996, 21, 477–481. [Google Scholar] [CrossRef]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef]
- Juven-Gershon, T.; Cheng, S.; Kadonaga, J.T. Rational design of a super core promoter that enhances gene expression. Nat. Methods 2006, 3, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Liu, L.Q.; Redden, H.; Alper, H. Tuning Gene Expression in Yarrowia lipolytica by a Hybrid Promoter Approach. Appl. Environ. Microbiol. 2011, 77, 7905–7914. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Q.T.; Zhang, Y.F.; Du, G.C.; Chen, J.; Kang, Z. Construction and Characterization of Broad-Spectrum Promoters for Synthetic Biology. ACS Synth. Biol. 2018, 7, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Curran, K.A.; Karim, A.S.; Gupta, A.; Alper, H.S. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab. Eng. 2013, 19, 88–97. [Google Scholar] [CrossRef]
- Guo, Z.J.; Sherman, F. Signals sufficient for 3’-end formation of yeast mRNA. Mol. Cell. Biol. 1996, 16, 2772–2776. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X.; Wei, L.N.; Sheng, Y.; Zhang, G.L. Yeast Synthetic Terminators: Fine Regulation of Strength through Linker Sequences. ChemBioChem 2019, 20, 2383–2389. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Ha, K.S.; Uhm, H.; Park, K.; Lee, J.Y.; Hohng, S.; Kang, C. Transcription reinitiation by recycling RNA polymerase that diffuses on DNA after releasing terminated RNA. Nat. Commun. 2020, 11, 450. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A., III; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343-U41. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef]
- Berrow, N.S.; Alderton, D.; Sainsbury, S.; Nettleship, J.; Assenberg, R.; Rahman, N.; Stuart, D.I.; Owens, R.J. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007, 35, 12. [Google Scholar] [CrossRef]
- Ma, H.; Kunes, S.; Schatz, P.J.; Botstein, D. Plasmid construction by homologous recombination in yeast. Gene 1987, 58, 201–216. [Google Scholar] [CrossRef]
- MacPherson, M.; Saka, Y. Short Synthetic Terminators for Assembly of Transcription Units in Vitro and Stable Chromosomal Integration in Yeast S. cerevisiae. ACS Synth. Biol. 2017, 6, 130–138. [Google Scholar] [CrossRef]
- Purnick, P.E.; Weiss, R. The second wave of synthetic biology: From modules to systems. Nat. Rev. Mol. Cell Biol. 2009, 10, 410–422. [Google Scholar] [CrossRef]
- MacDonald, J.T.; Barnes, C.; Kitney, R.I.; Freemont, P.S.; Stan, G.-B.V. Computational design approaches and tools for synthetic biology. Integr. Biol. 2011, 3, 97–108. [Google Scholar] [CrossRef]
- Basehoar, A.D.; Zanton, S.J.; Pugh, B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004, 117, 847. [Google Scholar] [CrossRef]
- Lee, S.; Hahn, S. Model for binding of transcription factor TFIIB to the TBP-DNA complex. Nature 1995, 376, 609–612. [Google Scholar] [CrossRef]
- Lagrange, T.; Kim, T.K.; Orphanides, G.; Ebright, Y.W.; Ebright, R.H.; Reinberg, D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: Organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl. Acad. Sci. USA 1996, 93, 10620–10625. [Google Scholar] [CrossRef]
- Cramer, P. Organization and regulation of gene transcription. Nature 2019, 573, 45–54. [Google Scholar] [CrossRef]
- Kadonaga, J.T. The DPE, a core promoter element for transcription by RNA polymerase II. Exp. Mol. Med. 2002, 34, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.Y.; Santoso, B.; Boulay, T.; Dong, E.; Ohler, U.; Kadonaga, J.T. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004, 18, 1606–1617. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Alper, H.S. Promoter engineering: Recent advances in controlling transcription at the most fundamental level. Biotechnol. J. 2013, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Juven-Gershon, T.; Kadonaga, J.T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010, 339, 225–229. [Google Scholar] [CrossRef]
- van Helden, J.; del Olmo, M.; Perez-Ortin, J.E. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 2000, 28, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Argaman, L.; Hershberg, R.; Vogel, J.; Bejerano, G.; Wagner, E.G.H.; Margalit, H.; Altuvia, S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001, 11, 941–950. [Google Scholar] [CrossRef]
- Curran, K.A.; Morse, N.J.; Markham, K.A.; Wagman, A.M.; Gupta, A.; Alper, H.S. Short Synthetic Terminators for Improved Heterologous Gene Expression in Yeast. ACS Synth. Biol. 2015, 4, 824–832. [Google Scholar] [CrossRef]
- Dye, M.J.; Proudfoot, N.J. Terminal exon definition occurs contrascriptionally and promotes termination of RNA polymerase II. Mol. Cell 1999, 3, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, S.; Young, E.M.; Jones, T.S.; Densmore, D.; Voigt, C.A. Genetic circuit design automation for yeast. Nat. Microbiol. 2020, 5, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Blount, B.; Weenink, T.; Vasylechko, S.; Ellis, T. Rational Diversification of a Promoter Providing Fine-Tuned Expression and Orthogonal Regulation for Synthetic Biology. PLoS ONE 2012, 7, e33279. [Google Scholar] [CrossRef]
- Kaiser, C.; Michaelis, S.; Mitchell, A. Methods in Yeast Genetics: A Cold Spring Harbor Course Manual; CHSL Press: Plainview, NY, USA, 1994. [Google Scholar]
- Raser, J.M.; O’Shea, E.K. Control of stochasticity in eukaryotic gene expression. Science 2004, 304, 1811–1814. [Google Scholar] [CrossRef]
- Harbison, C.T.; Gordon, D.B.; Lee, T.I.; Rinaldi, N.J.; Macisaac, K.D.; Danford, T.W.; Hannett, N.M.; Tagne, J.B.; Reynolds, D.B.; Yoo, J.; et al. Transcriptional regulatory code of a eukaryotic genome. Nature 2004, 431, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.Z.; Killion, P.J.; Iyer, V.R. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 2007, 39, 683–687. [Google Scholar] [CrossRef]
- Casini, A.; Christodoulou, G.; Freemont, P.S.; Baldwin, G.S.; Ellis, T.; MacDonald, J.T. R2oDNA Designer: Computational Design of Biologically Neutral Synthetic DNA Sequences. ACS Synth. Biol. 2014, 3, 525–528. [Google Scholar] [CrossRef]
- Zhang, G.; Cao, Q.; Liu, J.; Liu, B.; Li, J.; Li, C. Refactoring β-amyrin synthesis in Saccharomyces cerevisiae. AIChE J. 2015, 61, 3172–3179. [Google Scholar] [CrossRef]
- Li, X.; Wang, Z.; Zhang, G.; Yi, L. Improving lycopene production in Saccharomyces cerevisiae through optimizing pathway and chassis metabolism. Chem. Eng. Sci. 2019, 193, 364–369. [Google Scholar] [CrossRef]
- Wei, L.; Wang, Z.; Zhang, G.; Ye, B. Characterization of Terminators in Saccharomyces cerevisiae and an Exploration of Factors Affecting Their Strength. ChemBioChem 2017, 18, 2422–2427. [Google Scholar] [CrossRef]
- Cambray, G.; Guimaraes, J.C.; Mutalik, V.K.; Lam, C.; Mai, Q.-A.; Thimmaiah, T.; Carothers, J.M.; Arkin, A.P.; Endy, D. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 2013, 41, 5139–5148. [Google Scholar] [CrossRef] [PubMed]
- Yamanishi, M.; Ito, Y.; Kintaka, R.; Imamura, C.; Katahira, S.; Ikeuchi, A.; Moriya, H.; Matsuyama, T. A Genome-Wide Activity Assessment of Terminator Regions in Saccharomyces cerevisiae Provides a “Terminatome” Toolbox. ACS Synth. Biol. 2013, 2, 337–347. [Google Scholar] [CrossRef] [PubMed]
- West, S.; Proudfoot, N.J. Transcriptional Termination Enhances Protein Expression in Human Cells. Mol. Cell 2009, 33, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Jing, L.; Qiang, L. Can terminators be used as insulators into yeast synthetic gene circuits? J. Biol. Eng. 2016, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Mogno, I.; Vallania, F.; Mitra, R.D.; Cohen, B.A. TATA is a modular component of synthetic promoters. Genome Res. 2010, 20, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, F.; Kasahara, K.; Kokubo, T. Highly redundant function of multiple AT-rich sequences as core promoter elements in the TATA-less RPS5 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 2011, 39, 59–75. [Google Scholar] [CrossRef]
- Misawa, N.; Nakagawa, M.; Kobayashi, K.; Yamano, S.; Izawa, Y.; Nakamura, K.; Harashima, K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 1990, 172, 6704–6712. [Google Scholar] [CrossRef]
- Lubliner, S.; Keren, L.; Segal, E. Sequence features of yeast and human core promoters that are predictive of maximal promoter activity. Nucleic Acids Res. 2013, 41, 5569–5581. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).