Prognostic Impact of Dihydropyrimidine Dehydrogenase Germline Variants in Unresectable Non-Small Cell Lung Cancer Patients Treated with Platin-Based Chemotherapy
Abstract
1. Introduction
2. Results
2.1. Patients’ Characteristics
2.2. Efficacy Outcomes
2.3. Toxicity Outcomes
3. Discussion
4. Materials and Methods
4.1. Genotyping Studies
4.2. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef] [PubMed]
- Bjerring, A.W.; Fossa, S.D.; Haugnes, H.S.; Nome, R.; Stokke, T.M.; Haugaa, K.H.; Kiserud, C.E.; Edvardsen, T.; Sarvari, I. The cardiac impact of cisplatin-based chemotherapy in survivors of testicular cancer: A 30-year follow-up. Eur. Heart J. Cardiovasc. Imaging 2021, 22, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.R.; Vallis, K.A. Transition metal compounds as cancer radiosensitizers. Chem. Soc. Rev. 2019, 48, 540–557. [Google Scholar] [CrossRef] [PubMed]
- Trendowski, M.R.; El Charif, O.; Dinh, P.C., Jr.; Travis, L.B.; Dolan, M.E. Genetic and Modifiable Risk Factors Contributing to Cisplatin-induced Toxicities. Clin. Cancer Res. 2019, 25, 1147–1155. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, C.; Gao, X.; Yao, Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics 2022, 12, 2115–2132. [Google Scholar] [CrossRef] [PubMed]
- Muggia, F.M.; Bonetti, A.; Hoeschele, J.D.; Rozencweig, M.; Howell, S.B. Platinum Antitumor Complexes: 50 Years Since Barnett Rosenberg’s Discovery. J. Clin. Oncol. 2015, 33, 4219–4226. [Google Scholar] [CrossRef]
- Qi, L.; Luo, Q.; Zhang, Y.; Jia, F.; Zhao, Y.; Wang, F. Advances in Toxicological Research of the Anticancer Drug Cisplatin. Chem. Res. Toxicol. 2019, 32, 1469–1486. [Google Scholar] [CrossRef]
- Chong, S.X.; Au-Yeung, S.C.; To, K.K. Monofunctional Platinum (PtII) Compounds—Shifting the Paradigm in Designing New Pt-based Anticancer Agents. Curr. Med. Chem. 2016, 23, 1268–1285. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Deo, S.V.S.; Sharma, J.; Kumar, S. GLOBOCAN 2020 Report on Global Cancer Burden: Challenges and Opportunities for Surgical Oncologists. Ann. Surg. Oncol. 2022, 29, 6497–6500. [Google Scholar] [CrossRef]
- Nooreldeen, R.; Bach, H. Current and Future Development in Lung Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 8661. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.L.; Zhang, A.L.; Bruno, D.S.; Almeida, F.A. NSCLC in the Era of Targeted and Immunotherapy: What Every Pulmonologist Must Know. Diagnostics 2023, 13, 1117. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, L.E.L.; Menis, J.; Remon, J. First-line immune-chemotherapy combination for squamous NSCLC is already a reality. Transl. Lung Cancer Res. 2020, 9, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; Facchinetti, F.; Besse, B. The efficacy of immune checkpoint inhibitors in thoracic malignancies. Eur. Respir. Rev. 2021, 30, 200387. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Rodriguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Reck, M.; Remon, J.; Hellmann, M.D. First-Line Immunotherapy for Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2022, 40, 586–597. [Google Scholar] [CrossRef]
- Novello, S.; Kowalski, D.M.; Luft, A.; Gumus, M.; Vicente, D.; Mazieres, J.; Rodriguez-Cid, J.; Tafreshi, A.; Cheng, Y.; Lee, K.H.; et al. Pembrolizumab Plus Chemotherapy in Squamous Non-Small-Cell Lung Cancer: 5-Year Update of the Phase III KEYNOTE-407 Study. J. Clin. Oncol. 2023, 41, 1999–2006. [Google Scholar] [CrossRef]
- Lee, J.W.; Pussegoda, K.; Rassekh, S.R.; Monzon, J.G.; Liu, G.; Hwang, S.; Bhavsar, A.P.; Pritchard, S.; Ross, C.J.; Amstutz, U.; et al. Clinical Practice Recommendations for the Management and Prevention of Cisplatin-Induced Hearing Loss Using Pharmacogenetic Markers. Ther. Drug Monit. 2016, 38, 423–431. [Google Scholar] [CrossRef]
- Khrunin, A.V.; Moisseev, A.; Gorbunova, V.; Limborska, S. Genetic polymorphisms and the efficacy and toxicity of cisplatin-based chemotherapy in ovarian cancer patients. Pharm. J. 2010, 10, 54–61. [Google Scholar] [CrossRef]
- Chang, C.; Hu, Y.; Hogan, S.L.; Mercke, N.; Gomez, M.; O’Bryant, C.; Bowles, D.W.; George, B.; Wen, X.; Aleksunes, L.M.; et al. Pharmacogenomic Variants May Influence the Urinary Excretion of Novel Kidney Injury Biomarkers in Patients Receiving Cisplatin. Int. J. Mol. Sci. 2017, 18, 1333. [Google Scholar] [CrossRef]
- Windsor, R.E.; Strauss, S.J.; Kallis, C.; Wood, N.E.; Whelan, J.S. Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: A pilot study. Cancer 2012, 118, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Lim, H.S.; Shin, E.S.; Yoo, Y.K.; Park, Y.H.; Lee, J.E.; Jang, I.J.; Lee, D.H.; Lee, J.S. Comprehensive analysis of UGT1A polymorphisms predictive for pharmacokinetics and treatment outcome in patients with non-small-cell lung cancer treated with irinotecan and cisplatin. J. Clin. Oncol. 2006, 24, 2237–2244. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.H.; Yu, Z.; Zhang, T.T.; Shin, M.H.; Kim, H.N.; Choi, J.S. Influence of polymorphisms in MTHFR 677 C→T, TYMS 3R→2R and MTR 2756 A→G on NSCLC risk and response to platinum-based chemotherapy in advanced NSCLC. Pharmacogenomics 2011, 12, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Sosa Iglesias, V.; Giuranno, L.; Dubois, L.J.; Theys, J.; Vooijs, M. Drug Resistance in Non-Small Cell Lung Cancer: A Potential for NOTCH Targeting? Front. Oncol. 2018, 8, 267. [Google Scholar] [CrossRef]
- Kryczka, J.; Kryczka, J.; Czarnecka-Chrebelska, K.H.; Brzezianska-Lasota, E. Molecular Mechanisms of Chemoresistance Induced by Cisplatin in NSCLC Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 8885. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, C.Y.; Duan, F.G.; Fan, X.X.; Yao, X.J.; Parks, R.J.; Tang, Y.J.; Wang, M.F.; Liu, L.; Tsang, B.K.; et al. p53 sensitizes chemoresistant non-small cell lung cancer via elevation of reactive oxygen species and suppression of EGFR/PI3K/AKT signaling. Cancer Cell Int. 2019, 19, 188. [Google Scholar] [CrossRef]
- Perez-Ramirez, C.; Canadas-Garre, M.; Alnatsha, A.; Villar, E.; Valdivia-Bautista, J.; Faus-Dader, M.J.; Calleja-Hernandez, M.A. Pharmacogenetics of platinum-based chemotherapy: Impact of DNA repair and folate metabolism gene polymorphisms on prognosis of non-small cell lung cancer patients. Pharm. J. 2019, 19, 164–177. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Zou, T.; Cui, J.; Yin, J.; Zheng, W.; Jiang, W.; Zhou, H.; Liu, Z. Pharmacogenomics of platinum-based chemotherapy response in NSCLC: A genotyping study and a pooled analysis. Oncotarget 2016, 7, 55741–55756. [Google Scholar] [CrossRef]
- Yin, J.Y.; Huang, Q.; Zhao, Y.C.; Zhou, H.H.; Liu, Z.Q. Meta-analysis on pharmacogenetics of platinum-based chemotherapy in non small cell lung cancer (NSCLC) patients. PLoS ONE 2012, 7, e38150. [Google Scholar] [CrossRef]
- Smit, E.F.; Burgers, S.A.; Biesma, B.; Smit, H.J.; Eppinga, P.; Dingemans, A.M.; Joerger, M.; Schellens, J.H.; Vincent, A.; van Zandwijk, N.; et al. Randomized phase II and pharmacogenetic study of pemetrexed compared with pemetrexed plus carboplatin in pretreated patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 2009, 27, 2038–2045. [Google Scholar] [CrossRef]
- Takano, M.; Sugiyama, T. UGT1A1 polymorphisms in cancer: Impact on irinotecan treatment. Pharmgenom. Pers. Med. 2017, 10, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Yu, H.; Lu, L.; Jain, D.; Kidd, M.S.; Saif, M.W.; Chanock, S.J.; Hartge, P.; PanScan, C.; Risch, H.A. Genetic effects and modifiers of radiotherapy and chemotherapy on survival in pancreatic cancer. Pancreas 2011, 40, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, A.; Ruwali, M.; Pant, M.C.; Rahman, Q.; Parmar, D. Association of genetic variability in enzymes metabolizing chemotherapeutic agents with treatment response in head and neck cancer cases. Asia Pac. J. Clin. Oncol. 2017, 13, e11–e20. [Google Scholar] [CrossRef]
- Amstutz, U.; Henricks, L.M.; Offer, S.M.; Barbarino, J.; Schellens, J.H.M.; Swen, J.J.; Klein, T.E.; McLeod, H.L.; Caudle, K.E.; Diasio, R.B.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 210–216. [Google Scholar] [CrossRef]
- Diasio, R.B.; Offer, S.M. Testing for Dihydropyrimidine Dehydrogenase Deficiency to Individualize 5-Fluorouracil Therapy. Cancers 2022, 14, 3207. [Google Scholar] [CrossRef]
- Lunenburg, C.; van der Wouden, C.H.; Nijenhuis, M.; Crommentuijn-van Rhenen, M.H.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; Mulder, H.; Rongen, G.A.; van Schaik, R.H.N.; et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction of DPYD and fluoropyrimidines. Eur. J. Hum. Genet. 2020, 28, 508–517. [Google Scholar] [CrossRef] [PubMed]
- van Kuilenburg, A.B.; Stroomer, A.E.; Abeling, N.G.; van Gennip, A.H. A pivotal role for beta-aminoisobutyric acid and oxidative stress in dihydropyrimidine dehydrogenase deficiency? Nucleosides Nucleotides Nucleic Acids 2006, 25, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, R.; Furukawa, T.; Mitsuo, R.; Noguchi, T.; Kitazono, M.; Okumura, H.; Sumizawa, T.; Haraguchi, M.; Che, X.F.; Uchimiya, H.; et al. Thymidine phosphorylase inhibits apoptosis induced by cisplatin. Biochem. Biophys. Res. Commun. 2003, 301, 358–363. [Google Scholar] [CrossRef]
- Takizawa, M.; Kawakami, K.; Obata, T.; Matsumoto, I.; Ohta, Y.; Oda, M.; Sasaki, T.; Watanabe, G. In vitro sensitivity to platinum-derived drugs is associated with expression of thymidylate synthase and dihydropyrimidine dehydrogenase in human lung cancer. Oncol. Rep. 2006, 15, 1533–1539. [Google Scholar] [CrossRef]
- Huang, M.Y.; Wu, C.H.; Huang, C.M.; Chung, F.Y.; Huang, C.W.; Tsai, H.L.; Chen, C.F.; Lin, S.R.; Wang, J.Y. DPYD, TYMS, TYMP, TK1, and TK2 genetic expressions as response markers in locally advanced rectal cancer patients treated with fluoropyrimidine-based chemoradiotherapy. BioMed Res. Int. 2013, 2013, 931028. [Google Scholar] [CrossRef]
- Goto, T.; Shinmura, K.; Yokomizo, K.; Sakuraba, K.; Kitamura, Y.; Shirahata, A.; Saito, M.; Kigawa, G.; Nemoto, H.; Sanada, Y.; et al. Expression levels of thymidylate synthase, dihydropyrimidine dehydrogenase, and thymidine phosphorylase in patients with colorectal cancer. Anticancer Res. 2012, 32, 1757–1762. [Google Scholar]
- Vera-Puente, O.; Rodriguez-Antolin, C.; Salgado-Figueroa, A.; Michalska, P.; Pernia, O.; Reid, B.M.; Rosas, R.; Garcia-Guede, A.; SacristAn, S.; Jimenez, J.; et al. MAFG is a potential therapeutic target to restore chemosensitivity in cisplatin-resistant cancer cells by increasing reactive oxygen species. Transl. Res. 2018, 200, 1–17. [Google Scholar] [CrossRef]
- Cortes-Sempere, M.; de Miguel, M.P.; Pernia, O.; Rodriguez, C.; de Castro Carpeno, J.; Nistal, M.; Conde, E.; Lopez-Rios, F.; Belda-Iniesta, C.; Perona, R.; et al. IGFBP-3 methylation-derived deficiency mediates the resistance to cisplatin through the activation of the IGFIR/Akt pathway in non-small cell lung cancer. Oncogene 2013, 32, 1274–1283. [Google Scholar] [CrossRef] [PubMed]
- Vera, O.; Rodriguez-Antolin, C.; de Castro, J.; Karreth, F.A.; Sellers, T.A.; Ibanez de Caceres, I. An epigenomic approach to identifying differential overlapping and cis-acting lncRNAs in cisplatin-resistant cancer cells. Epigenetics 2018, 13, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Tserga, E.; Nandwani, T.; Edvall, N.K.; Bulla, J.; Patel, P.; Canlon, B.; Cederroth, C.R.; Baguley, D.M. The genetic vulnerability to cisplatin ototoxicity: A systematic review. Sci. Rep. 2019, 9, 3455. [Google Scholar] [CrossRef] [PubMed]
- Pussegoda, K.; Ross, C.J.; Visscher, H.; Yazdanpanah, M.; Brooks, B.; Rassekh, S.R.; Zada, Y.F.; Dube, M.P.; Carleton, B.C.; Hayden, M.R.; et al. Replication of TPMT and ABCC3 genetic variants highly associated with cisplatin-induced hearing loss in children. Clin. Pharmacol. Ther. 2013, 94, 243–251. [Google Scholar] [CrossRef]
- Thiesen, S.; Yin, P.; Jorgensen, A.L.; Zhang, J.E.; Manzo, V.; McEvoy, L.; Barton, C.; Picton, S.; Bailey, S.; Brock, P.; et al. TPMT, COMT and ACYP2 genetic variants in paediatric cancer patients with cisplatin-induced ototoxicity. Pharm. Genom. 2017, 27, 213–222. [Google Scholar] [CrossRef]
- Hagleitner, M.M.; Coenen, M.J.; Patino-Garcia, A.; de Bont, E.S.; Gonzalez-Neira, A.; Vos, H.I.; van Leeuwen, F.N.; Gelderblom, H.; Hoogerbrugge, P.M.; Guchelaar, H.J.; et al. Influence of genetic variants in TPMT and COMT associated with cisplatin induced hearing loss in patients with cancer: Two new cohorts and a meta-analysis reveal significant heterogeneity between cohorts. PLoS ONE 2014, 9, e115869. [Google Scholar] [CrossRef]
- Yang, J.J.; Lim, J.Y.; Huang, J.; Bass, J.; Wu, J.; Wang, C.; Fang, J.; Stewart, E.; Harstead, E.H.; E, S.; et al. The role of inherited TPMT and COMT genetic variation in cisplatin-induced ototoxicity in children with cancer. Clin. Pharmacol. Ther. 2013, 94, 252–259. [Google Scholar] [CrossRef]
- Bjorn, N.; Sigurgeirsson, B.; Svedberg, A.; Pradhananga, S.; Branden, E.; Koyi, H.; Lewensohn, R.; de Petris, L.; Apellaniz-Ruiz, M.; Rodriguez-Antona, C.; et al. Genes and variants in hematopoiesis-related pathways are associated with gemcitabine/carboplatin-induced thrombocytopenia. Pharm. J. 2020, 20, 179–191. [Google Scholar] [CrossRef]
- Kappenberg-Nitescu, D.C.; Luchian, I.; Martu, I.; Solomon, S.M.; Martu, S.; Pasarin, L.; Martu, A.; Sioustis, I.A.; Goriuc, A.; Tatarciuc, M. Periodontal effects of two innovative oral rinsing substances in oncologic patients. Exp. Ther. Med. 2021, 21, 98. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Alonso, R.; Queiruga, J.; Arias, P.; Del Monte, A.; Yuste, F.; Rodriguez-Antolin, C.; Losantos-Garcia, I.; Borobia, A.M.; Rodriguez-Novoa, S. Analytical validation of a laboratory-development multigene pharmacogenetic assay. Pharm. Genom. 2021, 31, 177–184. [Google Scholar] [CrossRef] [PubMed]

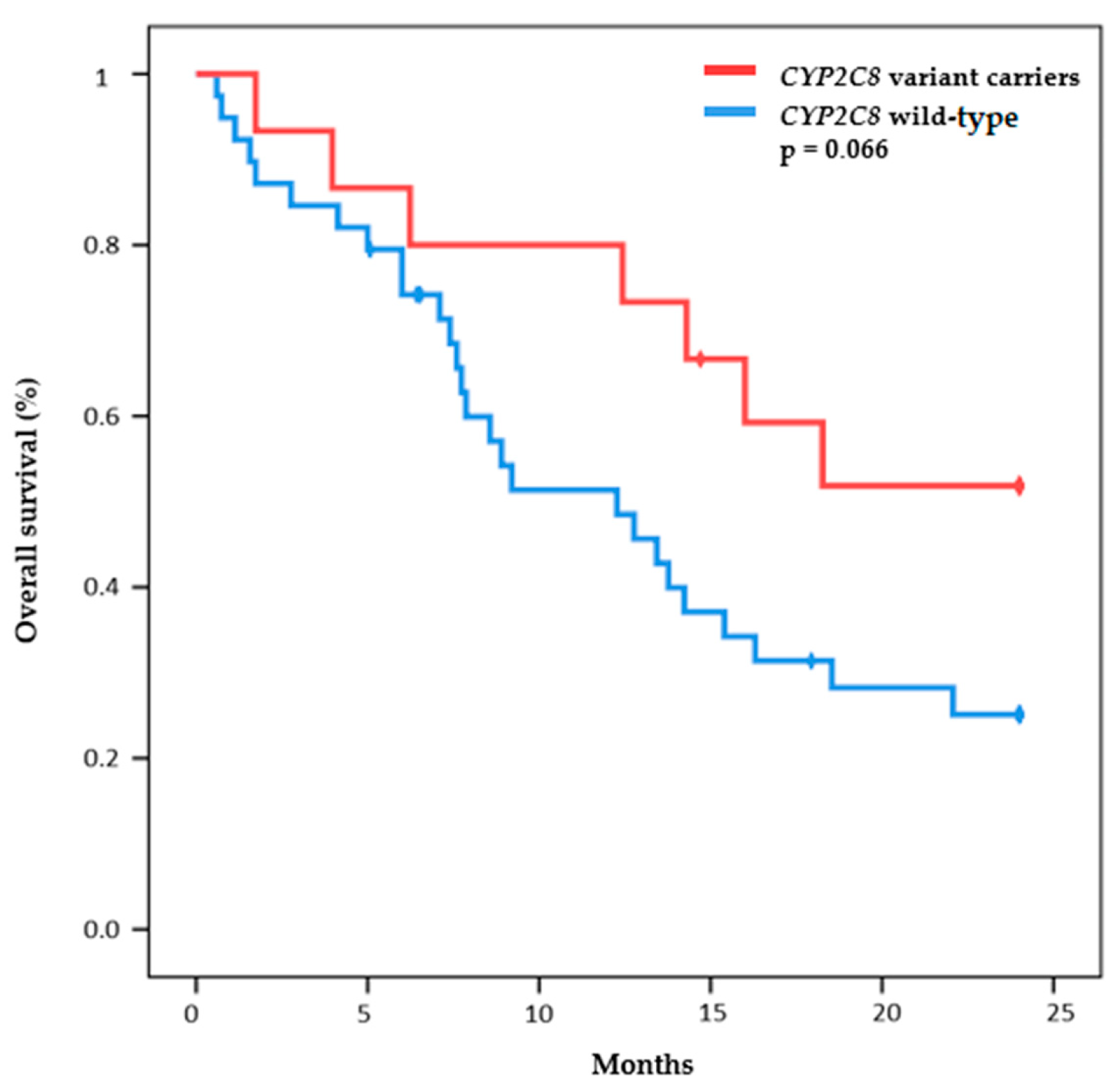
| n = 55 | |||
|---|---|---|---|
| Cisplatin-Based Chemotherapy (n = 22) | Carboplatin-Based Chemotherapy (n = 33) | ||
| Age (Average and range) | 61 (25–81) | ||
| Gender | |||
| Female | 20 (36%) | 12 | 8 |
| Male | 35 (64%) | 10 | 25 |
| Histological type | |||
| Adenocarcinoma | 34 (60%) | 15 | 19 |
| Squamous cell carcinoma | 17 (27.3%) | 6 | 11 |
| Other NSCLC | 4 (12.7%) | 1 | 3 |
| Disease Stage | |||
| IIIA | 5 (9.1%) | 4 | 1 |
| IIIB | 19 (34.5%) | 9 | 10 |
| IIIC | 1 (1.8%) | 0 | 1 |
| IV | 30 (54.5%) | 9 | 21 |
| Smoking status | |||
| Non-smoker | 4 (7.3%) | 1 | 3 |
| Ex-smoker | 30 (54.5%) | 12 | 18 |
| Smoker | 21 (38.2%) | 9 | 12 |
| ECOG performance status | |||
| 0 | 36 (65.5%) | 12 | 24 |
| 1 | 16 (29.1%) | 9 | 7 |
| 2 | 2 (3.6%) | 1 | 1 |
| 3 | 1 (1.8%) | 0 | 1 |
| n: 55 | |||||
|---|---|---|---|---|---|
| Gene | Status | Toxicity | Toxicity Grade ≥ 2 | ||
| Yes | No | Yes | No | ||
| UGT1A1 | WT * | 21 (84%) | 4 (16%) | 6 (24%) | 19 (76%) |
| Variant carrier | 23 (82.1) | 5 (17.9%) | 10 (35.7%) | 18 (64.3%) | |
| MTHFR | WT | 21 (84%) | 4 (16%) | 7 (28%) | 18 (72%) |
| Variant carrier | 23 (82.1%) | 5 (17.9%) | 9 (32.1%) | 19 (67.9%) | |
| CYP2C19 | WT | 21 (80.8%) | 5 (19.2%) | 17 (65.4%) | 9 (34.6%) |
| Variant carrier | 23 (82.1%) | 5 (17.9%) | 7 (25%) | 21 (75%) | |
| CYP3A5 | WT | 37 (84.1%) | 7 (15.9%) | 15 (34.1%) | 29 (65.9%) |
| Variant carrier | 7 (70%) | 3 (30%) | 1 (10%) | 9 (90%) | |
| CYP3A4 | WT | 39 (84.8%) | 7 (15.2%) | 13 (28.3%) | 33 (71.7%) |
| Variant carrier | 5 (71.4%) | 2 (28.6%) | 3 (42.9%) | 3 (57.1%) | |
| CYP2C9 | WT | 25 (78.1%) | 7 (21.9%) | 9 (28.1%) | 23 (71.9%) |
| Variant carrier | 19 (86.4%) | 3 (13.6%) | 7 (31.8%) | 15 (68.2%) | |
| CYP2C8 | WT | 32 (82.1%) | 7 (17.9%) | 14 (35.9%) | 25 (64.1%) |
| Variant carrier | 13 (81.3%) | 3 (18.7%) | 3 (18.8%) | 13 (81.3%) | |
| DPYD | WT | 39 (81.3%) | 9 (18.8%) | 15 (31.3%) | 33 (68.8%) |
| Variant carrier | 1 (16.7%) | 5 (83.3%) | 1 (16.7%) | 5 (83.3%) | |
| TPMT | WT | 41 (82%) | 9 (18%) | 17 (34%) | 33 (66%) |
| Variant carrier | 4 (80%) | 1 (20%) | 0 (0%) | 5 (100%) | |
| NUDT15 | WT | 44 (81.5%) | 10 (18.5%) | 17 (31.5%) | 37 (68.5%) |
| Variant carrier | 1 (100%) | 0 (0%) | 0 (0%) | 1 (100%) | |
| TFAP2B | WT | 33 (86.8%) | 5 (13.2%) | 11 (28.9%) | 27 (71.1%) |
| Variant carrier | 12 (70.6%) | 5 (29.4%) | 6 (35.3%) | 11 (64.7%) | |
| EPAS1 | WT | 25 (83.3%) | 5 (16.7%) | 8 (26.7%) | 22 (73.3%) |
| Variant carrier | 19 (79.2%) | 5 (20.8%) | 8 (33.3%) | 16 (66.7%) | |
| GM2A | WT | 42 (80.8%) | 10 (19.2%) | 15 (28.8%) | 37 (71.2%) |
| Variant carrier | 3 (100%) | 0 (0%) | 2 (66.7%) | 1 (33.3%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guijarro-Eguinoa, J.; Arjona-Hernandez, S.; Stewart, S.; Pernia, O.; Arias, P.; Losantos-García, I.; Rubio, T.; Burdiel, M.; Rodriguez-Antolin, C.; Cruz-Castellanos, P.; et al. Prognostic Impact of Dihydropyrimidine Dehydrogenase Germline Variants in Unresectable Non-Small Cell Lung Cancer Patients Treated with Platin-Based Chemotherapy. Int. J. Mol. Sci. 2023, 24, 9843. https://doi.org/10.3390/ijms24129843
Guijarro-Eguinoa J, Arjona-Hernandez S, Stewart S, Pernia O, Arias P, Losantos-García I, Rubio T, Burdiel M, Rodriguez-Antolin C, Cruz-Castellanos P, et al. Prognostic Impact of Dihydropyrimidine Dehydrogenase Germline Variants in Unresectable Non-Small Cell Lung Cancer Patients Treated with Platin-Based Chemotherapy. International Journal of Molecular Sciences. 2023; 24(12):9843. https://doi.org/10.3390/ijms24129843
Chicago/Turabian StyleGuijarro-Eguinoa, Javier, Sara Arjona-Hernandez, Stefan Stewart, Olga Pernia, Pedro Arias, Itsaso Losantos-García, Tania Rubio, Miranda Burdiel, Carlos Rodriguez-Antolin, Patricia Cruz-Castellanos, and et al. 2023. "Prognostic Impact of Dihydropyrimidine Dehydrogenase Germline Variants in Unresectable Non-Small Cell Lung Cancer Patients Treated with Platin-Based Chemotherapy" International Journal of Molecular Sciences 24, no. 12: 9843. https://doi.org/10.3390/ijms24129843
APA StyleGuijarro-Eguinoa, J., Arjona-Hernandez, S., Stewart, S., Pernia, O., Arias, P., Losantos-García, I., Rubio, T., Burdiel, M., Rodriguez-Antolin, C., Cruz-Castellanos, P., Higuera, O., Borobia, A. M., Rodriguez-Novoa, S., de Castro-Carpeño, J., Ibanez de Caceres, I., & Rosas-Alonso, R. (2023). Prognostic Impact of Dihydropyrimidine Dehydrogenase Germline Variants in Unresectable Non-Small Cell Lung Cancer Patients Treated with Platin-Based Chemotherapy. International Journal of Molecular Sciences, 24(12), 9843. https://doi.org/10.3390/ijms24129843





