The rs16969968 Tobacco Smoking-Related Single-Nucleotide Variant Is Associated with Clinical Markers in Patients with Severe COVID-19
Abstract
1. Introduction
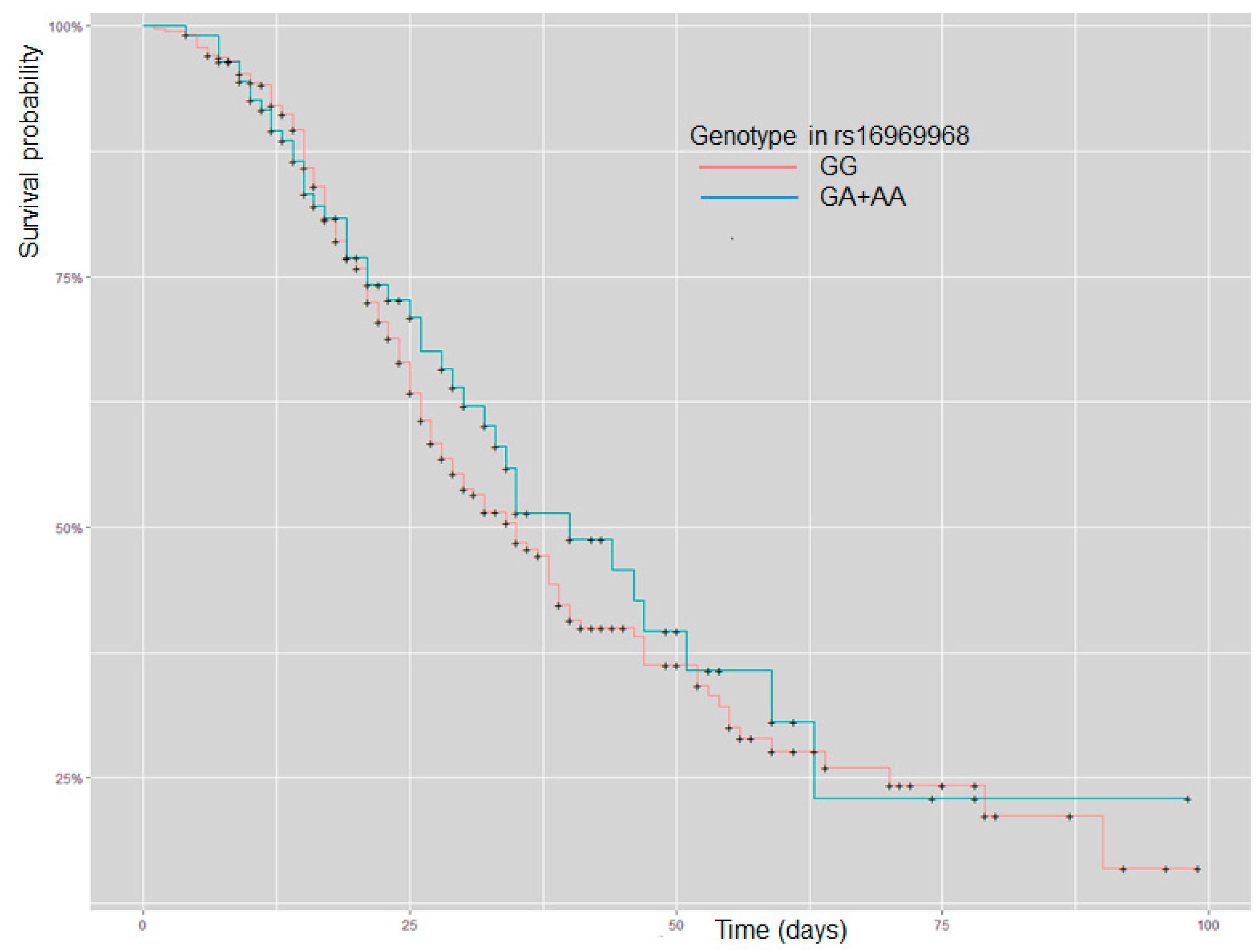
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; Elmahi, E.; et al. Dexamethasone in Hospitalized Patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [PubMed]
- Münzel, T.; Hahad, O.; Kuntic, M.; Keaney, J.F.; Deanfield, J.E.; Daiber, A. Effects of tobacco cigarettes, e-cigarettes, and waterpipe smoking on endothelial function and clinical outcomes. Eur. Heart J. 2020, 41, 4057–4070. [Google Scholar] [CrossRef] [PubMed]
- Luiz, C.; Almeida-Da-Silva, C.; Dakafay, H.M.; Liu, K.; Ojcius, D.M. Cigarette Smoke Stimulates SARS-CoV-2 Internalization by Activating AhR and Increasing ACE2 Expression in Human Gingival Epithelial Cells. Int. J. Mol. Sci. 2021, 22, 7669. [Google Scholar]
- Pérez-Morales, R.; González-Zamora, A.; González-Delgado, M.F.; Calleros Rincon, E.Y.; Olivas Calderon, E.H.; Martínez-Ramírez, O.C.; Rubio, J. CHRNA3 rs1051730 and CHRNA5 rs16969968 polymorphisms are associated with heavy smoking, lung cancer, and chronic obstructive pulmonary disease in a mexican population. Ann. Hum. Genet. 2018, 82, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Caliri, A.W.; Tommasi, S.; Besaratinia, A. Relationships among smoking, oxidative stress, inflammation, macromolecular damage, and cancer. Mutat. Res./Rev. Mutat. Res. 2021, 787, 108365. [Google Scholar] [CrossRef]
- Arcavi, L.; Benowitz, N.L. Cigarette Smoking and Infection. Arch. Intern. Med. 2004, 164, 2206–2216. [Google Scholar] [CrossRef]
- Lugg, S.T.; Scott, A.; Parekh, D.; Naidu, B.; Thickett, D.R. Cigarette smoke exposure and alveolar macrophages: Mechanisms for lung disease. Thorax 2022, 77, 94–101. [Google Scholar] [CrossRef]
- Gaiha, S.M.; Cheng, J.; Halpern-Felsher, B. Association between Youth Smoking, Electronic Cigarette Use, and COVID-19. J. Adolesc. Health 2020, 67, 519–523. [Google Scholar] [CrossRef]
- Fernández-Trujillo, L.; Sanchez, S.; Sangiovanni-Gonzalez, S.; Morales, E.; Velásquez, M.; Sua, L. Endobronchial ultrasound-guided Transbronchial needle aspiration (EBUS-TBNA) in simulator lesions of pulmonary pathology: A case report of pulmonary Myospherulosis. BMC Pulm. Med. 2020, 20, 246. [Google Scholar] [CrossRef]
- Pérez-Rubio, G.; Falfán-Valencia, R.; Fernández-López, J.C.; Ramírez-Venegas, A.; Hernández-Zenteno, R.D.J.; Flores-Trujillo, F.; Silva-Zolezzi, I. Diagnostics Genetic Factors Associated with COPD Depend on the Ancestral Caucasian/Amerindian Component in the Mexican Population. Diagnostics 2021, 11, 599. [Google Scholar] [CrossRef]
- Diabasana, Z.; Perotin, J.-M.; Belgacemi, R.; Ancel, J.; Mulette, P.; Launois, C.; Delepine, G.; Dubernard, X.; Mérol, J.C.; Ruaux, C.; et al. Chr15q25 Genetic Variant rs16969968 Alters Cell Differentiation in Respiratory Epithelia. Int. J. Mol. Sci. 2021, 22, 6657. [Google Scholar] [CrossRef] [PubMed]
- Improgo, M.R.D.; Scofield, M.D.; Tapper, A.R.; Gardner, P.D. The nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster: Dual role in nicotine addiction and lung cancer. Prog. Neurobiol. 2010, 92, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Busch, R.; Hobbs, B.D.; Zhou, J.; Castaldi, P.J.; McGeachie, M.J.; Hardin, M.E.; Hawrylkiewicz, I.; Sliwinski, P.; Yim, J.J.; Kim, W.J.; et al. Genetic association and risk scores in a chronic obstructive pulmonary disease meta-analysis of 16,707 subjects. Am. J. Respir. Cell Mol. Biol. 2017, 57, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Krais, A.M.; Hautefeuille, A.H.; Cros, M.P.; Krutovskikh, V.; Tournier, J.M.; Birembaut, P.; Thépot, A.; Paliwal, A.; Herceg, Z.; Boffetta, P.; et al. CHRNA5 as negative regulator of nicotine signaling in normal and cancer bronchial cells: Effects on motility, migration and p63 expression. Carcinogenesis 2011, 32, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.-C.; Zhou, C.-F.; Toloo, S.; Shen, T.; Tong, S.-L.; Zhu, Q.-X. Association of a disintegrin and metalloprotease 33 (ADAM33) gene polymorphisms with the risk of COPD: An updated meta-analysis of 2,644 cases and 4,804 controls. Mol. Biol. Rep. 2015, 42, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Wei, X.; Gong, C.; Wei, J.; Chen, Z.; Deng, J. A disintegrin and metalloprotease 33 (ADAM33) gene polymorphisms and the risk of asthma: A meta-analysis. Hum. Immunol. 2013, 74, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Anastassopoulou, C.; Gkizarioti, Z.; Patrinos, G.P.; Tsakris, A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum. Genom. 2020, 14, 40. [Google Scholar] [CrossRef]
- SeyedAlinaghi, S.A.; Mehrtak, M.; MohsseniPour, M.; Mirzapour, P.; Barzegary, A.; Habibi, P.; Moradmand-Badie, B.; Afsahi, A.M.; Karimi, A.; Heydari, M.; et al. Genetic susceptibility of COVID-19: A systematic review of current evidence. Eur. J. Med. Res. 2021, 26, 46. [Google Scholar] [CrossRef]
- Falvella, F.S.; Galvan, A.; Frullanti, E.; Spinola, M.; Calabrò, E.; Carbone, A.; Incarbone, M.; Santambrogio, L.; Pastorino, U.; Dragani, T.A. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin. Cancer Res. 2009, 15, 1837–1842. [Google Scholar] [CrossRef]
- Tournier, J.M.; Maouche, K.; Coraux, C.; Zahm, J.M.; Cloëz-Tayarani, I.; Nawrocki-Raby, B.; Bonnomet, A.; Burlet, H.; Lebargy, F.; Polette, M.; et al. α3α5β2-Nicotinic acetylcholine receptor contributes to the wound repair of the respiratory epithelium by modulating intracellular calcium in migrating cells. Am. J. Pathol. 2006, 168, 55–68. [Google Scholar] [CrossRef]
- Routhier, J.; Pons, S.; Freidja, M.L.; Dalstein, V.; Cutrona, J.; Jonquet, A.; Lalun, N.; Mérol, J.C.; Lathrop, M.; Stitzel, J.A.; et al. An innate contribution of human nicotinic receptor polymorphisms to COPD-like lesions. Nat. Commun. 2021, 12, 6384. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, R.A.; Fisher, A.J.; Borthwick, L.A. The Role of Epithelial Damage in the Pulmonary Immune Response. Cells 2021, 10, 2763. [Google Scholar] [CrossRef] [PubMed]
- Medjber, K.; Freidja, M.L.; Grelet, S.; Lorenzato, M.; Maouche, K.; Nawrocki-Raby, B.; Birembaut, P.; Polette, M.; Tournier, J.M. Role of nicotinic acetylcholine receptors in cell proliferation and tumour invasion in broncho-pulmonary carcinomas. Lung Cancer 2015, 87, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.L.; Bukey, B.R.; Geyer, A.J.; Willnauer, C.P.; Reynolds, P.R. Immunohistochemical detection and regulation of a 5 nicotinic acetylcholine receptor (nAChR) subunits by FoxA2 during mouse lung organogenesis. Respir. Res. 2011, 12, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.R.; Onaitis, M.W.; Rawlins, E.L.; Lu, Y.; Clark, C.P.; Xue, Y.; Randell, S.H.; Hogan, B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 12771–12775. [Google Scholar] [CrossRef] [PubMed]
- Diabasana, Z.; Perotin, J.M.; Belgacemi, R.; Ancel, J.; Mulette, P.; Delepine, G.; Gosset, P.; Maskos, U.; Polette, M.; Deslée, G.; et al. Nicotinic receptor subunits atlas in the adult human lung. Int. J. Mol. Sci. 2020, 21, 7446. [Google Scholar] [CrossRef]
- Yang, L.; Yang, Z.; Zuo, C.; Lv, X.; Liu, T.; Jia, C.; Chen, H. Epidemiological evidence for associations between variants in CHRNA genes and risk of lung cancer and chronic obstructive pulmonary disease. Front. Oncol. 2022, 12, 1001864. [Google Scholar] [CrossRef]
- Chen, L.S.; Baker, T.; Hung, R.J.; Horton, A.; Culverhouse, R.; Hartz, S.; Saccone, N.; Cheng, I.; Deng, B.; Han, Y.; et al. Genetic Risk Can Be Decreased: Quitting Smoking Decreases and Delays Lung Cancer for Smokers with High and Low CHRNA5 Risk Genotypes—A Meta-Analysis. EBioMedicine 2016, 11, 219–226. [Google Scholar] [CrossRef]
- Kaya, T.; Nalbant, A.; Kılıçcıoğlu, G.K.; Çayır, K.T.; Yaylacı, S.; Varım, C. The prognostic significance of erythrocyte sedimentation rate in COVID-19. Rev. Assoc. Med. Bras. 2021, 67, 1305–1310. [Google Scholar] [CrossRef]
- Kurt, C.; Altunçekiç Yildirim, A. Contribution of Erythrocyte Sedimentation Rate to Predict Disease Severity and Outcome in COVID-19 Patients. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 6510952. [Google Scholar] [CrossRef]
- Gallus, S.; Scala, M.; Possenti, I.; Jarach, C.M.; Clancy, L.; Fernandez, E.; Gorini, G.; Carreras, G.; Malevolti, M.C.; Commar, A.; et al. The role of smoking in COVID-19 progression: A comprehensive meta-analysis. Eur. Respir. Rev. 2023, 32, 220191. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Noubouossie, D.F.; Gandotra, S.; Cao, L. Elevated Plasma Fibrinogen Is Associated with Excessive Inflammation and Disease Severity in COVID-19 Patients. Front. Cell. Infect. Microbiol. 2021, 11, 734005. [Google Scholar] [CrossRef] [PubMed]
- Çekiç, D.; Emir Arman, M.; Cihad Genç, A.; İşsever, K.; Yıldırım, İ.; Bilal Genç, A.; Dheir, H.; Yaylacı, S. Predictive role of FAR ratio in COVID-19 patients. Int. J. Clin. Pract. 2021, 75, e14931. [Google Scholar] [CrossRef] [PubMed]
- Perez-Rubio, G.; Perez-Rodriguez, M.E.; Fernández-López, J.C.; Ramirez-Venegas, A.; Garcia-Colunga, J.; Avila-Moreno, F.; Camarena, A.; Sansores, R.H.; Falfan-Valencia, R. SNPs in NRXN1 and CHRNA5 are associated to smoking and regulation of GABAergic and glutamatergic pathways. Pharmacogenomics 2016, 17, 1145–1158. [Google Scholar] [CrossRef]
- Available online: https://www.ncbi.nlm.nih.gov/snp/rs16969968#variant_details (accessed on 20 May 2023).
- Wen, L.; Jiang, K.; Yuan, W.; Cui, W.; Li, M.D. Contribution of Variants in CHRNA5/A3/B4 Gene Cluster on Chromosome 15 to Tobacco Smoking: From Genetic Association to Mechanism. Mol. Neurobiol. 2016, 53, 472–484. [Google Scholar] [CrossRef]
- Wang, J.C.; Cruchaga, C.; Saccone, N.L.; Bertelsen, S.; Liu, P.; Budde, J.P.; Duan, W.; Fox, L.; Grucza, R.A.; Kern, J.; et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009, 18, 3125–3135. [Google Scholar] [CrossRef]
- Wang, J.C.; Spiegel, N.; Bertelsen, S.; Le, N.; McKenna, N.; Budde, J.P.; Harari, O.; Kapoor, M.; Brooks, A.; Hancock, D.; et al. Cis-regulatory variants affect CHRNA5 mRNA expression in populations of African and European ancestry. PLoS ONE 2013, 8, e80204. [Google Scholar] [CrossRef]
- Saber Cherif, L.; Diabasana, Z.; Perotin, J.M.; Ancel, J.; Petit, L.M.G.; Devilliers, M.A.; Bonnomet, A.; Lalun, N.; Delepine, G.; Maskos, U.; et al. The Nicotinic Receptor Polymorphism rs16969968 Is Associated with Airway Remodeling and Inflammatory Dysregulation in COPD Patients. Cells 2022, 11, 2937. [Google Scholar] [CrossRef]
| rs16969968/ CHRNA5 | Smokers (n = 257) | Non-Smokers (n = 660) | p-Value * |
| GG | 200 (77.8) | 537 (81.4) | 0.089 |
| GA | 49 (19.0) | 116 (17.6) | |
| AA | 8 (3.2) | 7 (1.0) | |
| G | 449 (87.4) | 1190 (90.2) | 0.096 |
| A | 65 (12.6) | 130 (9.8) | |
| rs3918396/ ADAM33 | Smokers (n = 257) | Non-Smokers (n = 654) | p-Value * |
| CC | 243 (94.6) | 612 (93.6) | 0.416 |
| CT | 14 (5.4) | 38 (5.8) | |
| TT | 0 (0) | 4 (0.6) | |
| C | 500 (97.3) | 1262 (96.5) | 0.479 |
| T | 14 (2.7) | 46 (3.5) |
| Variable | GA + AA (n = 180) | GG (n = 737) | p |
|---|---|---|---|
| Men (n, %) * | 119 (66.1) | 522 (70.8) | 0.251 |
| Age (years) ** | 61 (52–68) | 58 (50–68) | 0.267 |
| Days of hospitalization ** | 19 (13–33) | 20 (12–30) | 0.807 |
| BMI (kg/m2) ** | 29.4 (26.1–33.3) | 29.5 (26.1–33.4) | 0.924 |
| Days since symptoms onset ** | 9 (7–13) | 10 (7–14) | 0.522 |
| IMV requirement (n, %) * | 132 (73.3) | 578 (78.4) | 0.172 |
| IMV (days) ** | 13 (0–29) | 15 (5–25) | 0.880 |
| PaO2/FiO2 (mmHg) ** | 105 (73–173) | 120 (77–189) | 0.055 |
| Deaths (n, %) * | 66 (36.7) | 295 (40.0) | 0.458 |
| Treated with corticosteroids (%) | 84.9 | 78.5 | 0.620 |
| Comorbidities (n, %) * | |||
| DM type 2 | 44 (24.4) | 215 (29.2) | 0.241 |
| Smoking | 57 (31.7) | 200 (27.1) | 0.262 |
| Hypertension | 71 (39.4) | 244 (33.1) | 0.129 |
| Obesity | 78 (43.3) | 340 (46.1) | 0.553 |
| Chronic Lung Disease | 19 (10.6) | 61 (8.3) | 0.410 |
| Ischemic Heart Disease | 11 (6.1) | 25 (3.4) | 0.141 |
| Variable, n (%) | GA + AA (n = 180) | GG (n = 737) | p-Value * |
|---|---|---|---|
| Fever | 125 (69.4) | 526 (71.4) | 0.675 |
| Headache | 83 (46.1) | 316 (42.9) | 0.483 |
| Arthralgias | 111 (61.7) | 433 (58.8) | 0.529 |
| Myalgias | 120 (66.7) | 449 (60.9) | 0.180 |
| Dyspnea | 152 (84.4) | 624 (84.7) | 0.967 |
| Cough | 112 (62.2) | 510 (69.2) | 0.087 |
| Sore throat | 39 (21.7) | 188 (25.5) | 0.329 |
| Rhinorrhea | 29 (16.1) | 98 (13.3) | 0.390 |
| Ageusia | 20 (11.1) | 112 (15.2) | 0.200 |
| Anosmia | 5 (2.8) | 52 (7.1) | 0.050 |
| Thoracic pain | 8 (4.4) | 69 (9.4) | 0.047 |
| Vomit | 5 (2.8) | 18 (2.4) | 0.993 |
| Diarrhea | 22 (12.2) | 70 (9.5) | 0.341 |
| Variable | GA + AA | GG | p |
|---|---|---|---|
| WBC (×103/μL) | 9.8 (8.3–13.2) | 10.4 (8.0–13.5) | 0.505 |
| Lymphocytes (×103/μL) | 2 (0.7–6.3) | 2.1 (0.7–6.9) | 0.818 |
| Platelets (×103/μL) | 261 (213–327) | 258 (189–334) | 0.643 |
| LDH (UI/L) | 393 (290–490) | 387 (301–508) | 0.810 |
| D-Dimer (μg/mL) | 1.41 (0.77–3.26) | 1.67 (0.60–3.77) | 0.814 |
| ESR (mm/h) | 32 (23–40) | 26 (10–35) | 0.038 |
| CRP (mg/dL) | 12.0 (5.8–19.5) | 12.3 (6.6–20.9) | 0.726 |
| Fibrinogen (mg/dL) | 707 (575–781) | 666 (569–778) | 0.493 |
| Procalcitonin (ng/mL) | 0.24 (0.08–0.72) | 0.21 (0.09–0.65) | 0.941 |
| Ferritin (ng/mL) | 990 (669–2368) | 1064 (637–1892) | 0.518 |
| rs16969968 | HS-COV (n = 56) | LS-COV (n = 178) | p * | OR (CI, 95%) |
|---|---|---|---|---|
| GG | 39 (69.6) | 141 (79.2) | 0.034 | reference |
| GA | 13 (23.2) | 35 (19.6) | 1.34 (0. 64–2.78) | |
| AA | 4 (7.1) | 2 (1.1) | 7.23 (1.27–40.95) | |
| G | 91 (81.2) | 317 (89.0) | 0.046 | |
| A | 21 (18.7) | 39 (10.9) | 1.87 (1.05–3.34) | |
| GG + GA | 42 (92.8) | 176 (98.8) | 0.030 | |
| AA | 4 (7.1) | 2 (1.1) | 6.05 (1.40–25.88) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valencia-Pérez Rea, D.; Falfán-Valencia, R.; Fricke-Galindo, I.; Buendía-Roldán, I.; Chávez-Galán, L.; Nava-Quiroz, K.J.; Alanis-Ponce, J.; Pérez-Rubio, G. The rs16969968 Tobacco Smoking-Related Single-Nucleotide Variant Is Associated with Clinical Markers in Patients with Severe COVID-19. Int. J. Mol. Sci. 2023, 24, 9811. https://doi.org/10.3390/ijms24129811
Valencia-Pérez Rea D, Falfán-Valencia R, Fricke-Galindo I, Buendía-Roldán I, Chávez-Galán L, Nava-Quiroz KJ, Alanis-Ponce J, Pérez-Rubio G. The rs16969968 Tobacco Smoking-Related Single-Nucleotide Variant Is Associated with Clinical Markers in Patients with Severe COVID-19. International Journal of Molecular Sciences. 2023; 24(12):9811. https://doi.org/10.3390/ijms24129811
Chicago/Turabian StyleValencia-Pérez Rea, Daniela, Ramcés Falfán-Valencia, Ingrid Fricke-Galindo, Ivette Buendía-Roldán, Leslie Chávez-Galán, Karol J. Nava-Quiroz, Jesús Alanis-Ponce, and Gloria Pérez-Rubio. 2023. "The rs16969968 Tobacco Smoking-Related Single-Nucleotide Variant Is Associated with Clinical Markers in Patients with Severe COVID-19" International Journal of Molecular Sciences 24, no. 12: 9811. https://doi.org/10.3390/ijms24129811
APA StyleValencia-Pérez Rea, D., Falfán-Valencia, R., Fricke-Galindo, I., Buendía-Roldán, I., Chávez-Galán, L., Nava-Quiroz, K. J., Alanis-Ponce, J., & Pérez-Rubio, G. (2023). The rs16969968 Tobacco Smoking-Related Single-Nucleotide Variant Is Associated with Clinical Markers in Patients with Severe COVID-19. International Journal of Molecular Sciences, 24(12), 9811. https://doi.org/10.3390/ijms24129811










