Cm-p5 Peptide Dimers Inhibit Biofilms of Candida albicans Clinical Isolates, C. parapsilosis and Fluconazole-Resistant Mutants of C. auris
Abstract
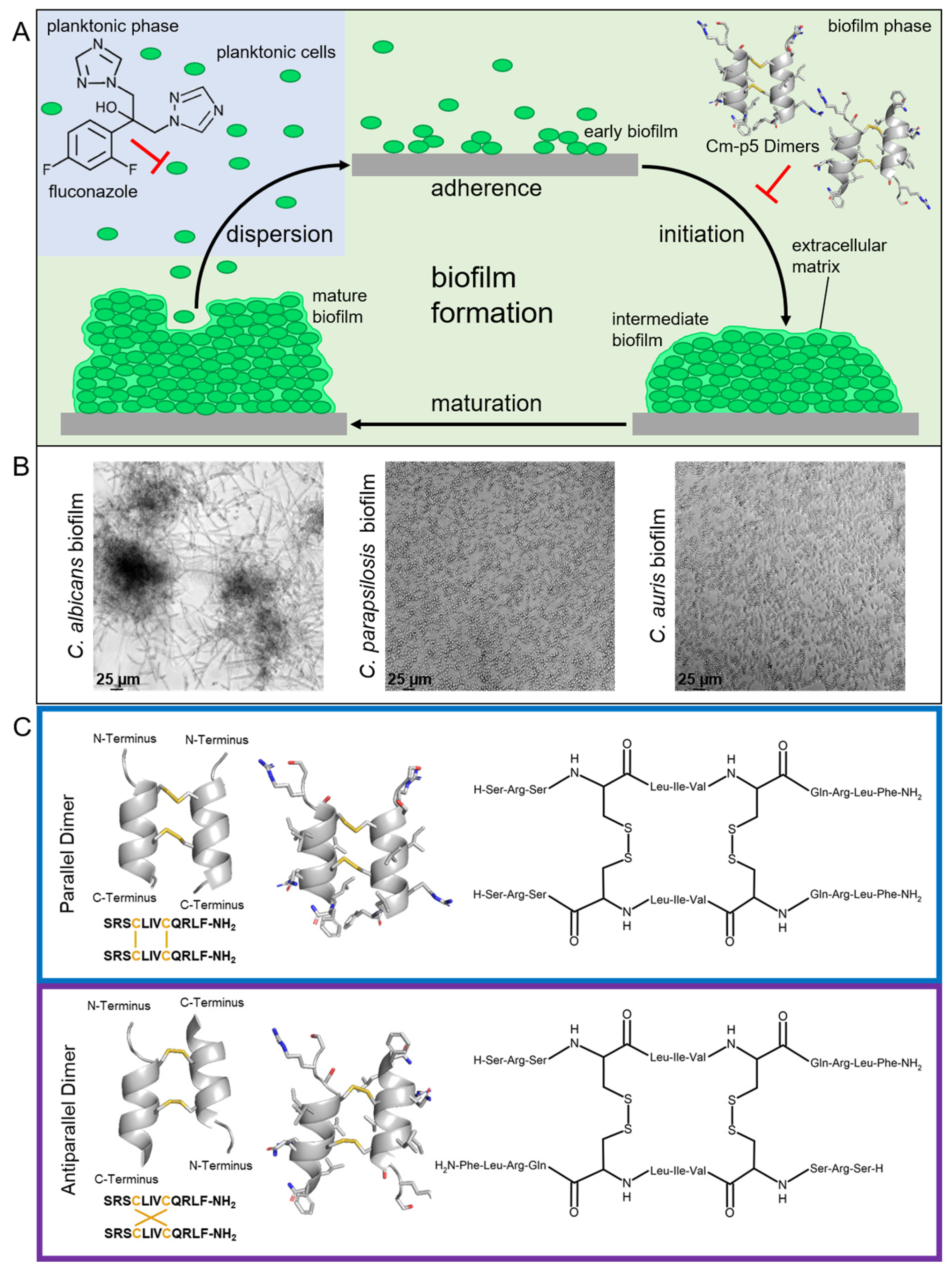
1. Introduction
2. Results
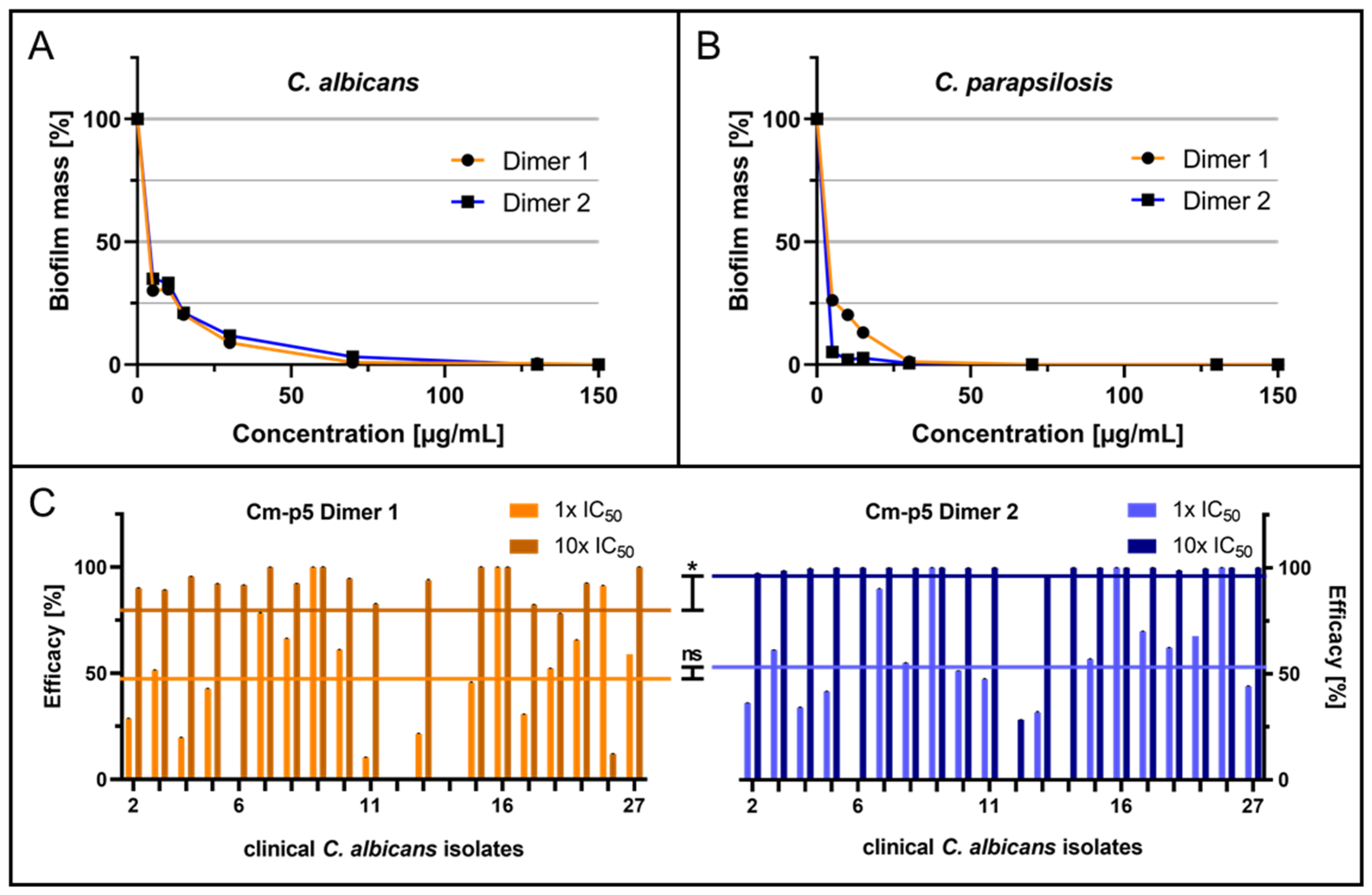
2.1. Anti-Biofilm Activity of Cm-p5 Dimeric Derivatives against C. albicans, C. parapsilosis and Invasive Clinical Isolates
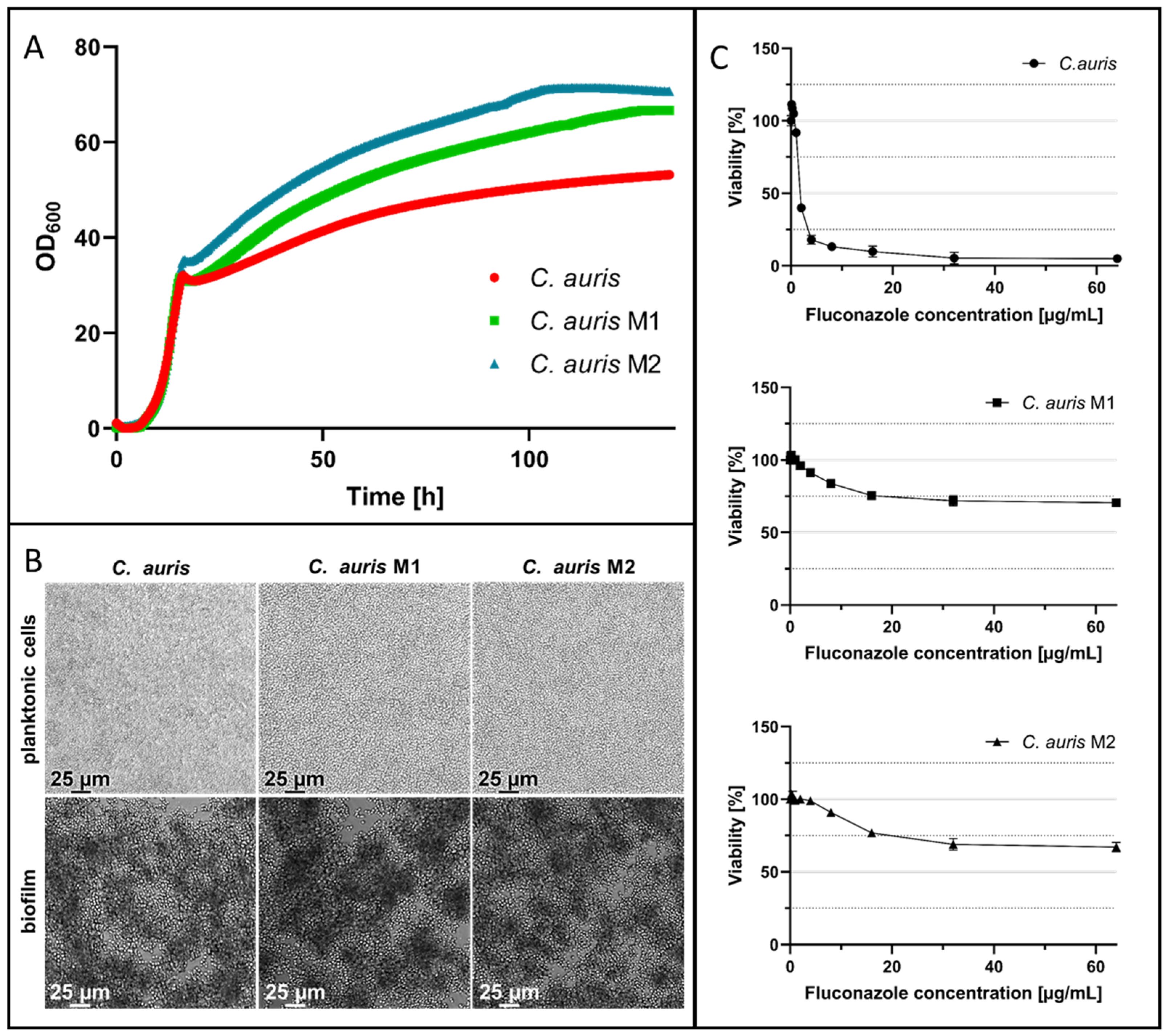
2.2. In Vitro Evolution of Fluconazole-Resistant C. auris
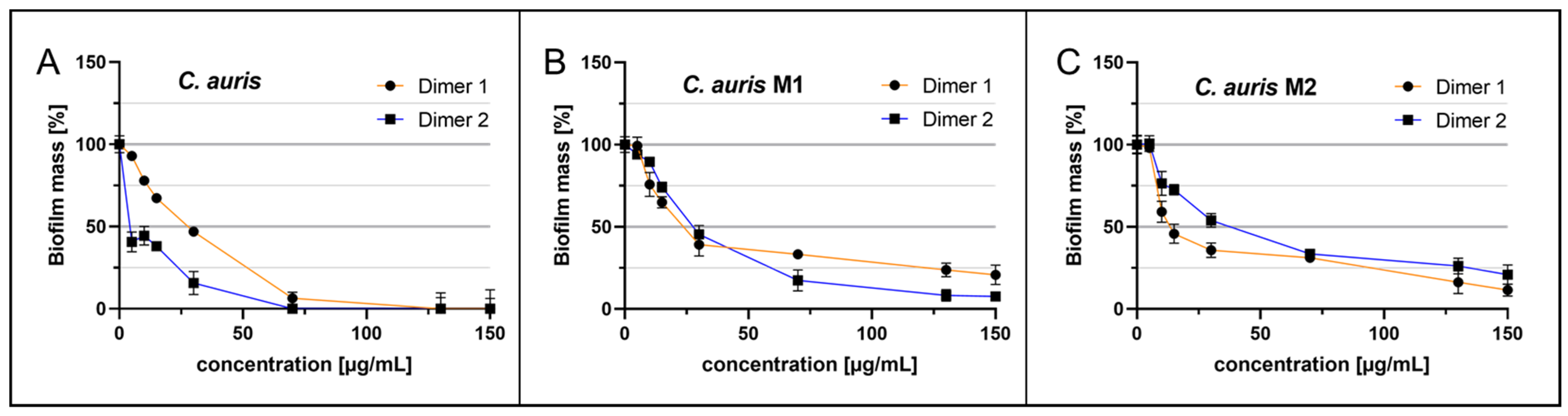
2.3. Anti-Biofilm Acitivity of Cm-p5 Dimeric Derivatives against Fluconazole-Resistant C. auris
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Microorganism Strains and Growth Conditions
4.3. In Vitro Evolution of Fluconazole-Resistant C. auris Strains
4.4. Viability Tests and Quantification
4.5. Biofilm Formation and Quantification
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R. Current Epidemiology of Candida Infection. Clin. Microbiol. Newsl. 2014, 36, 131–136. [Google Scholar] [CrossRef]
- Papon, N.; Courdavault, V.; Clastre, M.; Bennett, R.J. Emerging and Emerged Pathogenic Candida Species: Beyond the Candida albicans Paradigm. PLoS Pathog. 2013, 9, e1003550. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic Screens of a Candida albicans Homozygous Deletion Library Decouple Morphogenetic Switching and Pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: A 10.5-Year Analysis of Susceptibilities of Candida Species to Fluconazole and Voriconazole as Determined by CLSI Standardized Disk Diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef]
- Fox, E.P.; Nobile, C.J. A Sticky Situation: Untangling the Transcriptional Network Controlling Biofilm Development in Candida albicans. Transcription 2012, 3, 315–322. [Google Scholar] [CrossRef]
- Wall, G.; Montelongo-Jauregui, D.; Vidal Bonifacio, B.; Lopez-Ribot, J.L.; Uppuluri, P. Candida albicans Biofilm Growth and Dispersal: Contributions to Pathogenesis. Curr. Opin. Microbiol. 2019, 52, 1–6. [Google Scholar] [CrossRef]
- Finkel, J.S.; Mitchell, A.P. Genetic Control of Candida albicans Biofilm Development. Nat. Rev. Microbiol. 2011, 9, 109–118. [Google Scholar] [CrossRef]
- Liu, R.; Chen, X.; Falk, S.P.; Masters, K.S.; Weisblum, B.; Gellman, S.H. Nylon-3 Polymers Active against Drug-Resistant Candida albicans Biofilms. J. Am. Chem. Soc. 2015, 137, 2183–2186. [Google Scholar] [CrossRef]
- Vicente, F.E.M.; González-Garcia, M.; Diaz Pico, E.; Moreno-Castillo, E.; Garay, H.E.; Rosi, P.E.; Jimenez, A.M.; Campos-Delgado, J.A.; Rivera, D.G.; Chinea, G.; et al. Design of a Helical-Stabilized, Cyclic, and Nontoxic Analogue of the Peptide Cm-P5 with Improved Antifungal Activity. ACS Omega 2019, 4, 19081–19095. [Google Scholar] [CrossRef] [PubMed]
- Berkow, E.L.; Lockhart, S.R. Fluconazole Resistance in Candida Species: A Current Perspective. Infect. Drug. Resist. 2017, 10, 237–245. [Google Scholar] [CrossRef]
- Warrilow, A.G.; Nishimoto, A.T.; Parker, J.E.; Price, C.L.; Flowers, S.A.; Kelly, D.E.; Rogers, P.D.; Kelly, S.L. The Evolution of Azole Resistance in Candida albicans Sterol 14α-Demethylase (CYP51) through Incremental Amino Acid Substitutions. Antimicrob. Agents Chemother. 2019, 63, e02586-18. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, C.A. Cytochrome P-450-Dependent 14 Alpha-Sterol Demethylase of Candida albicans and Its Interaction with Azole Antifungals. Biochem. Soc. Trans. 1991, 19, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Heimark, L.; Shipkova, P.; Greene, J.; Munayyer, H.; Yarosh-Tomaine, T.; DiDomenico, B.; Hare, R.; Pramanik, B.N. Mechanism of Azole Antifungal Activity as Determined by Liquid Chromatographic/Mass Spectrometric Monitoring of Ergosterol Biosynthesis. J. Mass. Spectrom. 2002, 37, 265–269. [Google Scholar] [CrossRef]
- Cleveland, A.A.; Farley, M.M.; Harrison, L.H.; Stein, B.; Hollick, R.; Lockhart, S.R.; Magill, S.S.; Derado, G.; Park, B.J.; Chiller, T.M. Changes in Incidence and Antifungal Drug Resistance in Candidemia: Results from Population-Based Laboratory Surveillance in Atlanta and Baltimore, 2008–2011. Clin. Infect. Dis. 2012, 55, 1352–1361. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Rhomberg, P.R.; Messer, S.A.; Jones, R.N.; Castanheira, M. Isavuconazole, Micafungin, and 8 Comparator Antifungal Agents’ Susceptibility Profiles for Common and Uncommon Opportunistic Fungi Collected in 2013: Temporal Analysis of Antifungal Drug Resistance Using CLSI Species-Specific Clinical Breakpoints and Prop. Diagn. Microbiol. Infect. Dis. 2015, 82, 303–313. [Google Scholar] [CrossRef]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. Nov., a Novel Ascomycetous Yeast Isolated from the External Ear Canal of an Inpatient in a Japanese Hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef]
- Morales-López, S.E.; Parra-Giraldo, C.M.; Ceballos-Garzón, A.; Martínez, H.P.; Rodríguez, G.J.; Álvarez-Moreno, C.A.; Rodríguez, J.Y. Invasive Infections with Multidrug-Resistant Yeast Candida auris, Colombia. Emerg. Infect. Dis. 2017, 23, 162–164. [Google Scholar] [CrossRef]
- Vallabhaneni, S.; Baggs, J.; Tsay, S.; Srinivasan, A.R.; Jernigan, J.A.; Jackson, B.R. Trends in Antifungal Use in US Hospitals, 2006–2012. J. Antimicrob. Chemother. 2018, 73, 2867–2875. [Google Scholar] [CrossRef] [PubMed]
- Schelenz, S.; Hagen, F.; Rhodes, J.L.; Abdolrasouli, A.; Chowdhary, A.; Hall, A.; Ryan, L.; Shackleton, J.; Trimlett, R.; Meis, J.F.; et al. First Hospital Outbreak of the Globally Emerging Candida auris in a European Hospital. Antimicrob. Resist. Infect. Control. 2016, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Hamprecht, A.; Barber, A.E.; Mellinghoff, S.C.; Thelen, P.; Walther, G.; Yu, Y.; Neurgaonkar, P.; Dandekar, T.; Cornely, O.A.; Martin, R.; et al. Candida auris in Germany and Previous Exposure to Foreign Healthcare. Emerg. Infect. Dis. 2019, 25, 1763–1765. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, J.; Schrauzer, T.; Kirchhoff, L.; Meis, J.F.; Rath, P.M. Two Candida auris Cases in Germany with No Recent Contact to Foreign Healthcare—Epidemiological and Microbiological Investigations. J. Fungi 2021, 7, 380. [Google Scholar] [CrossRef] [PubMed]
- Di Pilato, V.; Codda, G.; Ball, L.; Giacobbe, D.R.; Willison, E.; Mikulska, M.; Magnasco, L.; Crea, F.; Vena, A.; Pelosi, P.; et al. Molecular Epidemiological Investigation of a Nosocomial Cluster of C. auris: Evidence of Recent Emergence in Italy and Ease of Transmission during the COVID-19 Pandemic. J. Fungi 2021, 7, 140. [Google Scholar] [CrossRef] [PubMed]
- Flowers, S.A.; Barker, K.S.; Berkow, E.L.; Toner, G.; Chadwick, S.G.; Gygax, S.E.; Morschhäuser, J.; Rogers, P.D. Gain-of-Function Mutations in UPC2 Are a Frequent Cause of ERG11 Upregulation in Azole-Resistant Clinical Isolates of Candida albicans. Eukaryot. Cell 2012, 11, 1289–1299. [Google Scholar] [CrossRef]
- Dunkel, N.; Liu, T.T.; Barker, K.S.; Homayouni, R.; Morschhäuser, J.; Rogers, P.D. A Gain-of-Function Mutation in the Transcription Factor Upc2p Causes Upregulation of Ergosterol Biosynthesis Genes and Increased Fluconazole Resistance in a Clinical Candida albicans Isolate. Eukaryot. Cell 2008, 7, 1180–1190. [Google Scholar] [CrossRef]
- Prasad, R.; Goffeau, A. Yeast ATP-Binding Cassette Transporters Conferring Multidrug Resistance. Annu. Rev. Microbiol. 2012, 66, 39–63. [Google Scholar] [CrossRef]
- Tsao, S.; Rahkhoodaee, F.; Raymond, M. Relative Contributions of the Candida albicans ABC Transporters Cdr1p and Cdr2p to Clinical Azole Resistance. Antimicrob. Agents Chemother. 2009, 53, 1344–1352. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Berman, J.; Novikov, A.; Bash, E.; Shachor-Meyouhas, Y.; Zakin, S.; Maor, Y.; Tarabia, J.; Schechner, V.; Adler, A.; et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg. Infect. Dis. 2017, 23, 195–203. [Google Scholar] [CrossRef]
- Kean, R.; Delaney, C.; Sherry, L.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R.; Williams, C.; Ramage, G. Transcriptome Assembly and Profiling of Candida auris Reveals Novel Insights into Biofilm-Mediated Resistance. mSphere 2018, 3, e00334–18. [Google Scholar] [CrossRef] [PubMed]
- Ostrowsky, B.; Greenko, J.; Adams, E.; Quinn, M.; O’Brien, B.; Chaturvedi, V.; Berkow, E.; Vallabhaneni, S.; Forsberg, K.; Chaturvedi, S.; et al. Candida auris Isolates Resistant to Three Classes of Antifungal Medications—New York, 2019. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Ciociola, T.; Giovati, L.; Conti, S.; Magliani, W.; Santinoli, C.; Polonelli, L. Natural and Synthetic Peptides with Antifungal Activity. Future Med. Chem. 2016, 8, 1413–1433. [Google Scholar] [CrossRef] [PubMed]
- Rautenbach, M.; Troskie, A.M.; Vosloo, J.A. Antifungal Peptides: To Be or Not to Be Membrane Active. Biochimie 2016, 130, 132–145. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Aoki, W.; Ueda, M. Characterization of Antimicrobial Peptides toward the Development of Novel Antibiotics. Pharmaceuticals 2013, 6, 1055–1081. [Google Scholar] [CrossRef] [PubMed]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef]
- Yeung, A.T.Y.; Gellatly, S.L.; Hancock, R.E.W. Multifunctional Cationic Host Defence Peptides and Their Clinical Applications. Cell. Mol. Life Sci. 2011, 68, 2161–2176. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- López-Abarrategui, C.; McBeth, C.; Mandal, S.M.; Sun, Z.J.; Heffron, G.; Alba-Menéndez, A.; Migliolo, L.; Reyes-Acosta, O.; García-Villarino, M.; Nolasco, D.O.; et al. Cm-P5: An Antifungal Hydrophilic Peptide Derived from the Coastal Mollusk Cenchritis muricatus (Gastropoda: Littorinidae). FASEB J. 2015, 29, 3315–3325. [Google Scholar] [CrossRef]
- Kubiczek, D.; Flaig, C.; Raber, H.; Dietz, S.; Kissmann, A.K.; Heerde, T.; Bodenberger, N.; Wittgens, A.; González-Garcia, M.; Kang, F.; et al. A Cerberus-Inspired Anti-Infective Multicomponent Gatekeeper Hydrogel against Infections with the Emerging “Superbug” Yeast Candida auris. Macromol. Biosci. 2020, 20, 2000005. [Google Scholar] [CrossRef] [PubMed]
- González-García, M.; Morales-Vicente, F.; Pico, E.D.; Garay, H.; Rivera, D.G.; Grieshober, M.; Olari, L.R.; Groß, R.; Conzelmann, C.; Krüger, F.; et al. Antimicrobial Activity of Cyclic-Monomeric and Dimeric Derivatives of the Snail-Derived Peptide Cm-P5 against Viral and Multidrug-Resistant Bacterial Strains. Biomolecules 2021, 11, 745. [Google Scholar] [CrossRef] [PubMed]
- Kubiczek, D.; Raber, H.; Gonzalez-García, M.; Morales-Vicente, F.; Staendker, L.; Otero-Gonzalez, A.J.; Rosenau, F. Derivates of the Antifungal Peptide CM-P5 Inhibit Development of Candida auris Biofilms in Vitro. Antibiotics 2020, 9, 363. [Google Scholar] [CrossRef]
- Häring, M.; Amann, V.; Kissmann, A.K.; Herberger, T.; Synatschke, C.; Kirsch-Pietz, N.; Perez-Erviti, J.A.; Otero-Gonzalez, A.J.; Morales-Vicente, F.; Andersson, J.; et al. Combination of Six Individual Derivatives of the Pom-1 Antibiofilm Peptide Doubles Their Efficacy against Invasive and Multi-Resistant Clinical Isolates of the Pathogenic Yeast Candida albicans. Pharmaceutics 2022, 14, 1332. [Google Scholar] [CrossRef] [PubMed]
- Bruno, M.; Kersten, S.; Bain, J.M.; Jaeger, M.; Rosati, D.; Kruppa, M.D.; Lowman, D.W.; Rice, P.J.; Graves, B.; Ma, Z.; et al. Transcriptional and Functional Insights into the Host Immune Response against the Emerging Fungal Pathogen Candida auris. Nat. Microbiol. 2020, 5, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Sabino, R.; Veríssimo, C.; Pereira, Á.A.; Antunes, F. Candida Auris, an Agent of Hospital-Associated Outbreaks: Which Challenging Issues Do We Need to Have in Mind? Microorganisms 2020, 8, 181. [Google Scholar] [CrossRef]
- CLSI M27-A3; Reference Method for Broth Dilution Antifungal Susceptibily Testing of Yeasts. CLSI: Wayne, PA, USA, 2008; Volume 28.
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The Clinical Impact of Bacterial Biofilms. Int. J. Oral. Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Welsh, R.M.; Bentz, M.L.; Shams, A.; Houston, H.; Lyons, A.; Rose, L.J.; Litvintseva, A.P. Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J. Clin. Microbiol. 2017, 55, 2996–3005. [Google Scholar] [CrossRef]
- Taff, H.T.; Mitchell, K.F.; Edward, J.A.; Andes, D.R. Mechanisms of Candida Biofilm Drug Resistance. Future Microbiol. 2013, 8, 1325–1337. [Google Scholar] [CrossRef]
- Nobile, C.J.; Johnson, A.D. Candida albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Davey, M.E.; O’toole, G.A. Microbial Biofilms: From Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef] [PubMed]
- Kolter, R.; Greenberg, E.P. Microbial Sciences: The Superficial Life of Microbes. Nature 2006, 441, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Quindós, G.; Marcos-Arias, C.; San-Millán, R.; Mateo, E.; Eraso, E. The Continuous Changes in the Aetiology and Epidemiology of Invasive Candidiasis: From Familiar Candida albicans to Multiresistant Candida auris. Int. Microbiol. 2018, 21, 107–119. [Google Scholar] [CrossRef]
- Enoch, D.A.; Yang, H.; Aliyu, S.H.; Micallef, C. The Changing Epidemiology of Invasive Fungal Infections. Methods Mol. Biol. 2017, 1508, 17–65. [Google Scholar] [CrossRef] [PubMed]
- Paramythiotou, E.; Frantzeskaki, F.; Flevari, A.; Armaganidis, A.; Dimopoulos, G. Invasive Fungal Infections in the ICU: How to Approach, How to Treat. Molecules 2014, 19, 1085–1119. [Google Scholar] [CrossRef]
- Tóth, R.; Nosek, J.; Mora-Montes, H.M.; Gabaldon, T.; Bliss, J.M.; Nosanchuk, J.D.; Turner, S.A.; Butler, G.; Vágvölgyi, C.; Gácser, A. Candida parapsilosis: From Genes to the Bedside. Clin. Microbiol. Rev. 2019, 32, e00111-18. [Google Scholar] [CrossRef]
- Harrington, R.; Kindermann, S.L.; Hou, Q.; Taylor, R.J.; Azie, N.; Horn, D.L. Candidemia and Invasive Candidiasis among Hospitalized Neonates and Pediatric Patients. Curr. Med. Res. Opin. 2017, 33, 1803–1812. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm Formation Mechanisms and Targets for Developing Antibiofilm Agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef]
- García, M.G.; Rodríguez, A.; Alba, A.; Vázquez, A.A.; Vicente, F.E.M.; Pérez-Erviti, J.; Spellerberg, B.; Stenger, S.; Grieshober, M.; Conzelmann, C.; et al. New Antibacterial Peptides from the Freshwater Mollusk Pomacea poeyana (Pilsbry, 1927). Biomolecules 2020, 10, 1437. [Google Scholar] [CrossRef]
- Raber, H.F.; Sejfijaj, J.; Kissmann, A.K.; Wittgens, A.; Gonzalez-Garcia, M.; Alba, A.; Vázquez, A.A.; Vicente, F.E.M.; Erviti, J.P.; Kubiczek, D.; et al. Antimicrobial Peptides Pom-1 and Pom-2 from Pomacea Poeyana Are Active against Candida auris, C. parapsilosis and C. albicans Biofilms. Pathogens 2021, 10, 496. [Google Scholar] [CrossRef]
- Amann, V.; Kissmann, A.K.; Krämer, M.; Krebs, I.; Perez-Erviti, J.A.; Otero-Gonzalez, A.J.; Morales-Vicente, F.; Rodríguez, A.; Ständker, L.; Weil, T.; et al. Increased Activities against Biofilms of the Pathogenic Yeast Candida albicans of Optimized Pom-1 Derivatives. Pharmaceutics 2022, 14, 318. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Chang, W.K.; Wimley, W.C.; Searson, P.C.; Hristova, K.; Merzlyakov, M. Characterization of Antimicrobial Peptide Activity by Electrochemical Impedance Spectroscopy. Biochim. Biophys. Acta 2008, 1778, 2430–2436. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mode of Action of Membrane Active Antimicrobial Peptides. Pept. Sci. 2004, 66, 236–248. [Google Scholar] [CrossRef]
- Bruder, S.; Reifenrath, M.; Thomik, T.; Boles, E.; Herzog, K. Parallelised Online Biomass Monitoring in Shake Flasks Enables Efficient Strain and Carbon Source Dependent Growth Characterisation of Saccharomyces cerevisiae. Microb. Cell. Fact. 2016, 15, 127. [Google Scholar] [CrossRef] [PubMed]
- Fai, P.B.; Grant, A. A Rapid Resazurin Bioassay for Assessing the Toxicity of Fungicides. Chemosphere 2009, 74, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Ramage, G.; Kean, R.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R. Biofilm-Forming Capability of Highly Virulent, Multidrug-Resistant Candida auris. Emerg. Infect. Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef]
- Jose, A.; Coco, B.J.; Milligan, S.; Young, B.; Lappin, D.F.; Bagg, J.; Murray, C.; Ramage, G. Reducing the Incidence of Denture Stomatitis: Are Denture Cleansers Sufficient? J. Prosthodont. 2010, 19, 252–257. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm Formation as Microbial Development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Negri, M.; Gonçalves, V.; Silva, S.; Henriques, M.; Azeredo, J.; Oliveira, R. Crystal Violet Staining to Quantify Candida Adhesion to Epithelial Cells. Br. J. Biomed. Sci. 2010, 67, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Rajendran, R.; Lappin, D.F.; Borghi, E.; Perdoni, F.; Falleni, M.; Tosi, D.; Smith, K.; Williams, C.; Jones, B.; et al. Biofilms Formed by Candida albicans Bloodstream Isolates Display Phenotypic and Transcriptional Heterogeneity That Are Associated with Resistance and Pathogenicity. BMC Microbiol. 2014, 14, 182. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amann, V.; Kissmann, A.-K.; Mildenberger, V.; Krebs, I.; Perez-Erviti, J.A.; Martell-Huguet, E.M.; Otero-Gonzalez, A.J.; Morales-Vicente, F.; Rodríguez-Castaño, G.P.; Firacative, C.; et al. Cm-p5 Peptide Dimers Inhibit Biofilms of Candida albicans Clinical Isolates, C. parapsilosis and Fluconazole-Resistant Mutants of C. auris. Int. J. Mol. Sci. 2023, 24, 9788. https://doi.org/10.3390/ijms24129788
Amann V, Kissmann A-K, Mildenberger V, Krebs I, Perez-Erviti JA, Martell-Huguet EM, Otero-Gonzalez AJ, Morales-Vicente F, Rodríguez-Castaño GP, Firacative C, et al. Cm-p5 Peptide Dimers Inhibit Biofilms of Candida albicans Clinical Isolates, C. parapsilosis and Fluconazole-Resistant Mutants of C. auris. International Journal of Molecular Sciences. 2023; 24(12):9788. https://doi.org/10.3390/ijms24129788
Chicago/Turabian StyleAmann, Valerie, Ann-Kathrin Kissmann, Vanessa Mildenberger, Imke Krebs, Julio A. Perez-Erviti, Ernesto M. Martell-Huguet, Anselmo J. Otero-Gonzalez, Fidel Morales-Vicente, Gina P. Rodríguez-Castaño, Carolina Firacative, and et al. 2023. "Cm-p5 Peptide Dimers Inhibit Biofilms of Candida albicans Clinical Isolates, C. parapsilosis and Fluconazole-Resistant Mutants of C. auris" International Journal of Molecular Sciences 24, no. 12: 9788. https://doi.org/10.3390/ijms24129788
APA StyleAmann, V., Kissmann, A.-K., Mildenberger, V., Krebs, I., Perez-Erviti, J. A., Martell-Huguet, E. M., Otero-Gonzalez, A. J., Morales-Vicente, F., Rodríguez-Castaño, G. P., Firacative, C., Rodríguez, A., Ständker, L., Weil, T., Spellerberg, B., Stenger, S., & Rosenau, F. (2023). Cm-p5 Peptide Dimers Inhibit Biofilms of Candida albicans Clinical Isolates, C. parapsilosis and Fluconazole-Resistant Mutants of C. auris. International Journal of Molecular Sciences, 24(12), 9788. https://doi.org/10.3390/ijms24129788









