Characterization of the Clinical Significance and Immunological Landscapes of a Novel TMEMs Signature in Hepatocellular Carcinoma and the Contribution of TMEM201 to Hepatocarcinogenesis
Abstract
1. Introduction
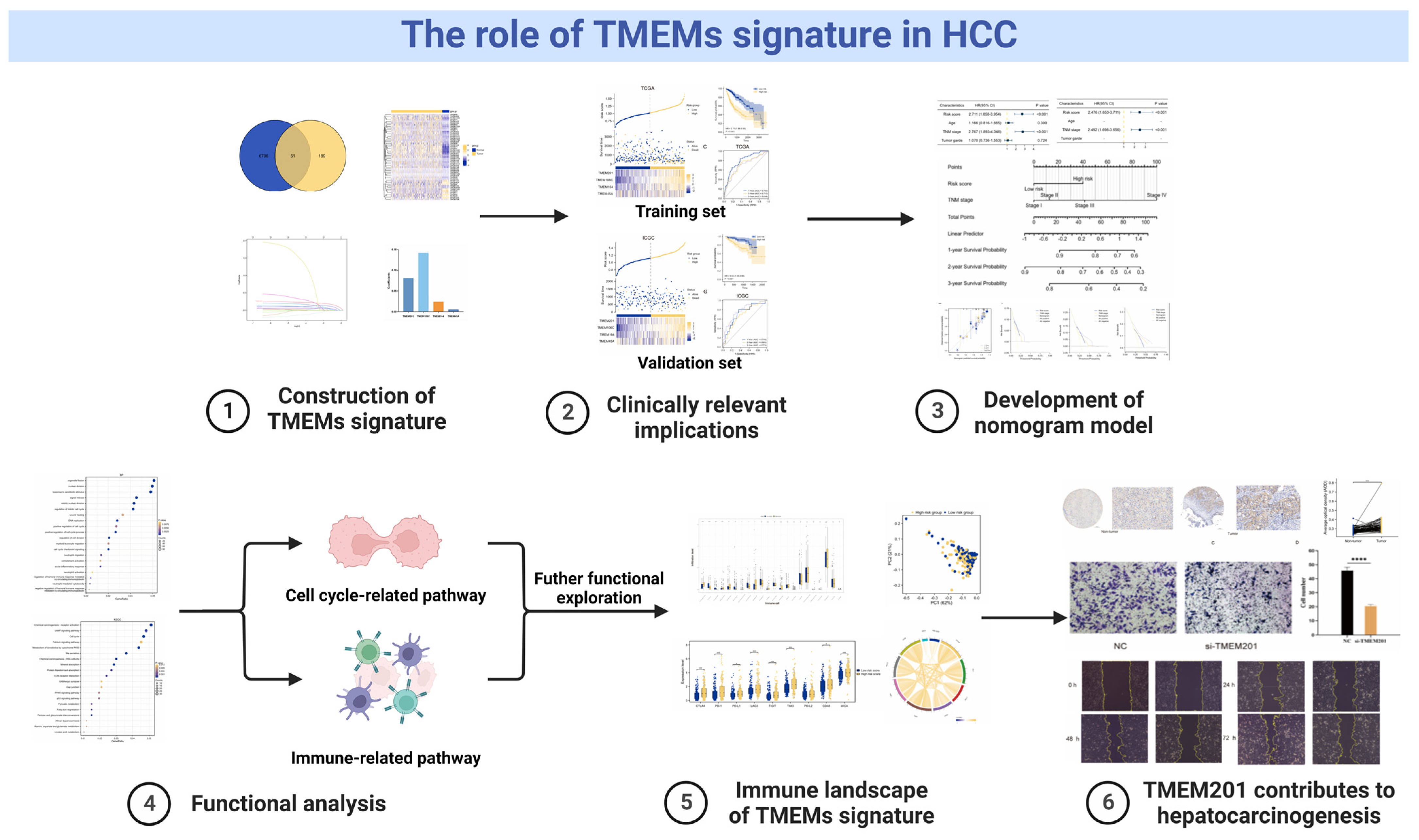
2. Results
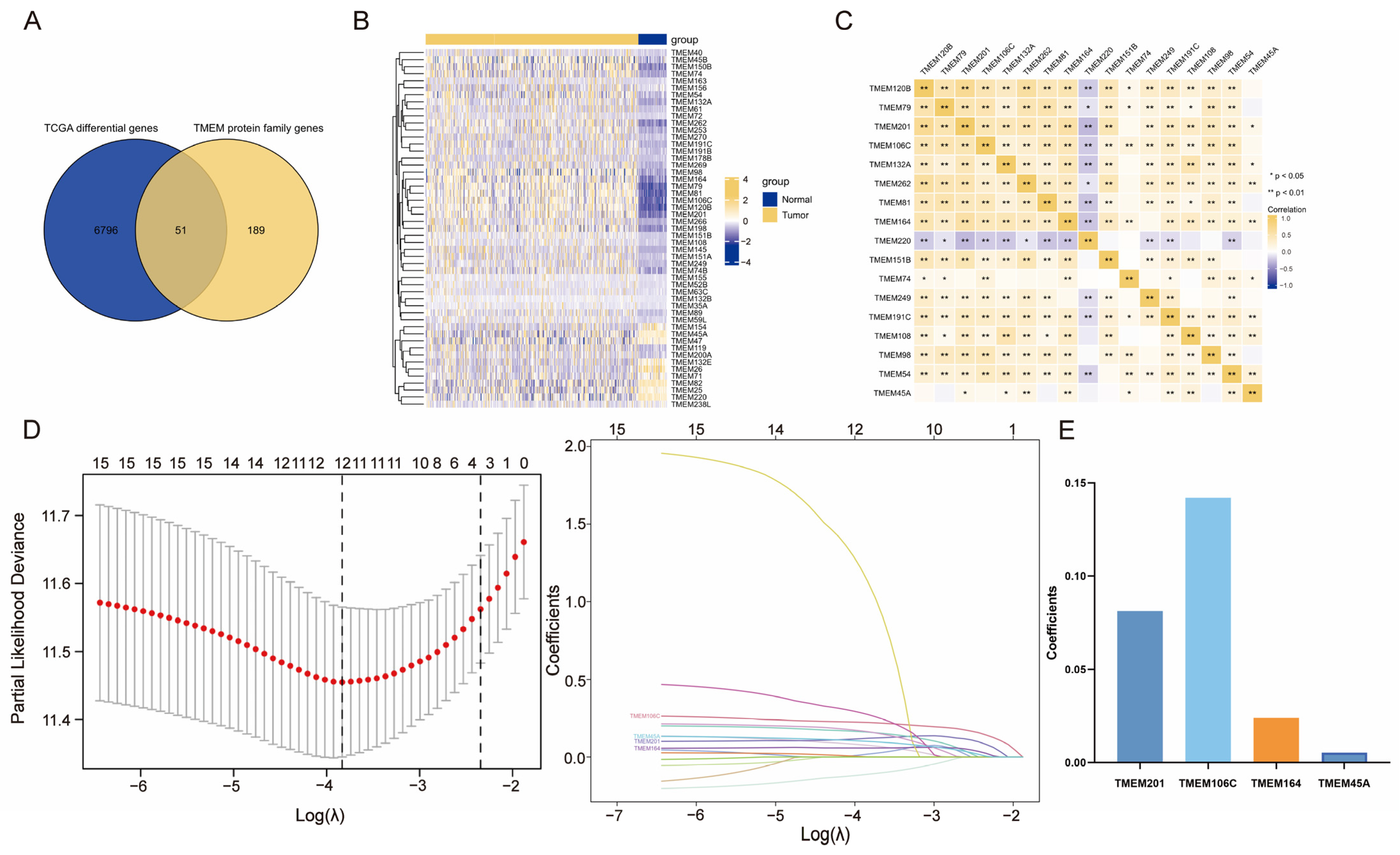
2.1. Investigation of Differentially Expressed TMEMs
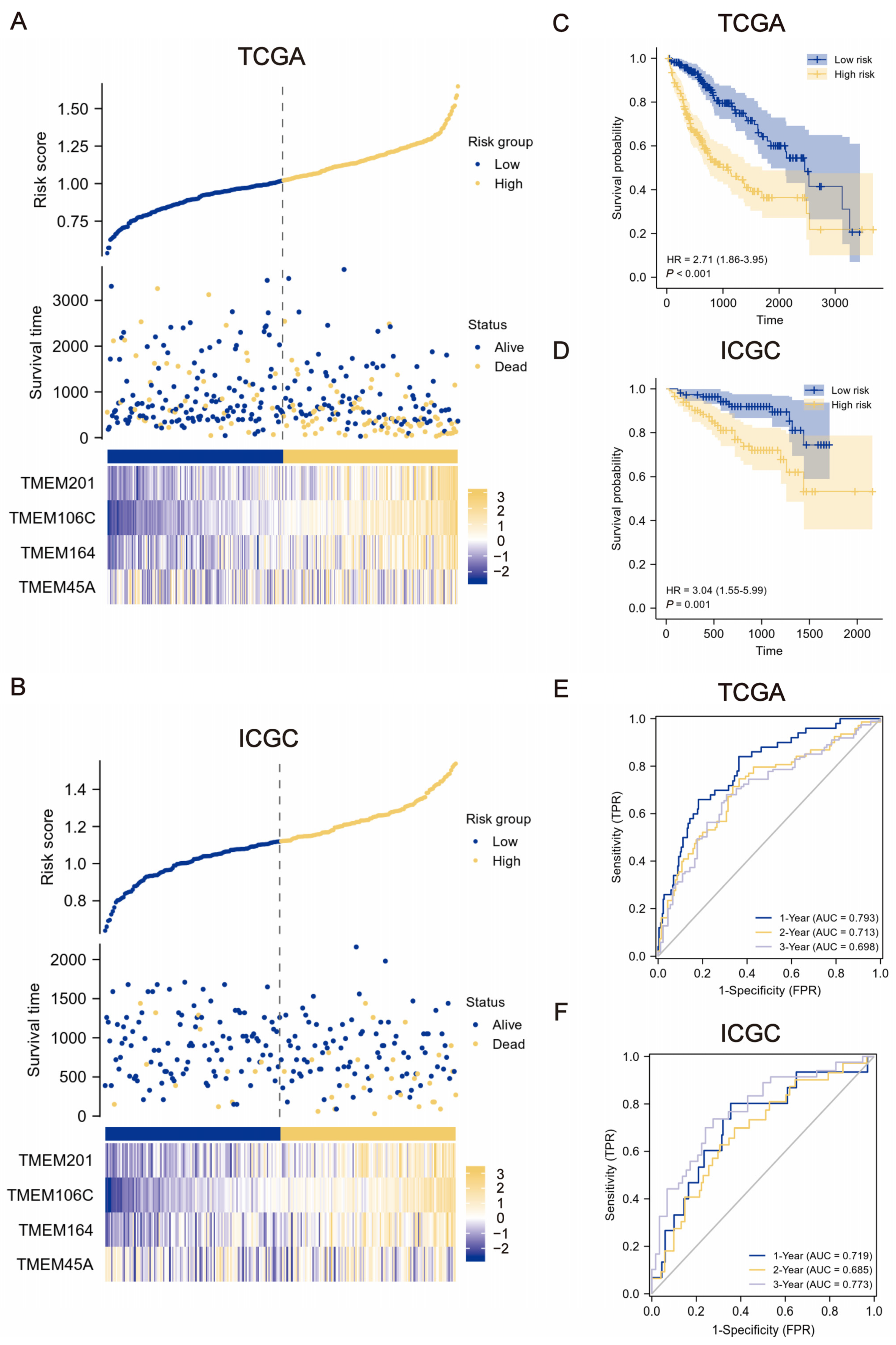
2.2. Construction and Verification of TMEMs Signature
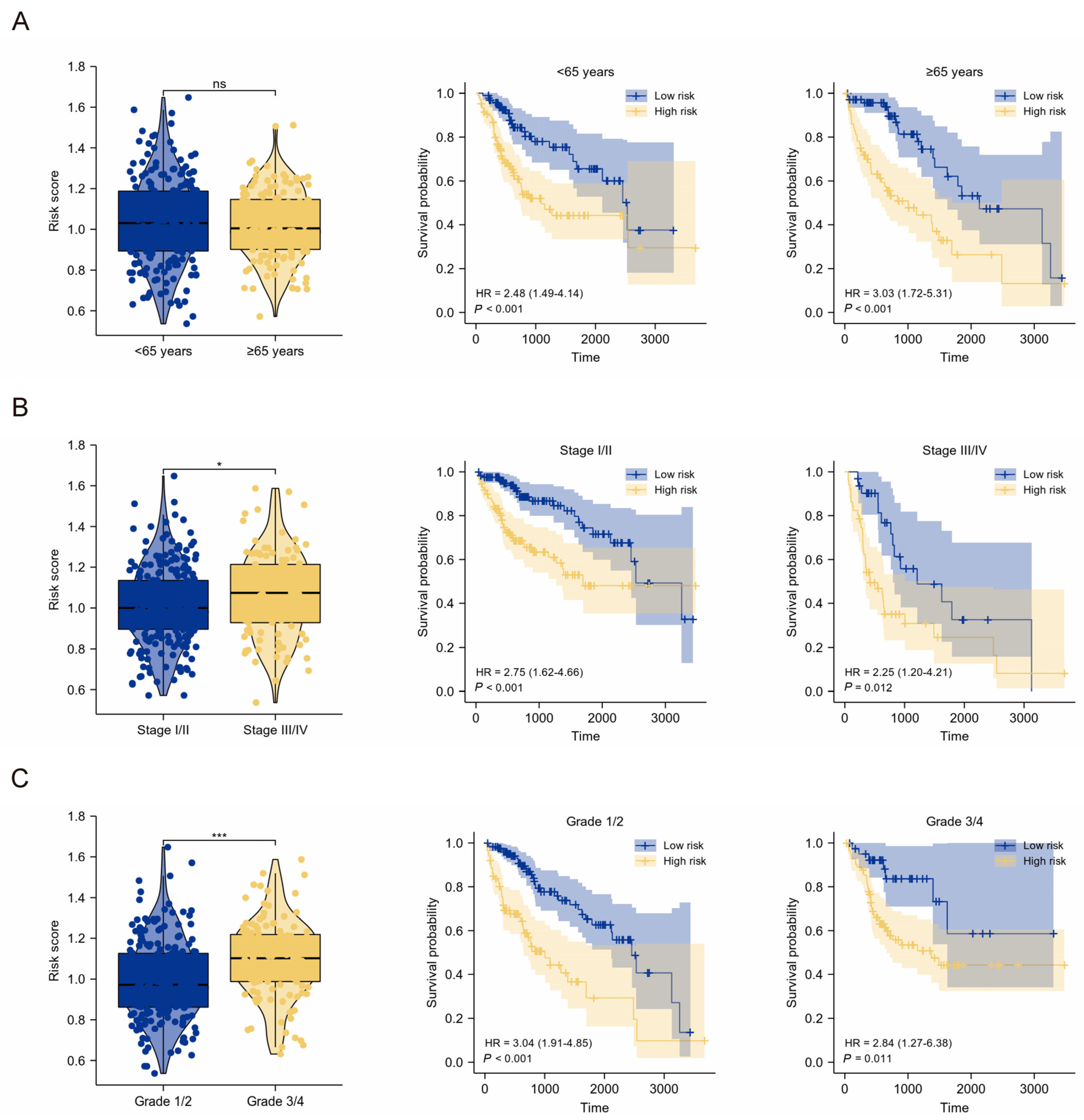
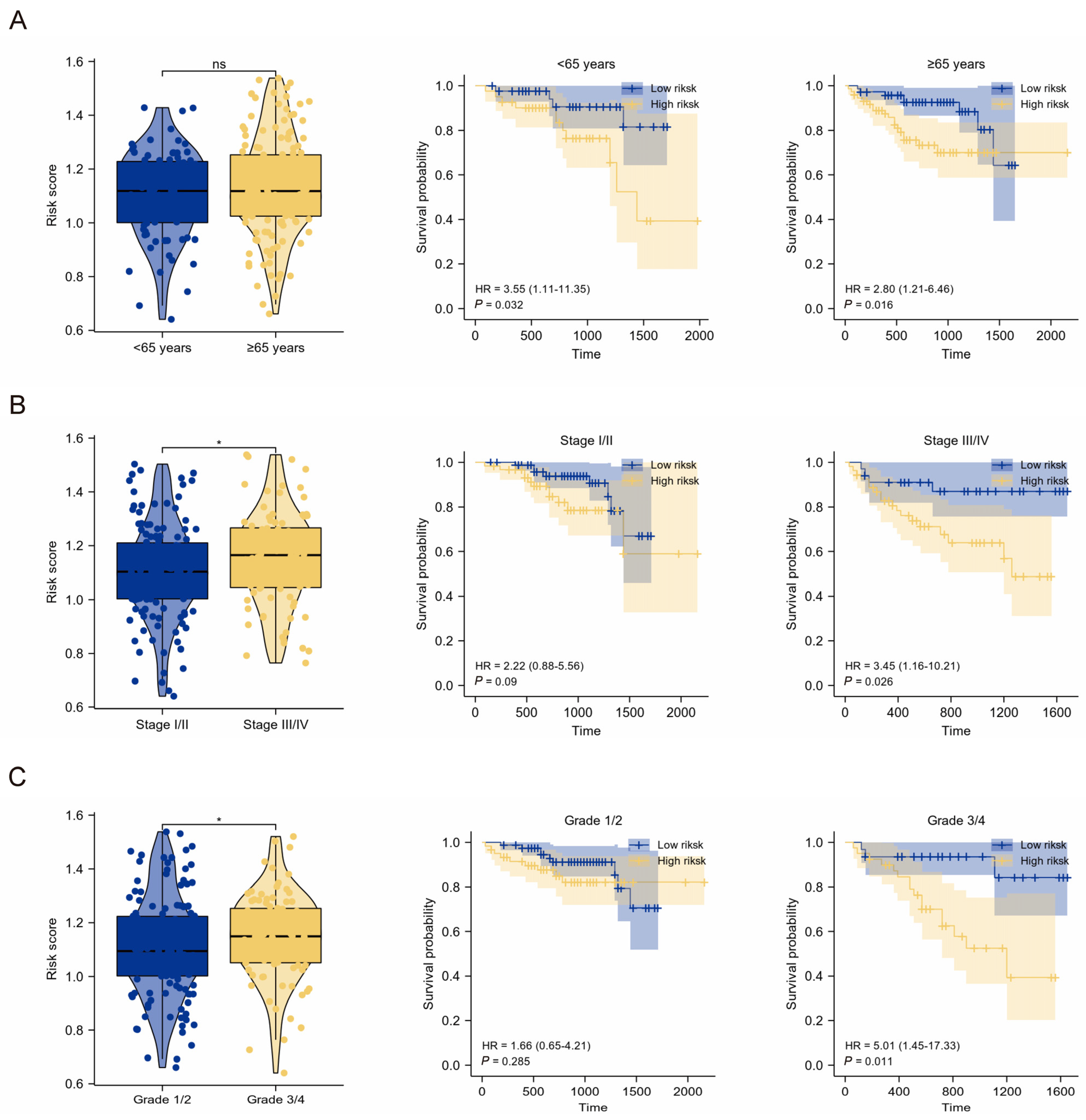
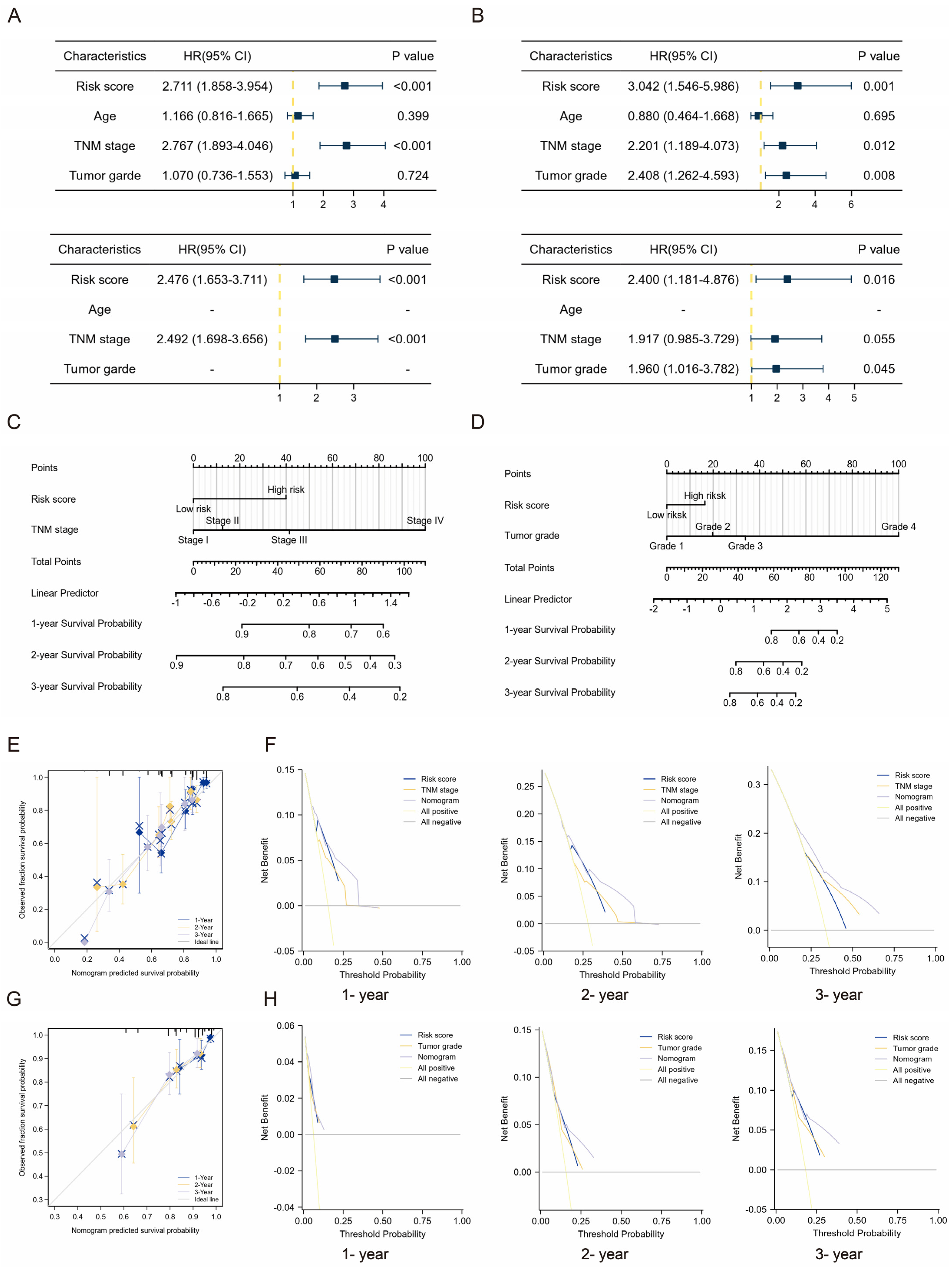
2.3. Clinical Relevance of the TMEMs Signature
2.4. Clinical Application of TMEMs Signature
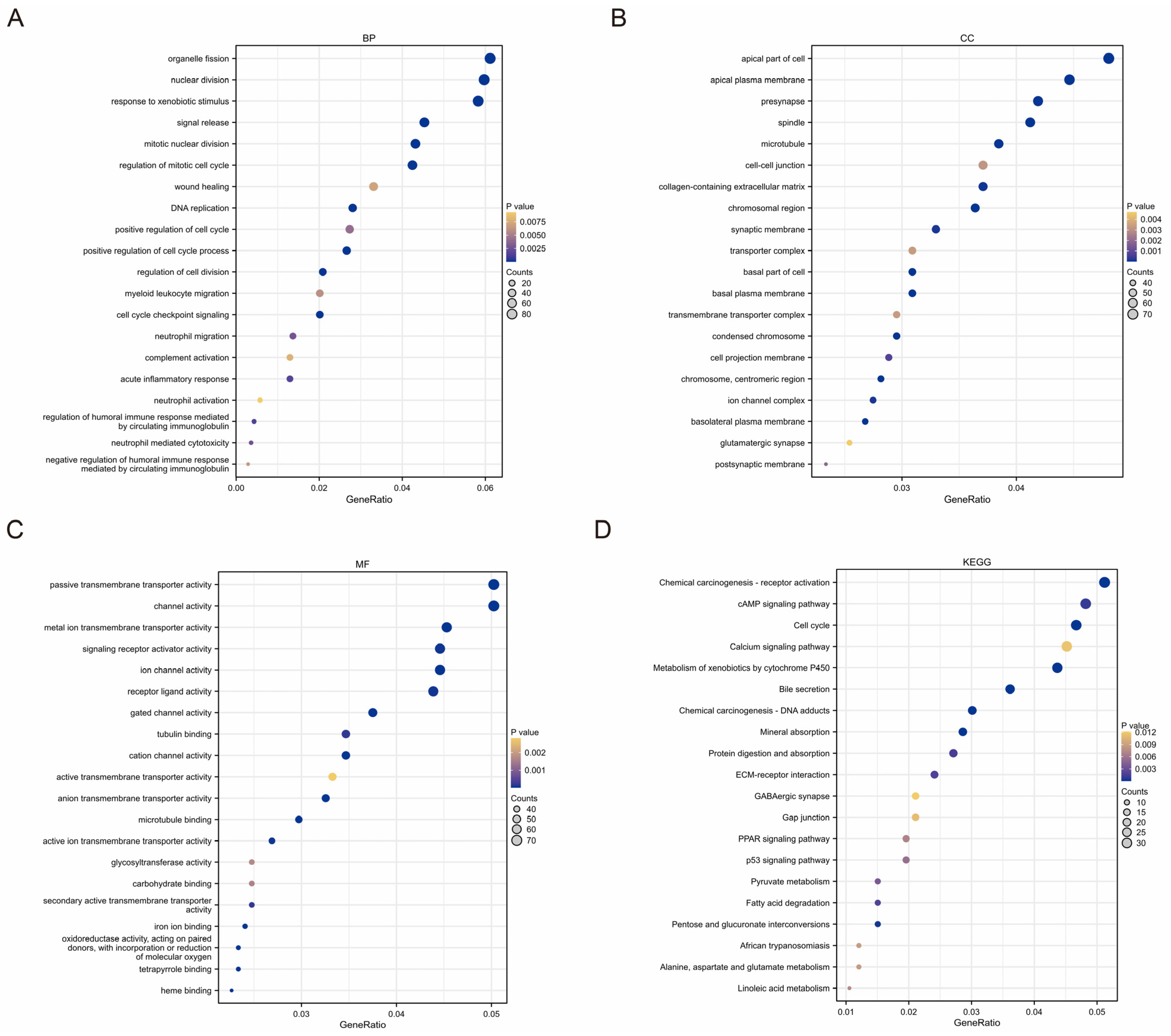
2.5. Functional Analyses Based on TMEMs Signature
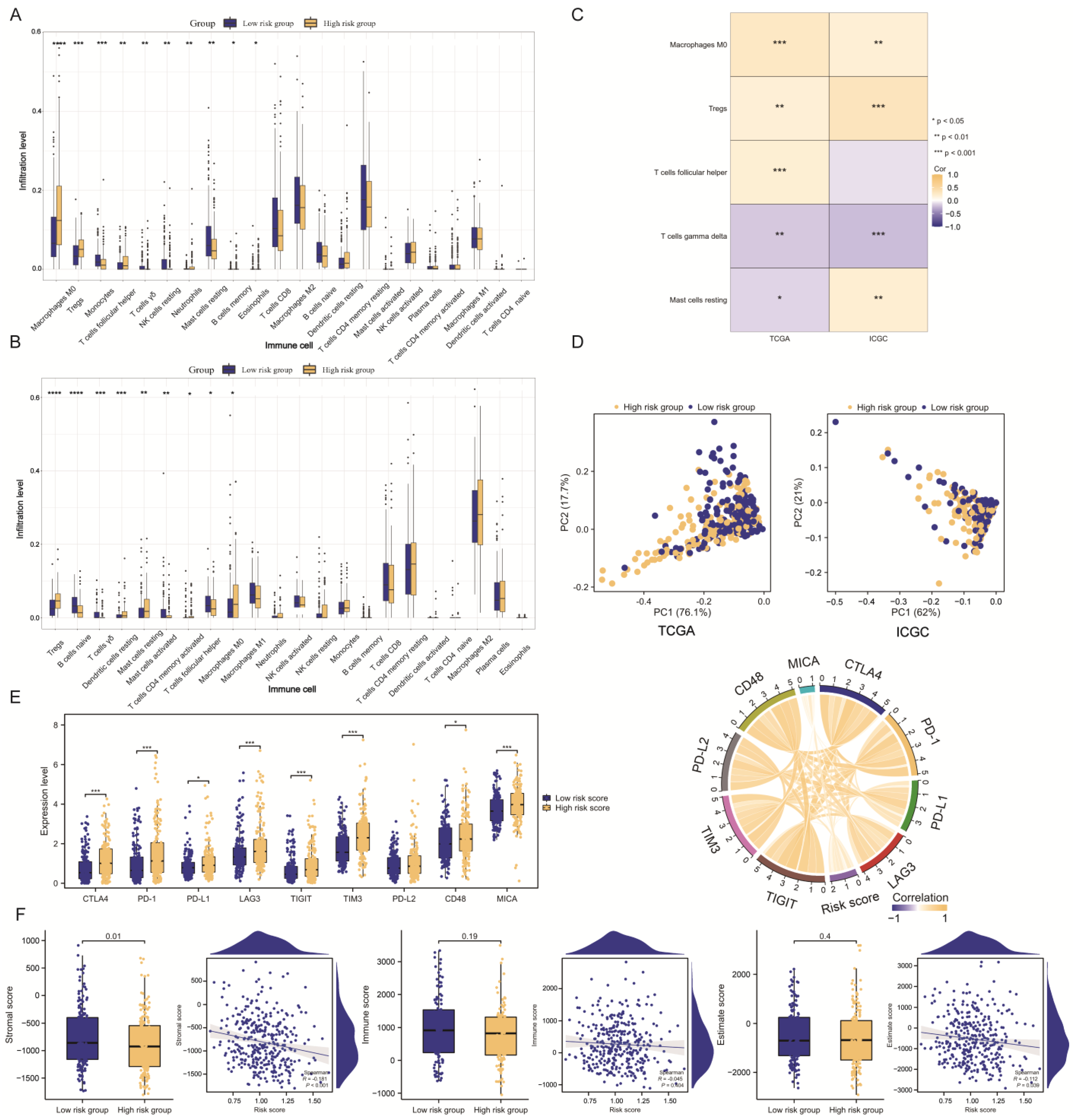
2.6. HCC Immune Landscape Affected by TMEMs Signature
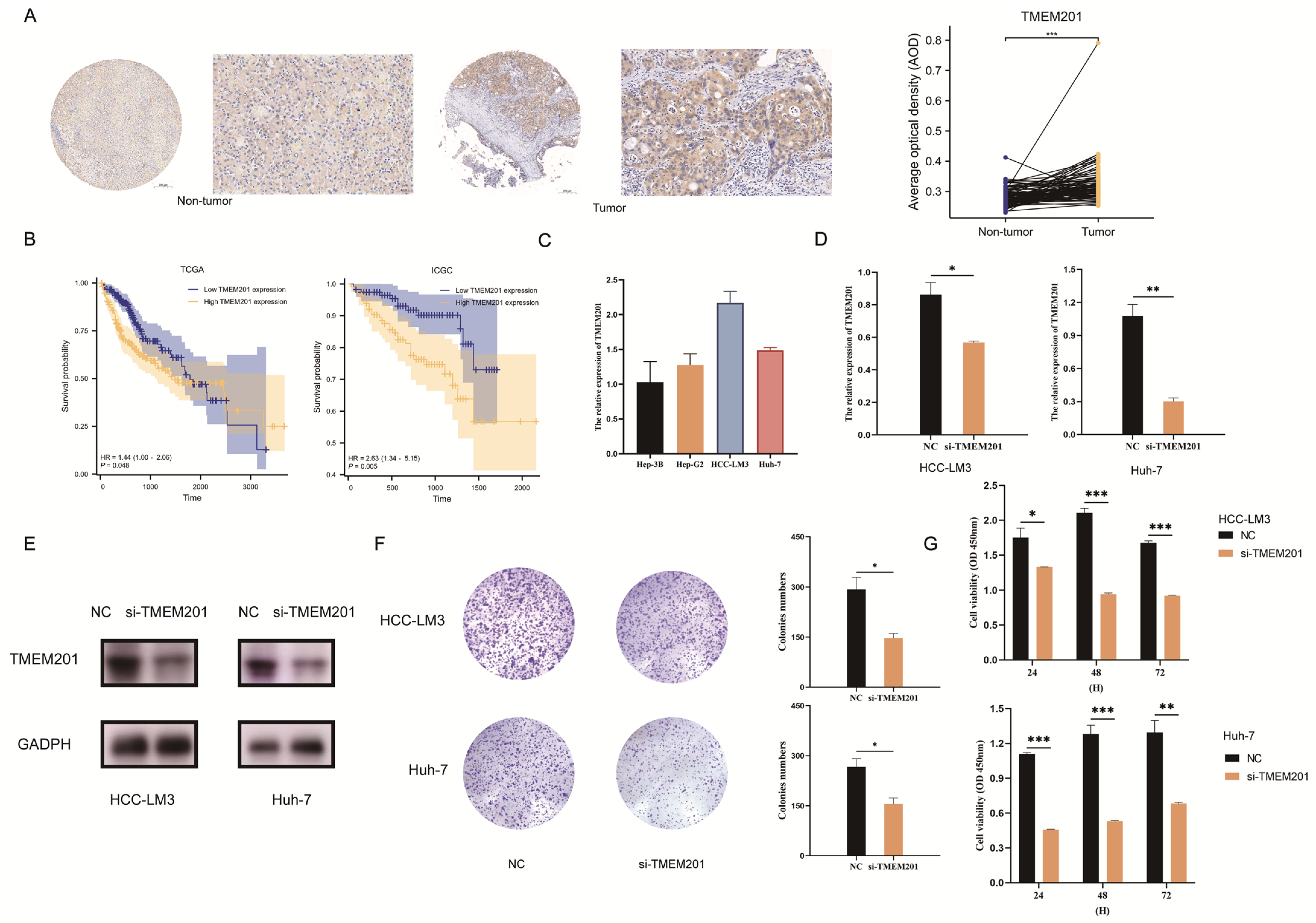
2.7. TMEM201 in HCC Development
3. Discussion
4. Methods and Materials
4.1. Data Sources
4.2. Identification of TMEMs in HCC
4.3. Construction and Validation of TMEMs Signature
4.4. Functional Enrichment Analysis
4.5. Immune Landscapes of TMEMs Signature
4.6. Cell Culture and Transfection
4.7. Immunohistochemistry (IHC) for Tissue Microarrays (TAM)
4.8. Assessment of Cell Proliferation and Evaluation of Cell Migration
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef]
- Brown, Z.J.; Tsilimigras, D.I.; Ruff, S.M.; Mohseni, A.; Kamel, I.R.; Cloyd, J.M.; Pawlik, T.M. Management of Hepatocellular Carcinoma: A Review. JAMA Surg. 2023, 158, 410. [Google Scholar] [CrossRef]
- Chen, L.; Fan, Z.; Zhao, Y.; Yang, H.; Lv, G. Genetic factors in the clinical predictive model for hepatocellular carcinoma: Evidence from genetic association analyses. J. Hepatol. 2023, 78, 3. [Google Scholar] [CrossRef]
- Chan, L.K.; Tsui, Y.M.; Ho, D.W.; Ng, I.O. Cellular heterogeneity and plasticity in liver cancer. Semin. Cancer Biol. 2022, 82, 134–149. [Google Scholar] [CrossRef]
- Marx, S.; Dal Maso, T.; Chen, J.W.; Bury, M.; Wouters, J.; Michiels, C.; Le Calvé, B. Transmembrane (TMEM) protein family members: Poorly characterized even if essential for the metastatic process. Semin. Cancer Biol. 2020, 60, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Han, Y.; Jin, W.; Tian, M.; Chen, P.; Min, J.; Hu, H.; Xu, B.; Zhu, W.; Xiong, L.; et al. Overexpression and biological function of TMEM48 in non-small cell lung carcinoma. Tumor. Biol. 2016, 37, 2575–2586. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Fen, Y.; Lin, X.; Ma, T.; Cai, H.; Ding, H. Overexpression of MAC30 is Resistant to Platinum-Based Chemotherapy in Patients With Non-Small Cell Lung Cancer. Technol. Cancer Res. Treat. 2016, 15, 815–820. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, K.; Zhang, Y.; Xu, H.; Zhang, X.; Wang, L.; Fan, C.; Jiang, G.; Wang, E. TMEM17 promotes malignant progression of breast cancer via AKT/GSK3β signaling. Cancer Manag. Res. 2018, 10, 2419–2428. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Han, G.; Lu, R.; Guan, S.; Wang, Y.; Ju, L.; Chen, L.; Shao, J.; Bian, Z. Transmembrane protein 106C promotes the development of hepatocellular carcinoma. J. Cell Biochem. 2020, 121, 4484–4495. [Google Scholar] [CrossRef]
- Sun, Y.; Deng, J.; Xia, P.; Chen, W.; Wang, L. The Expression of TMEM74 in Liver Cancer and Lung Cancer Correlating With Survival Outcomes. Appl. Immunohistochem. Mol. Morphol. 2019, 27, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Herman, J.G.; Linghu, E.; Guo, M. Epigenetic silencing of TMEM176A promotes esophageal squamous cell cancer development. Oncotarget 2017, 8, 70035–70048. [Google Scholar] [CrossRef] [PubMed]
- Bu, Y.; Liu, F.; Jia, Q.A.; Yu, S.N. Decreased Expression of TMEM173 Predicts Poor Prognosis in Patients with Hepatocellular Carcinoma. PLoS ONE 2016, 11, e0165681. [Google Scholar] [CrossRef] [PubMed]
- Doolan, P.; Clynes, M.; Kennedy, S.; Mehta, J.P.; Germano, S.; Ehrhardt, C.; Crown, J.; O’Driscoll, L. TMEM25, REPS2 and Meis 1: Favourable prognostic and predictive biomarkers for breast cancer. Tumor. Biol. 2009, 30, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.T.; Lo, C.M.; Guo, D.Y.; Qi, X.; Li, C.X.; Geng, W.; Liu, X.B.; Ling, C.C.; Ma, Y.Y.; Yeung, W.H.; et al. Identification of transmembrane protein 98 as a novel chemoresistance-conferring gene in hepatocellular carcinoma. Mol. Cancer Ther. 2014, 13, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Cheng, Y.; Zhang, Y.; Mo, X.; Li, T.; Liu, Y.; Wang, P.; Pan, W.; Chen, Y.; Xue, Y.; et al. The Secreted Form of Transmembrane Protein 98 Promotes the Differentiation of T Helper 1 Cells. J. Interferon Cytokine Res. 2015, 35, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Castet, F.; Heikenwalder, M.; Maini, M.K.; Mazzaferro, V.; Pinato, D.J.; Pikarsky, E.; Zhu, A.X.; Finn, R.S. Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2022, 19, 151–172. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.D.; Li, H.L.; Huang, M.S.; Yang, W.Z.; Yin, G.W.; Zhong, B.Y.; Sun, J.H.; Jin, Z.C.; Chen, J.J.; Ge, N.J.; et al. Transarterial chemoembolization with PD-(L)1 inhibitors plus molecular targeted therapies for hepatocellular carcinoma (CHANCE001). Signal Transduct. Target Ther. 2023, 8, 58. [Google Scholar] [CrossRef]
- Schmit, K.; Michiels, C. TMEM Proteins in Cancer: A Review. Front Pharmacol. 2018, 9, 1345. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, F.; Yang, M.; Duan, J.; Lai, J.; Sun, S.; Fan, S. LINC00238 inhibits hepatic carcinoma progression by activating TMEM106C-mediated apoptosis pathway. Mol. Med. Rep. 2021, 24, 757. [Google Scholar] [CrossRef]
- Flamant, L.; Roegiers, E.; Pierre, M.; Hayez, A.; Sterpin, C.; De Backer, O.; Arnould, T.; Poumay, Y.; Michiels, C. TMEM45A is essential for hypoxia-induced chemoresistance in breast and liver cancer cells. BMC Cancer 2012, 12, 391. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zhang, Y.; Wang, H.; Kan, W.; Guo, H.; Liu, Y.; Zang, Y.; Li, J. Inner nuclear membrane protein TMEM201 promotes breast cancer metastasis by positive regulating TGFβ signaling. Oncogene 2022, 41, 647–656. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Wang, Y.; Li, C.; Xie, Y.; Klionsky, D.J.; Kang, R.; Tang, D. TMEM164 is a new determinant of autophagy-dependent ferroptosis. Autophagy 2023, 19, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Dhar, P.; Wu, J.D. NKG2D and its ligands in cancer. Curr. Opin. Immunol. 2018, 51, 55–61. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Zhang, J.; Bajari, R.; Andric, D.; Gerthoffert, F.; Lepsa, A.; Nahal-Bose, H.; Stein, L.D.; Ferretti, V. The International Cancer Genome Consortium Data Portal. Nat. Biotechnol. 2019, 37, 367–369. [Google Scholar] [CrossRef]
- Swain, S.M.; Tang, G.; Brauer, H.A.; Goerlitz, D.S.; Lucas, P.C.; Robidoux, A.; Harris, B.T.; Bandos, H.; Ren, Y.; Geyer, C.E., Jr.; et al. NSABP B-41, a Randomized Neoadjuvant Trial: Genes and Signatures Associated with Pathologic Complete Response. Clin. Cancer Res. 2020, 26, 4233–4241. [Google Scholar] [CrossRef]
- Linghang, Q.; Yiyi, X.; Guosheng, C.; Kang, X.; Jiyuan, T.; Xiong, L.; Guangzhong, W.; Shuiqing, L.; Yanju, L. Effects of Atractylodes Oil on Inflammatory Response and Serum Metabolites in Adjuvant Arthritis Rats. Biomed. Pharmacother. 2020, 127, 110130. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Lou, Y.; Lu, J.; Fan, X.; Zhu, Q.; Sun, H. Characterization of the Clinical Significance and Immunological Landscapes of a Novel TMEMs Signature in Hepatocellular Carcinoma and the Contribution of TMEM201 to Hepatocarcinogenesis. Int. J. Mol. Sci. 2023, 24, 10285. https://doi.org/10.3390/ijms241210285
Chen D, Lou Y, Lu J, Fan X, Zhu Q, Sun H. Characterization of the Clinical Significance and Immunological Landscapes of a Novel TMEMs Signature in Hepatocellular Carcinoma and the Contribution of TMEM201 to Hepatocarcinogenesis. International Journal of Molecular Sciences. 2023; 24(12):10285. https://doi.org/10.3390/ijms241210285
Chicago/Turabian StyleChen, Desheng, Yichao Lou, Jing Lu, Xuhui Fan, Qi Zhu, and Hongcheng Sun. 2023. "Characterization of the Clinical Significance and Immunological Landscapes of a Novel TMEMs Signature in Hepatocellular Carcinoma and the Contribution of TMEM201 to Hepatocarcinogenesis" International Journal of Molecular Sciences 24, no. 12: 10285. https://doi.org/10.3390/ijms241210285
APA StyleChen, D., Lou, Y., Lu, J., Fan, X., Zhu, Q., & Sun, H. (2023). Characterization of the Clinical Significance and Immunological Landscapes of a Novel TMEMs Signature in Hepatocellular Carcinoma and the Contribution of TMEM201 to Hepatocarcinogenesis. International Journal of Molecular Sciences, 24(12), 10285. https://doi.org/10.3390/ijms241210285




