Alterations of miRNA Expression in Diffuse Hyperplastic Perilobar Nephroblastomatosis: Mapping the Way to Understanding Wilms’ Tumor Development and Differential Diagnosis
Abstract
1. Introduction
2. Results
3. Discussion
3.1. Clinical Relevance
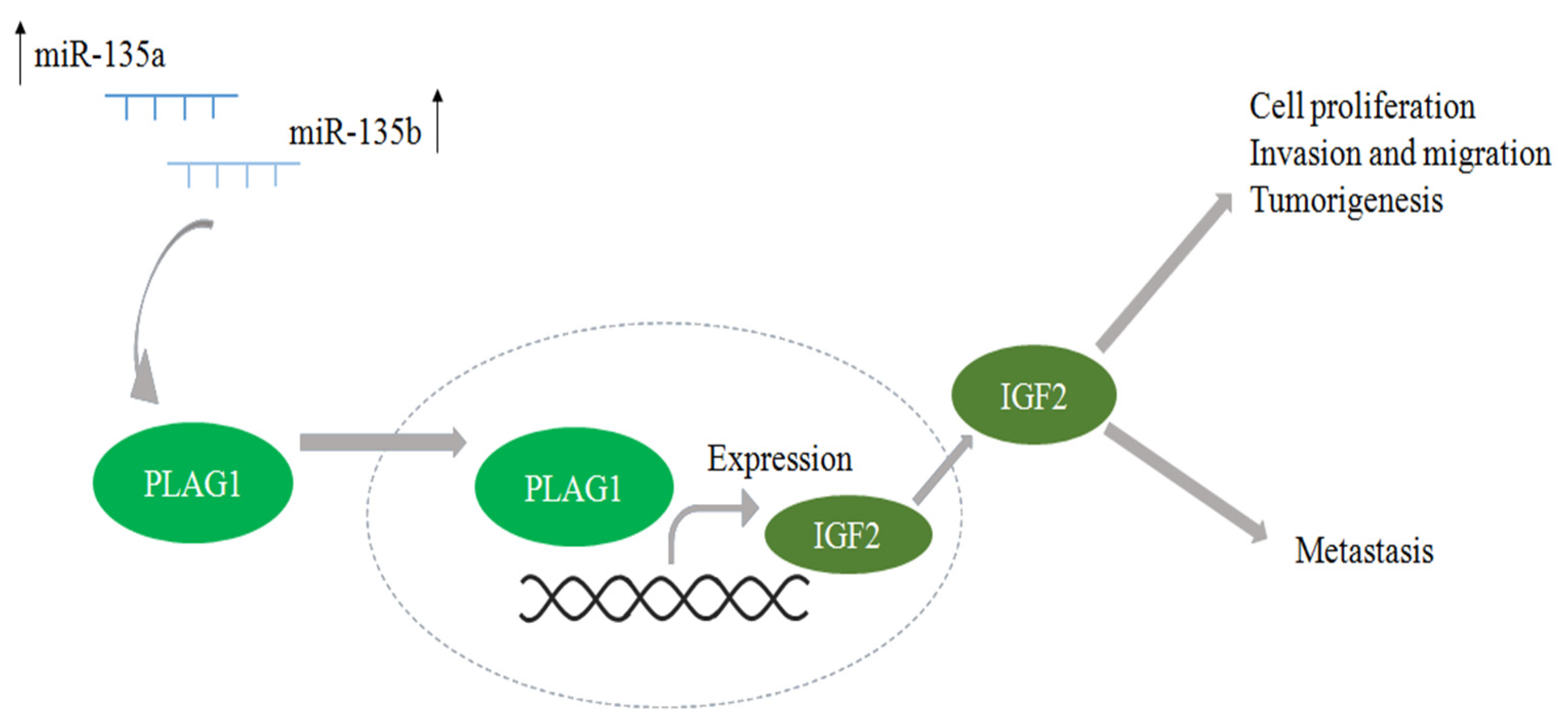
3.2. Results in Literature Context
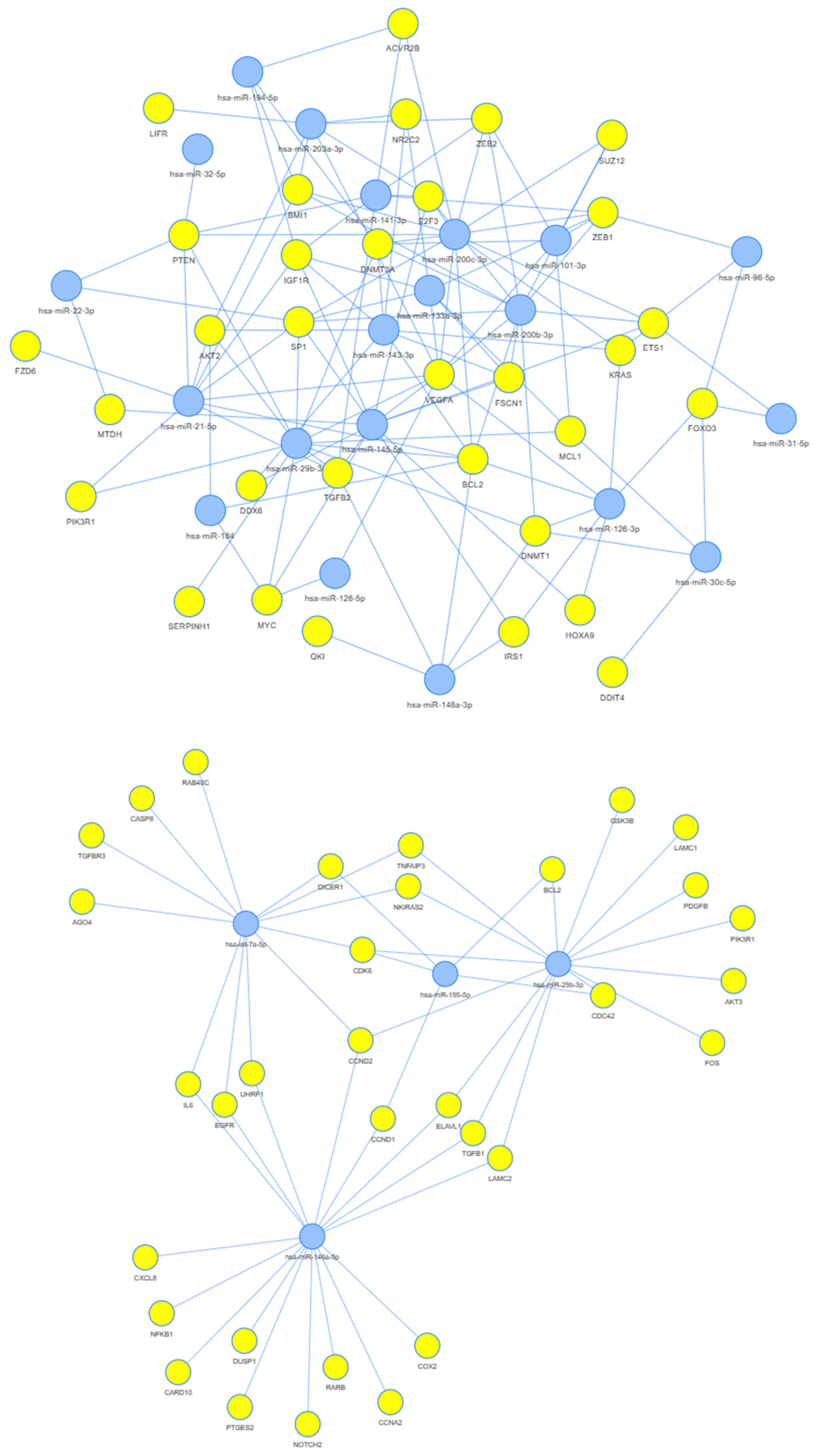
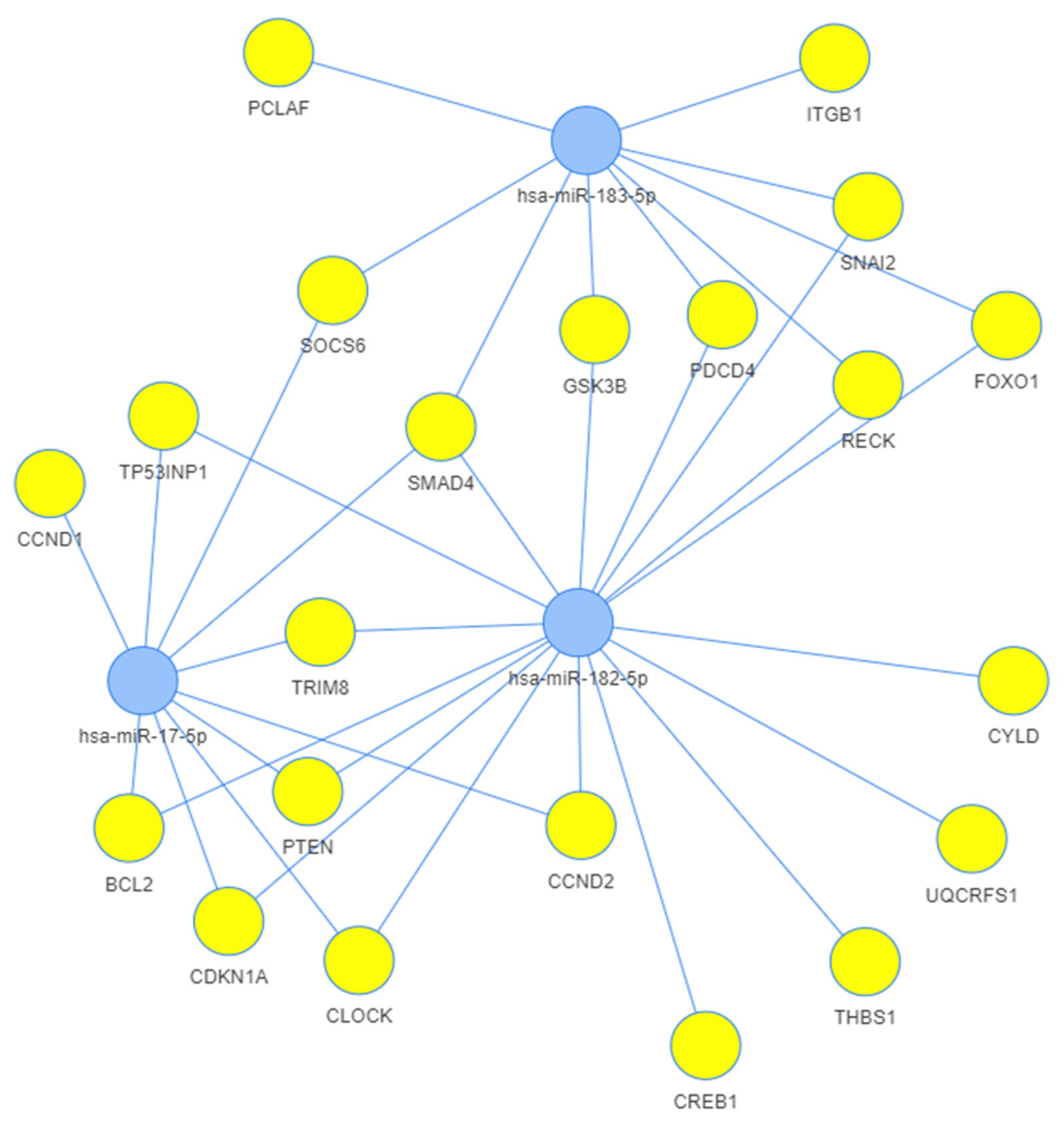
3.3. Network Analysis
4. Materials and Methods
4.1. Patients
4.2. Sample Handling
4.3. RNA Extraction and Reverse Transcription
4.4. Studying miRNA Expression Levels with qRT-PCR Arrays
4.5. Network Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nakata, K.; Colombet, M.; Stiller, C.A.; Pritchard-Jones, K.; Steliarova-Foucher, E. IICC-3 Contributors Incidence of Childhood Renal Tumours: An International Population-Based Study. Int. J. Cancer 2020, 147, 3313–3327. [Google Scholar] [CrossRef]
- Khan, A.; Feulefack, J.; Sergi, C.M. Exposure to Pesticides and Pediatric Wilms’ Tumor. A Meta-Analysis on Pre-Conception and Pregnancy Parental Exposure with an IARC/WHO Commentary. Hum. Exp. Toxicol. 2022, 41, 9603271221136211. [Google Scholar] [CrossRef]
- Srinivasan, A.S.; Saade-Lemus, S.; Servaes, S.E.; Acord, M.R.; Reid, J.R.; Anupindi, S.A.; States, L.J. Imaging Surveillance for Children with Predisposition to Renal Tumors. Pediatr. Radiol. 2019, 49, 1453–1462. [Google Scholar] [CrossRef]
- Turner, J.T.; Brzezinski, J.; Dome, J.S. Wilms Tumor Predisposition. In GeneReviews; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, DC, USA, 2003. [Google Scholar]
- Sánta, F.; Semjén, D.; Kuthi, L. Hereditary renal tumor syndromes. Orv. Hetil. 2023, 164, 363–375. [Google Scholar] [CrossRef]
- Graf, N.; Bergeron, C.; Brok, J.; de Camargo, B.; Chowdhury, T.; Furtwängler, R.; Gessler, M.; Godzinski, J.; Pritchard-Jones, K.; Ramirez-Villar, G.L.; et al. Fifty Years of Clinical and Research Studies for Childhood Renal Tumors within the International Society of Pediatric Oncology (SIOP). Ann. Oncol. 2021, 32, 1327–1331. [Google Scholar] [CrossRef] [PubMed]
- Popov, S.D.; Sebire, N.J.; Vujanic, G.M. Wilms’ Tumour—Histology and Differential Diagnosis. In Wilms Tumor; van den Heuvel-Eibrink, M.M., Ed.; Codon Publications: Brisbane, Australia, 2016; ISBN 9780994438119. [Google Scholar]
- Ehrlich, P.F.; Tornwall, B.; Chintagumpala, M.M.; Chi, Y.-Y.; Hoffer, F.A.; Perlman, E.J.; Kalapurakal, J.A.; Warwick, A.; Shamberger, R.C.; Khanna, G.; et al. Kidney Preservation and Wilms Tumor Development in Children with Diffuse Hyperplastic Perilobar Nephroblastomatosis: A Report from the Children’s Oncology Group Study AREN0534. Ann. Surg. Oncol. 2022, 29, 3252–3261. [Google Scholar] [CrossRef]
- Stabouli, S.; Printza, N.; Dotis, J.; Matis, A.; Koliouskas, D.; Gombakis, N.; Papachristou, F. Perilobar Nephroblastomatosis: Natural History and Management. Case Rep. Pediatr. 2014, 2014, 756819. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, J.K.; Chi, Y.-Y.; Smith, E.A.; Servaes, S.; Hoffer, F.A.; Mullen, E.A.; Perlman, E.J.; Tornwall, B.; Ehrlich, P.F.; Geller, J.I.; et al. Imaging Characteristics of Nephrogenic Rests Versus Small Wilms Tumors: A Report From the Children’s Oncology Group Study AREN03B2. AJR Am. J. Roentgenol. 2020, 214, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Charlton, J.; Irtan, S.; Bergeron, C.; Pritchard-Jones, K. Bilateral Wilms Tumour: A Review of Clinical and Molecular Features. Expert Rev. Mol. Med. 2017, 19, e8. [Google Scholar] [CrossRef]
- Santín, S.; Fraga, G.; Ruíz, P.; Pardo, N.; Torrent, M.; Martí, T.; Ballarín, J.; Ars, E.; Torra, R. WT1 Mutations May Be a Cause of Severe Renal Failure due to Nephroblastomatosis in Wilms’ Tumor Patients. Clin. Nephrol. 2011, 76, 244–248. [Google Scholar] [CrossRef]
- Traub, F.; Sickmann, K.; Tessema, M.; Wilkens, L.; Kreipe, H.H.; Kamino, K. Nephroblastomatosis and Loss of WT1 Expression Associated with Trisomy 13. Virchows Arch. 2006, 448, 214–217. [Google Scholar] [CrossRef]
- Wegert, J.; Ishaque, N.; Vardapour, R.; Geörg, C.; Gu, Z.; Bieg, M.; Ziegler, B.; Bausenwein, S.; Nourkami, N.; Ludwig, N.; et al. Mutations in the SIX1/2 Pathway and the DROSHA/DGCR8 miRNA Microprocessor Complex Underlie High-Risk Blastemal Type Wilms Tumors. Cancer Cell 2015, 27, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Bánki, T.; Drost, J.; van den Heuvel-Eibrink, M.M.; Mavinkurve-Groothuis, A.M.C.; de Krijger, R.R. Somatic, Genetic and Epigenetic Changes in Nephrogenic Rests and Their Role in the Transformation to Wilms Tumors, a Systematic Review. Cancers 2023, 15, 1363. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, D.M.; Tayeb, M.; Ho, J. MicroRNAs in Kidney Development and Disease. JCI Insight 2022, 7, e158277. [Google Scholar] [CrossRef]
- Liu, M.; Roth, A.; Yu, M.; Morris, R.; Bersani, F.; Rivera, M.N.; Lu, J.; Shioda, T.; Vasudevan, S.; Ramaswamy, S.; et al. The IGF2 Intronic miR-483 Selectively Enhances Transcription from IGF2 Fetal Promoters and Enhances Tumorigenesis. Genes Dev. 2013, 27, 2543–2548. [Google Scholar] [CrossRef] [PubMed]
- Kort, E.J.; Farber, L.; Tretiakova, M.; Petillo, D.; Furge, K.A.; Yang, X.J.; Cornelius, A.; Teh, B.T. The E2F3-Oncomir-1 Axis Is Activated in Wilms’ Tumor. Cancer Res. 2008, 68, 4034–4038. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J.; Backes, C.; Nourkami-Tutdibi, N.; Leidinger, P.; Deutscher, S.; Beier, M.; Gessler, M.; Graf, N.; Lenhof, H.-P.; Keller, A.; et al. Treatment-Independent miRNA Signature in Blood of Wilms Tumor Patients. BMC Genom. 2012, 13, 379. [Google Scholar] [CrossRef]
- Buglyó, G.; Magyar, Z.; Görbe, É.R.; Bánusz, R.; Csóka, M.; Micsik, T.; Berki, Z.; Varga, P.; Sápi, Z.; Nagy, B. Quantitative RT-PCR-Based miRNA Profiling of Blastemal Wilms’ Tumors from Formalin-Fixed Paraffin-Embedded Samples. J. Biotechnol. 2019, 298, 11–15. [Google Scholar] [CrossRef]
- Buglyó, G.; Magyar, Z.; Görbe, É.R.; Bánusz, R.; Csóka, M.; Micsik, T.; Mezei, M.; Yani, J.A.S.; Varga, P.; Sápi, Z.; et al. miRNA Profiling of Hungarian Regressive Wilms’ Tumor Formalin-Fixed Paraffin-Embedded (FFPE) Samples by Quantitative Real-Time Polymerase Chain Reaction (RT-PCR). Med. Sci. Monit. 2021, 27, e932731-1. [Google Scholar] [CrossRef]
- Beckwith, J.B. Nephrogenic Rests and the Pathogenesis of Wilms Tumor: Developmental and Clinical Considerations. Am. J. Med. Genet. 1998, 79, 268–273. [Google Scholar] [CrossRef]
- Perlman, E.J.; Faria, P.; Soares, A.; Hoffer, F.; Sredni, S.; Ritchey, M.; Shamberger, R.C.; Green, D.; Beckwith, J.B. National Wilms Tumor Study Group Hyperplastic Perilobar Nephroblastomatosis: Long-Term Survival of 52 Patients. Pediatr. Blood Cancer 2006, 46, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, U.; Das, S.; Vesely, P.; Alzoughbi, W.; Fröhlich, L.F.; Chowdhury, P.; Leuschner, I.; Hoefler, G.; Guertl, B. miR-192, miR-194, miR-215, miR-200c and miR-141 Are Downregulated and Their Common Target ACVR2B Is Strongly Expressed in Renal Childhood Neoplasms. Carcinogenesis 2012, 33, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Gadd, S.L.; Huang, C.-C.; Lu, Y.; Huff, V.; Perlman, E.J. Abstract 5353: Comprehensive Ggnomic Analysis of Diffuse Hyperplastic Perilobar Nephroblastomatosis (DHPLN). Cancer Res. 2011, 71, 5353. [Google Scholar] [CrossRef]
- Leichter, A.L.; Sullivan, M.J.; Eccles, M.R.; Chatterjee, A. MicroRNA Expression Patterns and Signalling Pathways in the Development and Progression of Childhood Solid Tumours. Mol. Cancer 2017, 16, 15. [Google Scholar] [CrossRef]
- Hohenstein, P.; Pritchard-Jones, K.; Charlton, J. The Yin and Yang of Kidney Development and Wilms’ Tumors. Genes Dev. 2015, 29, 467–482. [Google Scholar] [CrossRef]
- Juma, A.R.; Damdimopoulou, P.E.; Grommen, S.V.H.; Van de Ven, W.J.M.; De Groef, B. Emerging Role of PLAG1 as a Regulator of Growth and Reproduction. J. Endocrinol. 2016, 228, R45–R56. [Google Scholar] [CrossRef] [PubMed]
- Iacona, J.R.; Lutz, C.S. miR-146a-5p: Expression, Regulation, and Functions in Cancer. Wiley Interdiscip. Rev. RNA 2019, 10, e1533. [Google Scholar] [CrossRef]
- Hermann, H.; Runnel, T.; Aab, A.; Baurecht, H.; Rodriguez, E.; Magilnick, N.; Urgard, E.; Šahmatova, L.; Prans, E.; Maslovskaja, J.; et al. miR-146b Probably Assists miRNA-146a in the Suppression of Keratinocyte Proliferation and Inflammatory Responses in Psoriasis. J. Invest. Dermatol. 2017, 137, 1945–1954. [Google Scholar] [CrossRef]
- Akpa, M.M.; Iglesias, D.; Chu, L.; Thiébaut, A.; Jentoft, I.; Hammond, L.; Torban, E.; Goodyer, P.R. Wilms Tumor Suppressor, WT1, Cooperates with MicroRNA-26a and MicroRNA-101 to Suppress Translation of the Polycomb Protein, EZH2, in Mesenchymal Stem Cells. J. Biol. Chem. 2016, 291, 3785–3795. [Google Scholar] [CrossRef]
- Yan, S.; Wang, H.; Chen, X.; Liang, C.; Shang, W.; Wang, L.; Li, J.; Xu, D. MiR-182-5p Inhibits Colon Cancer Tumorigenesis, Angiogenesis, and Lymphangiogenesis by Directly Downregulating VEGF-C. Cancer Lett. 2020, 488, 18–26. [Google Scholar] [CrossRef]
- Wang, F.; Wu, D.; Xu, Z.; Chen, J.; Zhang, J.; Li, X.; Chen, S.; He, F.; Xu, J.; Su, L.; et al. miR-182-5p Affects Human Bladder Cancer Cell Proliferation, Migration and Invasion through Regulating Cofilin 1. Cancer Cell Int. 2019, 19, 42. [Google Scholar] [CrossRef]
- Wang, A.; Jin, C.; Li, H.; Qin, Q.; Li, L. LncRNA ADAMTS9-AS2 Regulates Ovarian Cancer Progression by Targeting miR-182-5p/FOXF2 Signaling Pathway. Int. J. Biol. Macromol. 2018, 120, 1705–1713. [Google Scholar] [CrossRef]
- Roser, A.-E.; Caldi Gomes, L.; Halder, R.; Jain, G.; Maass, F.; Tönges, L.; Tatenhorst, L.; Bähr, M.; Fischer, A.; Lingor, P. miR-182-5p and miR-183-5p Act as GDNF Mimics in Dopaminergic Midbrain Neurons. Mol. Ther. Nucleic Acids 2018, 11, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Amatya, V.J.; Kushitani, K.; Kai, Y.; Kambara, T.; Takeshima, Y. miR-182 and miR-183 Promote Cell Proliferation and Invasion by Targeting FOXO1 in Mesothelioma. Front. Oncol. 2018, 8, 446. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Chen, D.; Li, Y.; Zhang, E.; Yu, Z.; Chen, T.; Jiang, Z.; Ni, L.; Yang, S.; Gui, Y.; et al. microRNA-184 Functions as Tumor Suppressor in Renal Cell Carcinoma. Exp. Ther. Med. 2015, 9, 961–966. [Google Scholar] [CrossRef]
- Khalilian, S.; Abedinlou, H.; Hussen, B.M.; Imani, S.Z.H.; Ghafouri-Fard, S. The Emerging Role of miR-20b in Human Cancer and Other Disorders: Pathophysiology and Therapeutic Implications. Front. Oncol. 2022, 12, 985457. [Google Scholar] [CrossRef]
- Nguyen, T.T.P.; Suman, K.H.; Nguyen, T.B.; Nguyen, H.T.; Do, D.N. The Role of miR-29s in Human Cancers—An Update. Biomedicines 2022, 10, 2121. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.T.; Bossone, S.A.; Zhu, G.; McDonald, G.A.; Sukhatme, V.P. Sp1 Is a Critical Regulator of the Wilms’ Tumor-1 Gene. J. Biol. Chem. 1997, 272, 2901–2913. [Google Scholar] [CrossRef]
- Hong, B.; Dong, R. Research Advances in the Targeted Therapy and Immunotherapy of Wilms Tumor: A Narrative Review. Transl. Cancer Res. 2021, 10, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Wünsch, L.; Flemming, P.; Glüer, S. Expression of MIB and BCL-2 in Patients with Nephrogenic Rests with and without Associated Wilms’ Tumors. Eur. J. Pediatr. Surg. 2001, 11, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Desai, K.; Kim, J.; Zhou, Q.; Guo, L.; Xiao, X.; Zhang, Y.; Zhou, L.; Yuksel, A.; Catchpoole, D.R.; et al. Wilms Tumor Mutational Subclasses Converge to Drive Overexpression. medRxiv 2023. [Google Scholar] [CrossRef]
- Sakairi, T.; Abe, Y.; Kopp, J.B. TGF-beta1 Reduces Wilms’ Tumor Suppressor Gene Expression in Podocytes. Nephrol. Dial. Transplant. 2011, 26, 2746–2752. [Google Scholar] [CrossRef]
- Shi, Q.; Wu, H.; Li, Y.; Shen, L.; Tian, X.; Lin, T.; Wei, G. Inhibition of Wilms’ Tumor Proliferation and Invasion by Blocking TGF-β Receptor I in the TGF-β/Smad Signaling Pathway. BioMed Res. Int. 2020, 2020, 8039840. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.H.; Kupa, J.; Duffy, K.A.; Kalish, J.M. Diagnosis and Management of Beckwith-Wiedemann Syndrome. Front. Pediatr. 2019, 7, 562. [Google Scholar] [CrossRef]
- Li, Q.; Li, M.; Zheng, K.; Li, H.; Yang, H.; Ma, S.; Zhong, M. Detection of microRNA Expression Levels Based on Microarray Analysis for Classification of Idiopathic Pulmonary Fibrosis. Exp. Ther. Med. 2020, 20, 3096–3103. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.; Greytak, S.; Odeh, H.; Guan, P.; Powers, J.; Bavarva, J.; Moore, H.M. Deleterious Effects of Formalin-Fixation and Delays to Fixation on RNA and miRNA-Seq Profiles. Sci. Rep. 2019, 9, 6980. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Licursi, V.; Conte, F.; Fiscon, G.; Paci, P. MIENTURNET: An Interactive Web Tool for microRNA-Target Enrichment and Network-Based Analysis. BMC Bioinform. 2019, 20, 545. [Google Scholar] [CrossRef]
- Li, J.; Han, X.; Wan, Y.; Zhang, S.; Zhao, Y.; Fan, R.; Cui, Q.; Zhou, Y. TAM 2.0: Tool for MicroRNA Set Analysis. Nucleic Acids Res. 2018, 46, W180–W185. [Google Scholar] [CrossRef]




| miRNA | DHPLN | WT, Weighted Average | WT—DHPLN |
|---|---|---|---|
| hsa-let-7a-5p | −0.08 | −0.93 | −0.85 |
| hsa-let-7b-5p | 0.18 | −0.92 | −1.10 |
| hsa-let-7c-5p | −0.2 | −1.14 | −0.94 |
| hsa-let-7f-5p | −0.41 | −1.02 | −0.61 |
| hsa-miR-100-5p | −0.43 | −0.13 | 0.30 |
| hsa-miR-101-3p | −1.44 | −1.50 | −0.06 |
| hsa-miR-106b-5p | −0.14 | 1.02 | 1.16 |
| hsa-miR-125a-5p | 0.02 | 0.00 | −0.02 |
| hsa-miR-125b-5p | −0.69 | 0.01 | 0.70 |
| hsa-miR-126-3p | −2.32 | −1.95 | 0.37 |
| hsa-miR-126-5p | −1.96 | −1.71 | 0.25 |
| hsa-miR-128-3p | 0.02 | 1.38 | 1.36 |
| hsa-miR-133a-3p | −2.56 | −1.28 | 1.28 |
| hsa-miR-135a-5p | 1.46 | 0.49 | −0.97 |
| hsa-miR-135b-5p | 2.2 | 0.00 | −2.20 |
| hsa-miR-143-3p | −1.83 | −1.54 | 0.29 |
| hsa-miR-141-3p | −5.51 | −2.49 | 3.02 |
| hsa-miR-145-5p | −1.96 | −1.85 | 0.11 |
| hsa-miR-146a-5p | −0.13 | −2.11 | −1.98 |
| hsa-miR-146b-5p | 0.64 | 0.72 | 0.08 |
| hsa-miR-148a-3p | −1.17 | −1.05 | 0.12 |
| hsa-miR-15a-5p | −1.2 | −0.67 | 0.53 |
| hsa-miR-15b-5p | −0.26 | 1.31 | 1.57 |
| hsa-miR-16-5p | −0.99 | 0.08 | 1.07 |
| hsa-miR-17-5p | −0.21 | 0.63 | 0.84 |
| hsa-miR-17-3p | −0.36 | 0.70 | 1.06 |
| hsa-miR-181a-5p | 1.43 | 1.37 | −0.06 |
| hsa-miR-181b-5p | 1.62 | 2.18 | 0.56 |
| hsa-miR-182-5p | −1.61 | 4.51 | 6.12 |
| hsa-miR-183-5p | −1.26 | 3.80 | 5.06 |
| hsa-miR-184 | −4.74 | −1.09 | 3.65 |
| hsa-miR-194-5p | −5.04 | −3.68 | 1.36 |
| hsa-miR-195-5p | −0.97 | −2.12 | −1.15 |
| hsa-miR-196a-5p | 0.42 | 1.97 | 1.55 |
| hsa-miR-19b-3p | −1.05 | 0.77 | 1.82 |
| hsa-miR-200b-3p | −4.72 | −4.06 | 0.66 |
| hsa-miR-200c-3p | −4.21 | −2.72 | 1.49 |
| hsa-miR-203a-3p | −4.17 | −3.67 | 0.50 |
| hsa-miR-205-5p | −0.47 | 1.67 | 2.14 |
| hsa-miR-20a-5p | −0.26 | 0.18 | 0.44 |
| hsa-miR-20b-5p | −2.48 | 0.11 | 2.59 |
| hsa-miR-21-5p | −0.97 | −1.93 | −0.96 |
| hsa-miR-218-5p | −1.25 | 1.22 | 2.47 |
| hsa-miR-22-3p | −2.36 | −3.26 | −0.90 |
| hsa-miR-221-3p | −0.72 | −0.91 | −0.19 |
| hsa-miR-222-3p | −0.47 | −0.73 | −0.26 |
| hsa-miR-223-3p | −0.56 | 1.25 | 1.81 |
| hsa-miR-224-5p | −1.51 | 1.89 | 3.40 |
| hsa-miR-23b-3p | −0.33 | −0.22 | 0.11 |
| hsa-miR-24-3p | −0.59 | −0.27 | 0.32 |
| hsa-miR-25-3p | 0.39 | 2.27 | 1.88 |
| hsa-miR-26a-5p | −0.39 | −0.69 | −0.30 |
| hsa-miR-26b-5p | −0.25 | −0.86 | −0.61 |
| hsa-miR-27a-3p | −0.79 | −1.19 | −0.40 |
| hsa-miR-27b-3p | −0.69 | −1.22 | −0.53 |
| hsa-miR-296-5p | 0.75 | 1.02 | 0.27 |
| hsa-miR-29b-3p | −1.89 | −4.28 | −2.39 |
| hsa-miR-30c-5p | −1.76 | −1.84 | −0.08 |
| hsa-miR-31-5p | −1.54 | −2.18 | −0.64 |
| hsa-miR-3163 | −4.16 | 0.47 | 4.63 |
| hsa-miR-32-5p | −1.3 | −1.27 | 0.03 |
| hsa-miR-330-3p | −0.6 | 0.86 | 1.46 |
| hsa-miR-331-3p | −0.72 | 0.44 | 1.16 |
| hsa-miR-34a-5p | 0.35 | 0.57 | 0.22 |
| hsa-miR-34b-3p | 0.1 | −1.37 | −1.47 |
| hsa-miR-34c-5p | −2.28 | −0.55 | 1.73 |
| hsa-miR-361-5p | −0.3 | 0.68 | 0.98 |
| hsa-miR-365a-3p | −0.52 | −1.37 | −0.85 |
| hsa-miR-374b-5p | 0.22 | −1.24 | −1.46 |
| hsa-miR-375 | −2.69 | −0.07 | 2.62 |
| hsa-miR-425-5p | 0.07 | 0.63 | 0.56 |
| hsa-miR-449a | 1.24 | 1.14 | −0.10 |
| hsa-miR-455-5p | −2.71 | −0.77 | 1.94 |
| hsa-miR-494-3p | −0.88 | 0.94 | 1.82 |
| hsa-miR-616-3p | 1.04 | 1.06 | 0.02 |
| hsa-miR-7-5p | 0 | −0.79 | −0.79 |
| hsa-miR-9-3p | −2.12 | −1.74 | 0.38 |
| hsa-miR-92a-3p | 0.09 | 0.83 | 0.74 |
| hsa-miR-93-5p | 0.29 | 1.08 | 0.79 |
| hsa-miR-96-5p | −1.25 | −1.62 | −0.37 |
| hsa-miR-99a-5p | −1.22 | −0.50 | 0.72 |
| hsa-miR-99b-5p | −0.03 | 1.05 | 1.08 |
| Patient ID | Age (Years) | Sex | Date of Surgery | Laterality |
|---|---|---|---|---|
| 1 | 1 | female | 2007 * | bilateral |
| 2 | 4 | male | 1991 | bilateral |
| 3 | 4 | male | 2014 | right |
| 4 | 1 | male | 2016 | bilateral |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Csók, Á.; Micsik, T.; Magyar, Z.; Tornóczky, T.; Kuthi, L.; Nishi, Y.; Szirák, K.; Csóka, M.; Ottóffy, G.; Soltész, B.; et al. Alterations of miRNA Expression in Diffuse Hyperplastic Perilobar Nephroblastomatosis: Mapping the Way to Understanding Wilms’ Tumor Development and Differential Diagnosis. Int. J. Mol. Sci. 2023, 24, 8793. https://doi.org/10.3390/ijms24108793
Csók Á, Micsik T, Magyar Z, Tornóczky T, Kuthi L, Nishi Y, Szirák K, Csóka M, Ottóffy G, Soltész B, et al. Alterations of miRNA Expression in Diffuse Hyperplastic Perilobar Nephroblastomatosis: Mapping the Way to Understanding Wilms’ Tumor Development and Differential Diagnosis. International Journal of Molecular Sciences. 2023; 24(10):8793. https://doi.org/10.3390/ijms24108793
Chicago/Turabian StyleCsók, Ádám, Tamás Micsik, Zsófia Magyar, Tamás Tornóczky, Levente Kuthi, Yumika Nishi, Krisztina Szirák, Monika Csóka, Gábor Ottóffy, Beáta Soltész, and et al. 2023. "Alterations of miRNA Expression in Diffuse Hyperplastic Perilobar Nephroblastomatosis: Mapping the Way to Understanding Wilms’ Tumor Development and Differential Diagnosis" International Journal of Molecular Sciences 24, no. 10: 8793. https://doi.org/10.3390/ijms24108793
APA StyleCsók, Á., Micsik, T., Magyar, Z., Tornóczky, T., Kuthi, L., Nishi, Y., Szirák, K., Csóka, M., Ottóffy, G., Soltész, B., Balogh, I., & Buglyó, G. (2023). Alterations of miRNA Expression in Diffuse Hyperplastic Perilobar Nephroblastomatosis: Mapping the Way to Understanding Wilms’ Tumor Development and Differential Diagnosis. International Journal of Molecular Sciences, 24(10), 8793. https://doi.org/10.3390/ijms24108793






