Sex-Dependent Transcriptional Changes in Response to Stress in Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Pilot Project
Abstract
1. Introduction
2. Results
2.1. Participant Characteristics
2.2. Transcriptomic Changes between Maximal Exertion (T1) and Baseline before Exercise Challenge (T0) Stratified by Sex
Cell Type Abundance Changes (between T1 and T0) Stratified by Sex
2.3. Transcriptomic Changes between 4 h after Maximal Exertion (T2) and Maximal Exertion (T1) Stratified by Sex
2.3.1. Cell Type Abundance Changes (between T2 and T1)
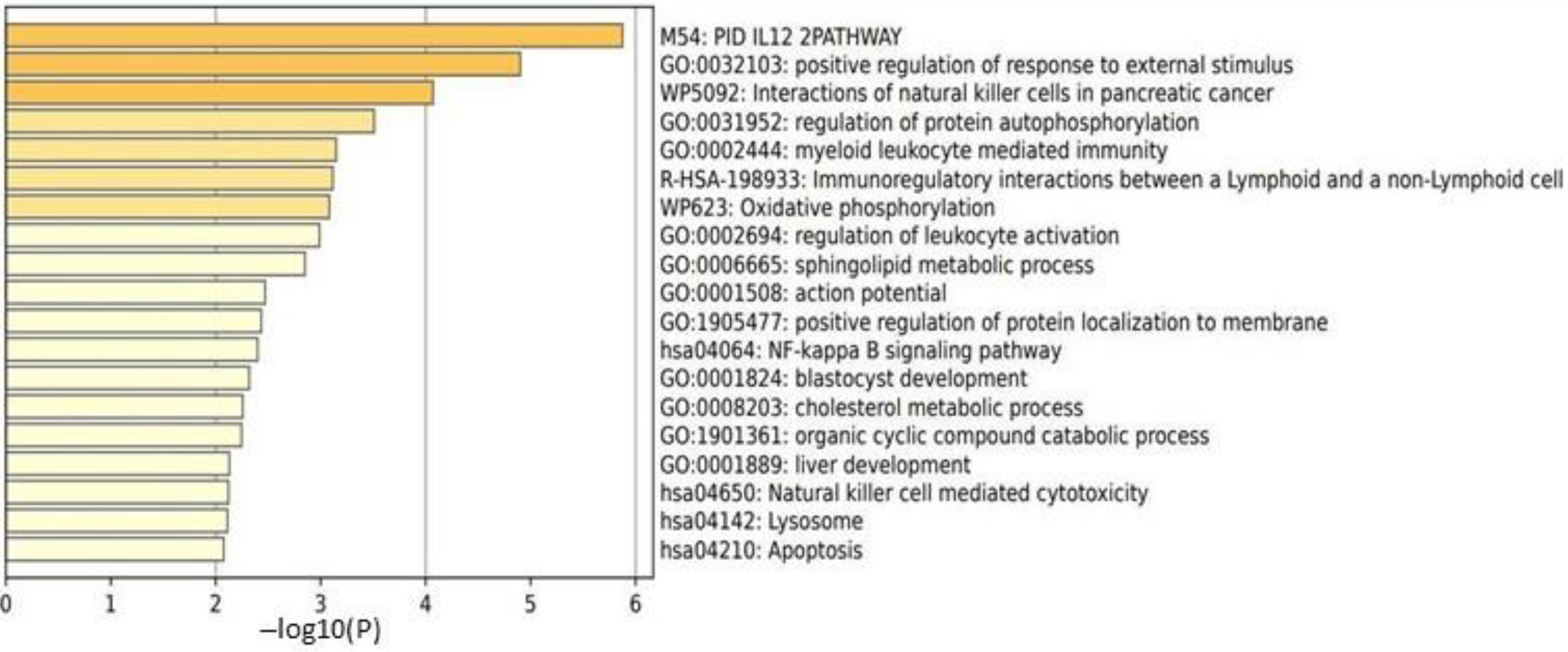
2.3.2. Male HC vs. Female HC Functional Pathway Analysis (between T2 and T1)
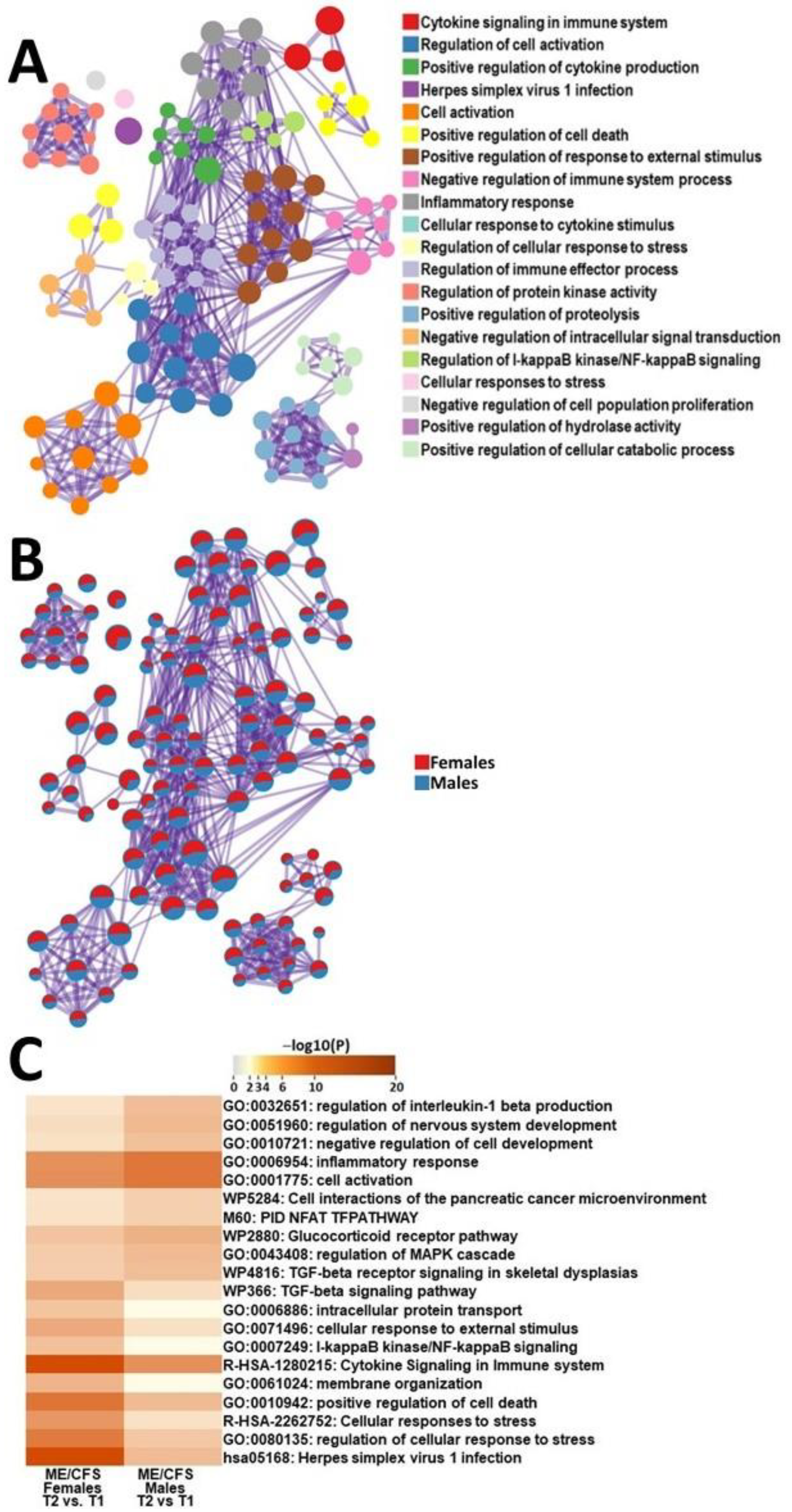
2.3.3. Male ME/CFS vs. Female ME/CFS Functional Pathway Analysis (between T2 and T1)
2.4. Transcriptomic Changes between 4 h after Maximal Exertion (T2) and Baseline (T0) Stratified by Sex
2.4.1. Cell Type Abundance Changes (between T2 and T0)
2.4.2. Male HC vs. Female HC Functional Pathway Analysis (between T2 and T0)
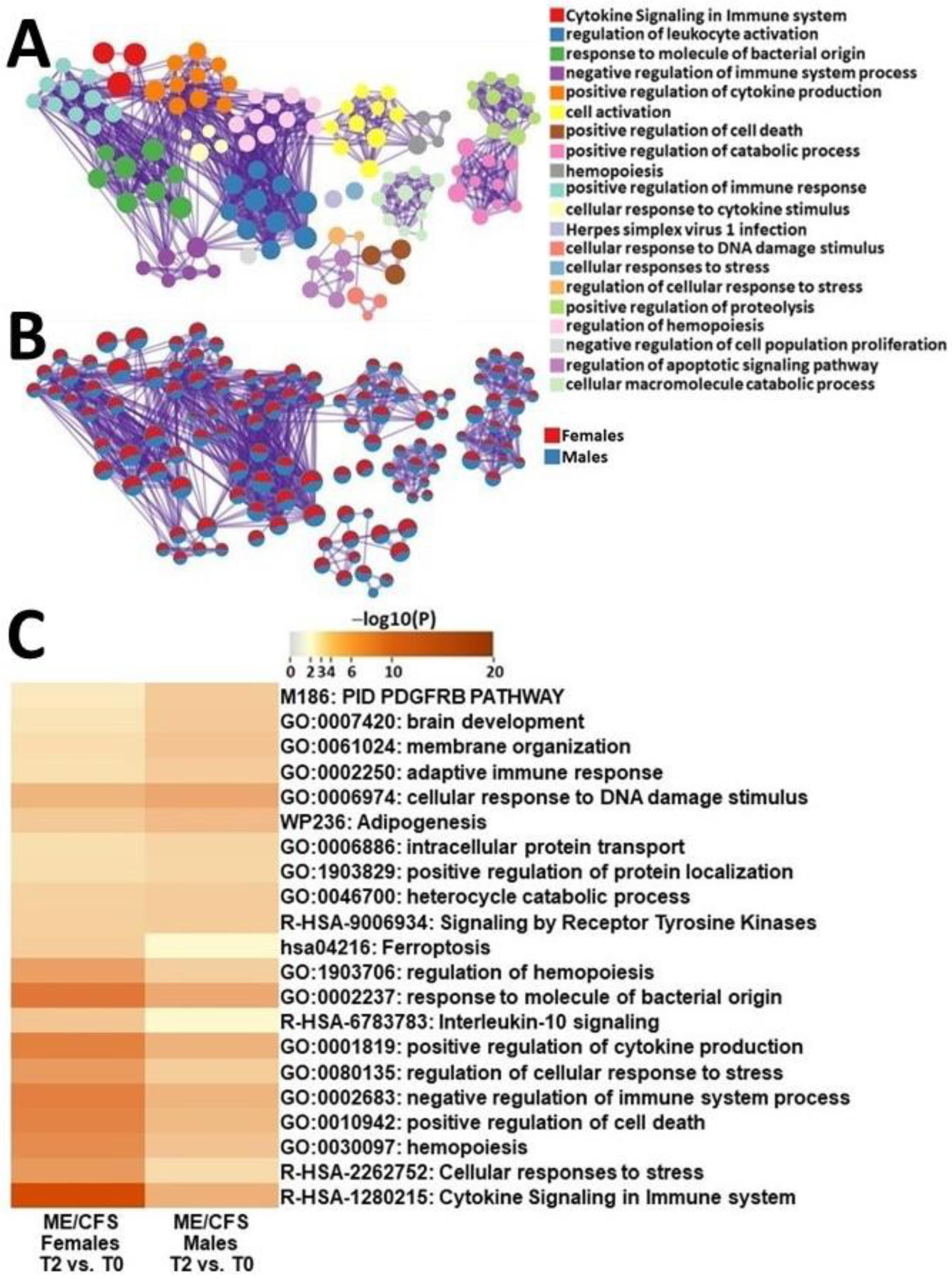
2.4.3. Male ME/CFS vs. Female ME/CFS Functional Pathway Analysis (between T2 and T0)
2.5. Nanostring Validation
3. Discussion
3.1. Transcriptomic Changes between Maximal Exertion (T1) and Baseline before Exercise (T0) in ME/CFS Patients and HCs
3.2. Transcriptomic Changes between 4 h after Maximal Exertion (T2) and Maximal Exertion (T1) in ME/CFS Patients and HCs
3.3. Transcriptomic Changes between 4 h after Maximal Exertion (T2) and Maximal Exertion (T0) in ME/CFS Patients and HCs
4. Materials and Methods
4.1. Cohort
4.2. PBMC Isolation and RNA Extraction
4.3. RNA Sequencing
4.4. RNA-Seq Analysis
4.5. Validation of RNA-Seq Results
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Committee on the Diagnostic Criteria for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome; Board on the Health of Select Populations; Institute of Medicine. Beyond Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Redefining an Illness. Mil. Med. 2015, 180, 721–723. [Google Scholar] [CrossRef]
- Bateman, L.; Bested, A.C.; Bonilla, H.F.; Chheda, B.V.; Chu, L.; Curtin, J.M.; Dempsey, T.T.; Dimmock, M.E.; Dowell, T.G.; Felsenstein, D.; et al. Myalgic Encephalomyelitis/Chronic Fatigue Syn-drome: Essentials of Diagnosis and Management. Mayo Clin. Proc. 2021, 96, 2861–2878. [Google Scholar] [CrossRef] [PubMed]
- Pendergrast, T.; Brown, A.; Sunnquist, M.; Jantke, R.; Newton, J.L.; Strand, E.B.; A Jason, L. Housebound versus nonhousebound patients with myalgic encephalomyelitis and chronic fatigue syndrome. Chronic Illn. 2016, 12, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Kedor, C.; Freitag, H.; Meyer-Arndt, L.; Wittke, K.; Hanitsch, L.G.; Zoller, T.; Steinbeis, F.; Haffke, M.; Rudolf, G.; Heidecker, B.; et al. A prospective observational study of post-COVID-19 chronic fatigue syndrome following the first pandemic wave in Germany and biomarkers associated with symptom severity. Nat. Commun. 2022, 13, 5104. [Google Scholar] [CrossRef] [PubMed]
- Faro, M.; Sàez-Francás, N.; Castro-Marrero, J.; Aliste, L.; de Sevilla, T.F.; Alegre, J. Gender Differences in Chronic Fatigue Syndrome. Reumatol. Clín. 2016, 12, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Pelà, G.; Goldoni, M.; Solinas, E.; Cavalli, C.; Tagliaferri, S.; Ranzieri, S.; Frizzelli, A.; Marchi, L.; Mori, P.A.; Majori, M.; et al. Sex-Related Differences in Long-COVID-19 Syndrome. J. Women’s Health 2022, 31, 620–630. [Google Scholar] [CrossRef]
- Kerr, J.R.; Petty, R.; Burke, B.; Gough, J.; Fear, D.; Sinclair, L.I.; Mattey, D.L.; Richards, S.C.M.; Montgomery, J.; Baldwin, D.A.; et al. Gene expression subtypes in patients with chronic fatigue syndrome/myalgic encephalomyelitis. J. Infect. Dis. 2008, 197, 1171–1184. [Google Scholar] [CrossRef]
- Brenu, E.W.; Ashton, K.J.; van Driel, M.; Staines, D.R.; Peterson, D.; Atkinson, G.M.; Marshall-Gradisnik, S.M. Cytotoxic lymphocyte microRNAs as prospective biomarkers for Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. J. Affect. Disord. 2012, 141, 261–269. [Google Scholar] [CrossRef]
- Nkiliza, A.; Parks, M.; Cseresznye, A.; Oberlin, S.; Evans, J.E.; Darcey, T.; Aenlle, K.; Niedospial, D.; Mullan, M.; Crawford, F.; et al. Sex-specific plasma lipid profiles of ME/CFS patients and their association with pain, fatigue, and cognitive symptoms. J. Transl. Med. 2021, 19, 370. [Google Scholar] [CrossRef]
- Cheema, A.K.; Sarria, L.; Bekheit, M.; Collado, F.; Almenar-Perez, E.; Martin-Martinez, E.; Alegre, J.; Castro-Marrero, J.; Fletcher, M.A.; Klimas, N.G.; et al. Unravelling myalgic encephalomy-elitis/chronic fatigue syndrome (ME/CFS): Gender-specific changes in the microRNA expression profiling in ME/CFS. J. Cell. Mol. Med. 2020, 24, 5865–5877. [Google Scholar] [CrossRef]
- Germain, A.; Giloteaux, L.; Moore, G.E.; Levine, S.M.; Chia, J.K.; Keller, B.A.; Stevens, J.; Franconi, C.J.; Mao, X.; Shungu, D.C.; et al. Plasma metabolomics reveals disrupted response and recovery following maximal exercise in myalgic encephalomyelitis/chronic fatigue syndrome. JCI Insight 2022, 7, e157621. [Google Scholar] [CrossRef] [PubMed]
- Van Booven, D.J.; Gamer, J.; Joseph, A.; Perez, M.; Zarnowski, O.; Pandya, M.; Collado, F.; Klimas, N.; Oltra, E.; Nathanson, L. Stress-Induced Transcriptomic Changes in Females with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Reveal Disrupted Immune Signatures. Int. J. Mol. Sci. 2023, 24, 2698. [Google Scholar] [CrossRef] [PubMed]
- Ware, J.E., Jr.; Sherbourne, C.D. The MOS 36-item short-form health survey (SF-36). I. Conceptual framework and item selection. Med. Care 1992, 30, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented re-source for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expres-sion from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Trinchieri, G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 2003, 3, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.G.; Holmes, T.H.; Anderson, J.N.; Maecker, H.T.; Rosenberg-Hasson, Y.; Valencia, I.J.; Chu, L.; Younger, J.W.; Tato, C.M.; Davis, M.M. Cytokine signature associated with disease severity in chronic fatigue syndrome patients. Proc. Natl. Acad. Sci. USA 2017, 114, E7150–E7158. [Google Scholar] [CrossRef]
- Russell, L.; Broderick, G.; Taylor, R.; Fernandes, H.; Harvey, J.; Barnes, Z.; Smylie, A.; Collado, F.; Balbin, E.G.; Katz, B.Z.; et al. Illness progression in chronic fatigue syndrome: A shifting immune baseline. BMC Immunol. 2016, 17, 3. [Google Scholar] [CrossRef]
- Fletcher, M.A.; Zeng, X.R.; Barnes, Z.; Levis, S.; Klimas, N.G. Plasma cytokines in women with chronic fatigue syndrome. J. Transl. Med. 2009, 7, 96. [Google Scholar] [CrossRef]
- Roerink, M.E.; Knoop, H.; Bronkhorst, E.M.; Mouthaan, H.A.; Hawinkels, L.J.A.C.; Joosten, L.A.B.; van der Meer, J.W.M. Cytokine signatures in chronic fatigue syndrome patients: A Case Control Study and the effect of anakinra treatment. J. Transl. Med. 2017, 15, 267. [Google Scholar] [CrossRef]
- Capuron, L.; Gumnick, J.F.; Musselman, D.L.; Lawson, D.H.; Reemsnyder, A.; Nemeroff, C.B.; Miller, A.H. Neurobehavioral effects of in-terferon-alpha in cancer patients: Phenomenology and paroxetine responsiveness of symptom dimensions. Neuropsychopharmacology 2002, 26, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Mandarano, A.H.; Maya, J.; Giloteaux, L.; Peterson, D.L.; Maynard, M.; Gottschalk, C.G.; Hanson, M.R. Myalgic encephalomyelitis/chronic fatigue syndrome patients exhibit altered T cell metabolism and cytokine associations. J. Clin. Investig. 2020, 130, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N.; Gurvich, C.; Huang, K.; Gooley, P.R.; Armstrong, C.W. The underlying sex differences in neuroendocrine adaptations relevant to Myalgic Encephalomyelitis Chronic Fatigue Syndrome. Front. Neuroendocr. 2022, 66, 100995. [Google Scholar] [CrossRef] [PubMed]
- GO:0032103. Available online: http://amigo.geneontology.org/amigo/term/GO:0032103 (accessed on 1 March 2023).
- Naviaux, R.K.; Naviaux, J.C.; Li, K.; Bright, A.T.; Alaynick, W.A.; Wang, L.; Baxter, A.; Nathan, N.; Anderson, W.; Gordon, E. Metabolic features of chronic fatigue syndrome. Proc. Natl. Acad. Sci. USA 2016, 113, E5472–E5480. [Google Scholar] [CrossRef]
- Yamano, E.; Sugimoto, M.; Hirayama, A.; Kume, S.; Yamato, M.; Jin, G.; Tajima, S.; Goda, N.; Iwai, K.; Fukuda, S.; et al. Index markers of chronic fatigue syndrome with dysfunction of TCA and urea cycles. Sci. Rep. 2016, 6, 34990. [Google Scholar] [CrossRef]
- Fluge, O.; Mella, O.; Bruland, O.; Risa, K.; Dyrstad, S.E.; Alme, K.; Rekeland, I.G.; Sapkota, D.; Rosland, G.V.; Fossa, A.; et al. Metabolic profiling indicates impaired pyruvate dehydro-genase function in myalgic encephalopathy/chronic fatigue syndrome. JCI Insight 2016, 1, e89376. [Google Scholar] [CrossRef]
- Caligiuri, M.A. Human natural killer cells. Blood 2008, 112, 461–469. [Google Scholar] [CrossRef]
- Eaton-Fitch, N.; du Preez, S.; Cabanas, H.; Staines, D.; Marshall-Gradisnik, S. A systematic review of natural killer cells profile and cytotoxic function in myalgic encephalomyelitis/chronic fatigue syndrome. Syst. Rev. 2019, 8, 279. [Google Scholar] [CrossRef]
- Paudel, S.; Ghimire, L.; Jin, L.; Jeansonne, D.; Jeyaseelan, S. Regulation of emergency granulopoiesis during infection. Front. Immunol. 2022, 13, 961601. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, B. The multi-differentiation potential of peripheral blood mononuclear cells. Stem Cell Res. Ther. 2012, 3, 48. [Google Scholar] [CrossRef]
- Rasa, S.; Nora-Krukle, Z.; Henning, N.; Eliassen, E.; Shikova, E.; Harrer, T.; Scheibenbogen, C.; Murovska, M.; Prusty, B.K.; European Network on ME/CFS (EUROMENE). Chronic viral infections in myalgic encephalomy-elitis/chronic fatigue syndrome (ME/CFS). J. Transl. Med. 2018, 16, 268. [Google Scholar] [CrossRef]
- Cohen, J.I. Herpesvirus latency. J. Clin. Investig. 2020, 130, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Sandu, I.; Cerletti, D.; Claassen, M.; Oxenius, A. Exhausted CD8+ T cells exhibit low and strongly inhibited TCR signaling during chronic LCMV infection. Nat. Commun. 2020, 11, 4454. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.D.; Lemle, M.D.; Komaroff, A.L.; Snyder, S.H. Redox imbalance links COVID-19 and myalgic encephalomyelitis/chronic fatigue syndrome. Proc. Natl. Acad. Sci. USA 2021, 118, e2024358118. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; Kubera, M.; Uytterhoeven, M.; Vrydags, N.; Bosmans, E. Increased plasma peroxides as a marker of oxidative stress in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Med. Sci Monit. 2011, 17, SC11–SC15. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal. Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Pai, S.; Thomas, R. Immune deficiency or hyperactivity-Nf-kappab illuminates autoimmunity. J. Autoimmun. 2008, 31, 245–251. [Google Scholar] [CrossRef]
- Maes, M.; Mihaylova, I.; Bosmans, E. Not in the mind of neurasthenic lazybones but in the cell nucleus: Patients with chronic fatigue syndrome have increased production of nuclear factor kappa beta. Neuro Endocrinol. Lett. 2007, 28, 456–462. [Google Scholar]
- Sweetman, E.; Ryan, M.; Edgar, C.; Mackay, A.; Vallings, R.; Tate, W. Changes in the transcriptome of circulating immune cells of a New Zealand cohort with myalgic encephalomyelitis/chronic fatigue syndrome. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738418820402. [Google Scholar] [CrossRef]
- Hornig, M.; Montoya, J.G.; Klimas, N.G.; Levine, S.; Felsenstein, D.; Bateman, L.; Peterson, D.L.; Gottschalk, C.G.; Schultz, A.F.; Che, X.; et al. Distinct plasma immune signatures in ME/CFS are present early in the course of illness. Sci. Adv. 2015, 1, e1400121. [Google Scholar] [CrossRef]
- Hornig, M.; Gottschalk, G.; Peterson, D.L.; Knox, K.K.; Schultz, A.F.; Eddy, M.L.; Che, X.; I Lipkin, W. Cytokine network analysis of cerebrospinal fluid in myalgic encephalomyelitis/chronic fatigue syndrome. Mol. Psychiatry 2015, 21, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Moneghetti, K.J.; Skhiri, M.; Contrepois, K.; Kobayashi, Y.; Maecker, H.; Davis, M.; Snyder, M.; Haddad, F.; Montoya, J.G. Value of Circulating Cytokine Profiling During Submaximal Exercise Testing in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Sci. Rep. 2018, 8, 2779. [Google Scholar] [CrossRef] [PubMed]
- Karshikoff, B.; Sundelin, T.; Lasselin, J. Role of Inflammation in Human Fatigue: Relevance of Multidimensional Assessments and Potential Neuronal Mechanisms. Front. Immunol. 2017, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Safieh-Garabedian, B.; Poole, S.; Allchorne, A.; Winter, J.; Woolf, C.J. Contribution of interleukin-1 beta to the inflamma-tion-induced increase in nerve growth factor levels and inflammatory hyperalgesia. Br. J. Pharmacol. 1995, 115, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.K.; Hoffman-Goetz, L.; Neves, C.D.C.; Lacerda, A.C.R.; Lage, V.K.S.; Soares, A.A.; Chaves, M.G.A.; Lima, L.P.; Silva, T.J.; Vieira, L.M.; et al. Exercise and the Immune System: Regulation, Integration, and Adaptation. Physiol. Rev. 2000, 80, 1055–1081. [Google Scholar] [CrossRef]
- Gersuk, G.M.; Westermark, B.; Mohabeer, A.J.; Challita, P.M.; Pattamakom, S.; Pattengale, P.K. Inhibition of human natural killer cell activity by platelet-derived growth factor (PDGF). III. Membrane binding studies and differential biological effect of recombinant PDGF isoforms. Scand. J. Immunol. 1991, 33, 521–532. [Google Scholar] [CrossRef]
- A Daynes, R.; Dowell, T.; A Araneo, B. Platelet-derived growth factor is a potent biologic response modifier of T cells. J. Exp. Med. 1991, 174, 1323–1333. [Google Scholar] [CrossRef]
- Tang, J.; Kozaki, K.; Farr, A.G.; Martin, P.J.; Lindahl, P.; Betsholtz, C.; Raines, E.W. The absence of platelet-derived growth factor-B in cir-culating cells promotes immune and inflammatory responses in atherosclerosis-prone ApoE-/- mice. Am. J. Pathol. 2005, 167, 901–912. [Google Scholar] [CrossRef]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef]
- Moore, K.W.; de Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef]
- Brenu, E.W.; van Driel, M.L.; Staines, D.R.; Ashton, K.J.; Ramos, S.B.; Keane, J.; Ramos, S.B.; Keane, J.; Klimas, N.G.; Marshall-Gradisnik, S.M. Immunological abnormalities as potential bi-omarkers in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. J. Transl. Med. 2011, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Barry, J.C.; Shakibakho, S.; Durrer, C.; Simtchouk, S.; Jawanda, K.K.; Cheung, S.T.; Mui, A.L.; Little, J.P. Hyporesponsiveness to the anti-inflammatory action of interleukin-10 in type 2 diabetes. Sci. Rep. 2016, 6, 21244. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhang, H.; Dauphars, D.J.; He, Y.-W. A Potential Role of Interleukin 10 in COVID-19 Pathogenesis. Trends Immunol. 2020, 42, 3–5. [Google Scholar] [CrossRef]
- Islam, H.; Chamberlain, T.C.; Mui, A.L.; Little, J.P. Elevated Interleukin-10 Levels in COVID-19: Potentiation of Pro-Inflammatory Responses or Impaired Anti-Inflammatory Action? Front. Immunol. 2021, 12, 677008. [Google Scholar] [CrossRef]
- Zhong, Q.; Sj, D.; Km, L.; Lamprecht, R.S.; Em, Z.; Ce, G.; Dn, P.; Aj, B.; Am, C. Faculty Opinions recommendation of Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2015, 149, 1060–1072. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.L.; Huang, Z.J.; Lin, Z.T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef]
- Anderson, G.; Maes, M. Mitochondria and immunity in chronic fatigue syndrome. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2020, 103, 109976. [Google Scholar] [CrossRef]
- Germain, A.; Barupal, D.K.; Levine, S.M.; Hanson, M.R. Comprehensive Circulatory Metabolomics in ME/CFS Reveals Disrupted Metabolism of Acyl Lipids and Steroids. Metabolites 2020, 10, 34. [Google Scholar] [CrossRef]
- Mascia, G.; Arbelo, E.; Porto, I.; Brugada, R.; Brugada, J. The arrhythmogenic right ventricular cardiomyopathy in comparison to the athletic heart. J. Cardiovasc. Electrophysiol. 2020, 31, 1836–1843. [Google Scholar] [CrossRef]
- McArdle, W.D.; Katch, F.I.; Katch, V.L. Exercise Physiology: Nutrition, Energy, and Human Performance; Lip-Pincott Williams & Wilkins: Philadelphia, PA, USA, 2001. [Google Scholar]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020, 2, lqaa078. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Holtzman, C.S.; Bhatia, S.; Cotler, J.; Jason, L.A. Assessment of Post-Exertional Malaise (PEM) in Patients with Myalgic Enceph-alomyelitis (ME) and Chronic Fatigue Syndrome (CFS): A Patient-Driven Survey. Diagnostics 2019, 9, 26. [Google Scholar] [CrossRef]
- Jason, L.A.; Holtzman, C.S.; Sunnquist, M.; Cotler, J. The development of an instrument to assess post-exertional malaise in pa-tients with myalgic encephalomyelitis and chronic fatigue syndrome. J. Health Psychol. 2021, 26, 238–248. [Google Scholar] [CrossRef]


| Category | ME/CFS Males | Healthy Controls | p-Value | |
|---|---|---|---|---|
| Age | 42.2 ± 4.25 | 45.3 ± 2.67 | 0.550 | |
| BMI | 25.8 ± 1.37 | 28.8 ± 0.89 | 0.115 | |
| Physical Health | ||||
| Physical Function | 52.9 ± 9.03 | 76.8 ± 7.73 | 0.050 * | |
| Role-Physical | 16.7 ± 11.24 | 91.1 ± 5.63 | <0.001 * | |
| Body pain | 65.2 ± 6.93 | 74.1 ± 6.62 | 0.371 | |
| General Health | 34.6 ± 7.16 | 73.1 ± 5.88 | <0.001 * | |
| Mental Health | ||||
| Vitality | 26.7 ± 7.24 | 65.7 ± 6.09 | <0.001 * | |
| Social Function | 36.5 ± 7.29 | 84.8 ± 4.76 | <0.001 * | |
| Role Emotional | 63.9 ± 12.62 | 92.8 ± 5.18 | 0.050 * | |
| Mental Health | 71.7 ± 5.54 | 83.1 ± 4.86 | 0.135 |
| T1 vs. T0 in Male ME/CFS Patients | ||
|---|---|---|
| Cell Type | p-Value | Fold Change |
| CD4+ Naive T cells | 0.050 * | −1.372 |
| NK cells | 0.019 * | 1.529 |
| T2 vs. T1 in Male ME/CFS Patients | ||
|---|---|---|
| Cell Type | p-Value | Fold Change |
| CD4+ Naive T cells | 0.002 * | 1.622 |
| NK cells | 0.008 * | −1.461 |
| Dendritic cells | 0.023 * | −1.587 |
| Eosinophils | 0.002 * | −4.066 |
| T2 vs. T1 in male HCs | ||
| CD8+ T cells | 0.001 * | 12.42 |
| CD4+ Naive T cells | <0.001 * | −2.133 |
| NK cells | 0.0003 * | 1.817 |
| Dendritic cells | 0.002 * | 2.171 |
| Eosinophils | <0.001 * | 4.063 |
| T2 vs. T0 in Male ME/CFS Patients | ||
|---|---|---|
| Cell Type | p-Value | Fold Change |
| CD4+ Memory T cells | 0.002 * | 1.622 |
| Dendritic cells | 0.116 | −1.659 |
| Eosinophils | 0.050 * | −2.731 |
| T2 vs. T0 in male HCs | ||
| CD4+ Naive T cells | 0.002 * | 1.715 |
| Dendritic cells | <0.001 * | −2.619 |
| Eosinophils | 0.002 * | −3.522 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gamer, J.; Van Booven, D.J.; Zarnowski, O.; Arango, S.; Elias, M.; Kurian, A.; Joseph, A.; Perez, M.; Collado, F.; Klimas, N.; et al. Sex-Dependent Transcriptional Changes in Response to Stress in Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Pilot Project. Int. J. Mol. Sci. 2023, 24, 10255. https://doi.org/10.3390/ijms241210255
Gamer J, Van Booven DJ, Zarnowski O, Arango S, Elias M, Kurian A, Joseph A, Perez M, Collado F, Klimas N, et al. Sex-Dependent Transcriptional Changes in Response to Stress in Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Pilot Project. International Journal of Molecular Sciences. 2023; 24(12):10255. https://doi.org/10.3390/ijms241210255
Chicago/Turabian StyleGamer, Jackson, Derek J. Van Booven, Oskar Zarnowski, Sebastian Arango, Mark Elias, Asha Kurian, Andrew Joseph, Melanie Perez, Fanny Collado, Nancy Klimas, and et al. 2023. "Sex-Dependent Transcriptional Changes in Response to Stress in Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Pilot Project" International Journal of Molecular Sciences 24, no. 12: 10255. https://doi.org/10.3390/ijms241210255
APA StyleGamer, J., Van Booven, D. J., Zarnowski, O., Arango, S., Elias, M., Kurian, A., Joseph, A., Perez, M., Collado, F., Klimas, N., Oltra, E., & Nathanson, L. (2023). Sex-Dependent Transcriptional Changes in Response to Stress in Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Pilot Project. International Journal of Molecular Sciences, 24(12), 10255. https://doi.org/10.3390/ijms241210255





