Circulating miRNAs Expression in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome
Abstract
1. Introduction
2. Results
2.1. Epidemiological and Clinical Features of ME/CFS and CTR Groups
2.2. Quantification of Pro-Inflammatory Cytokines in Plasma Samples
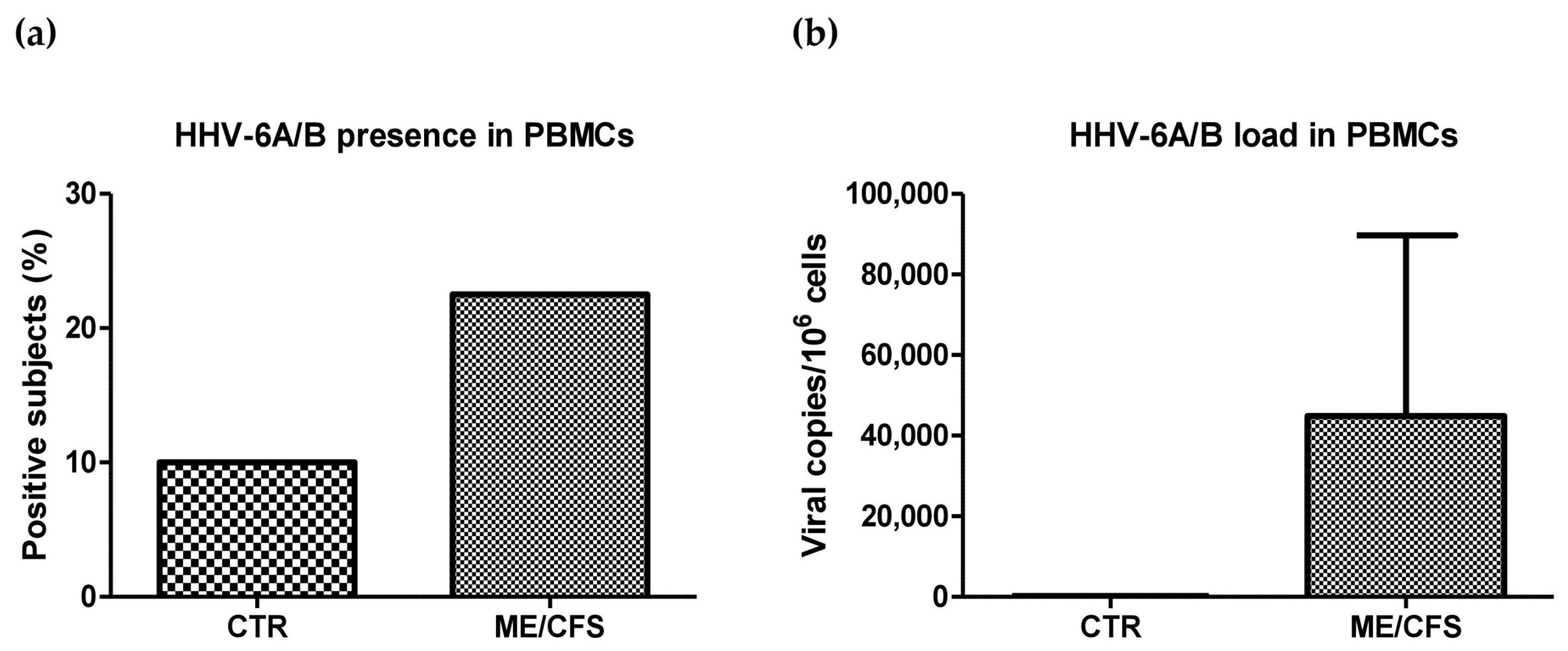
2.3. HHV-6A/B Presence and Load
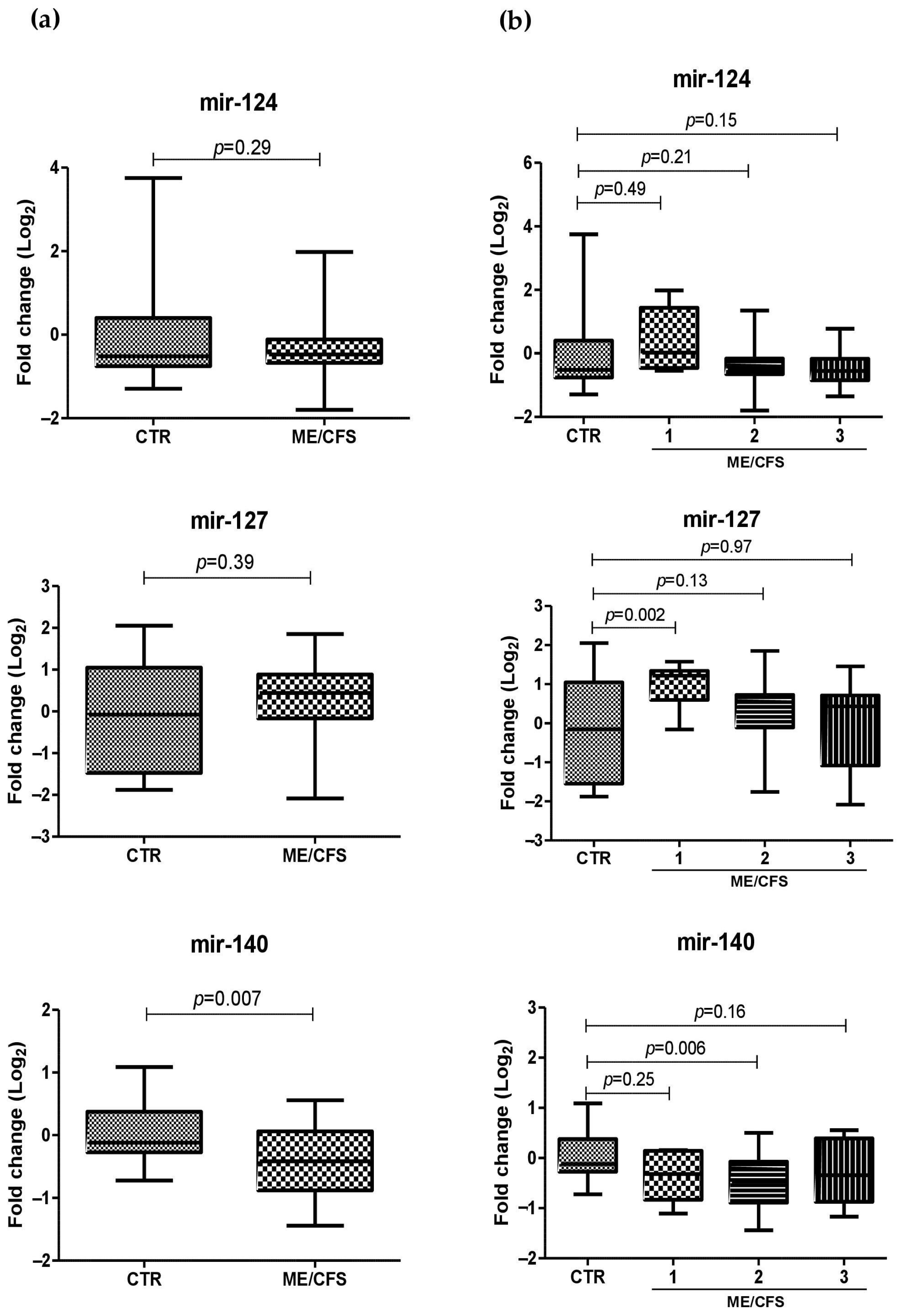
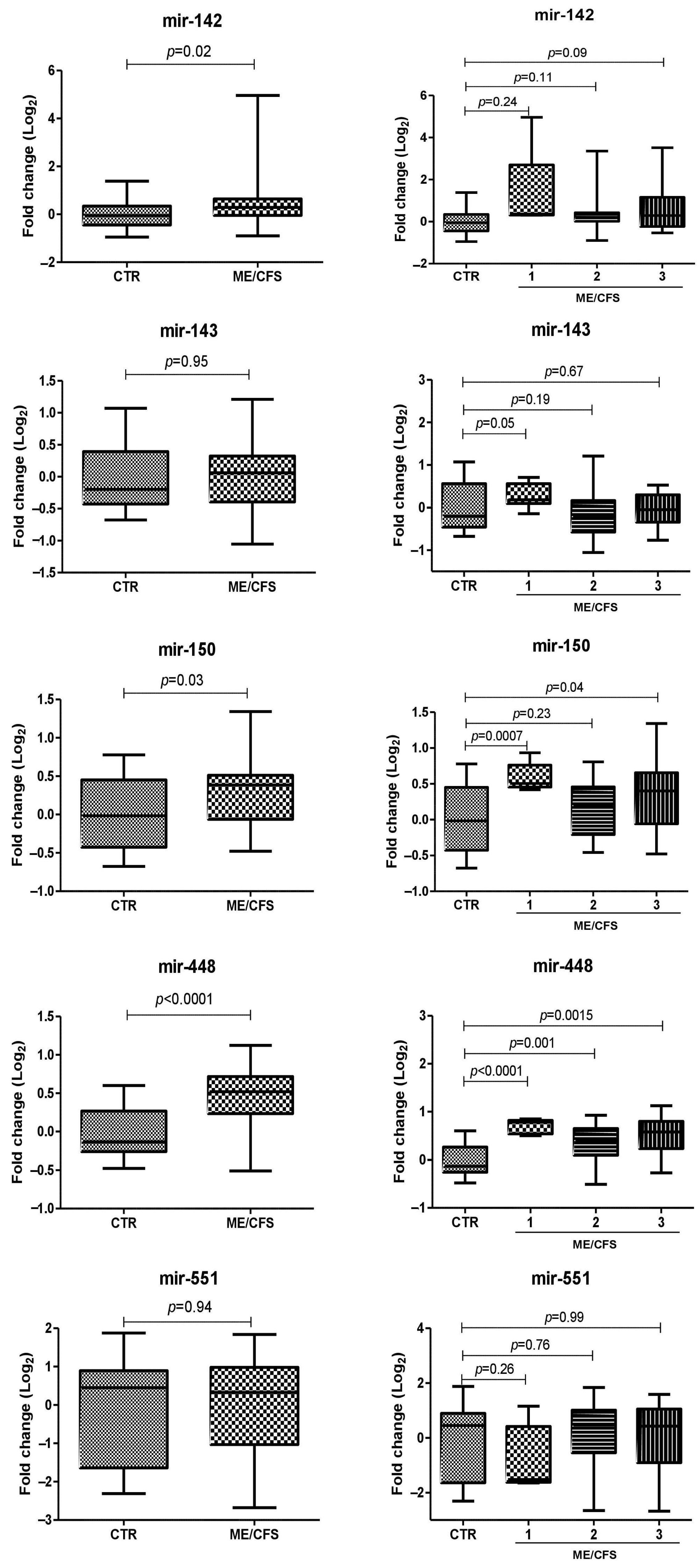
2.4. miRNA Plasma Levels in ME/CFS Patients
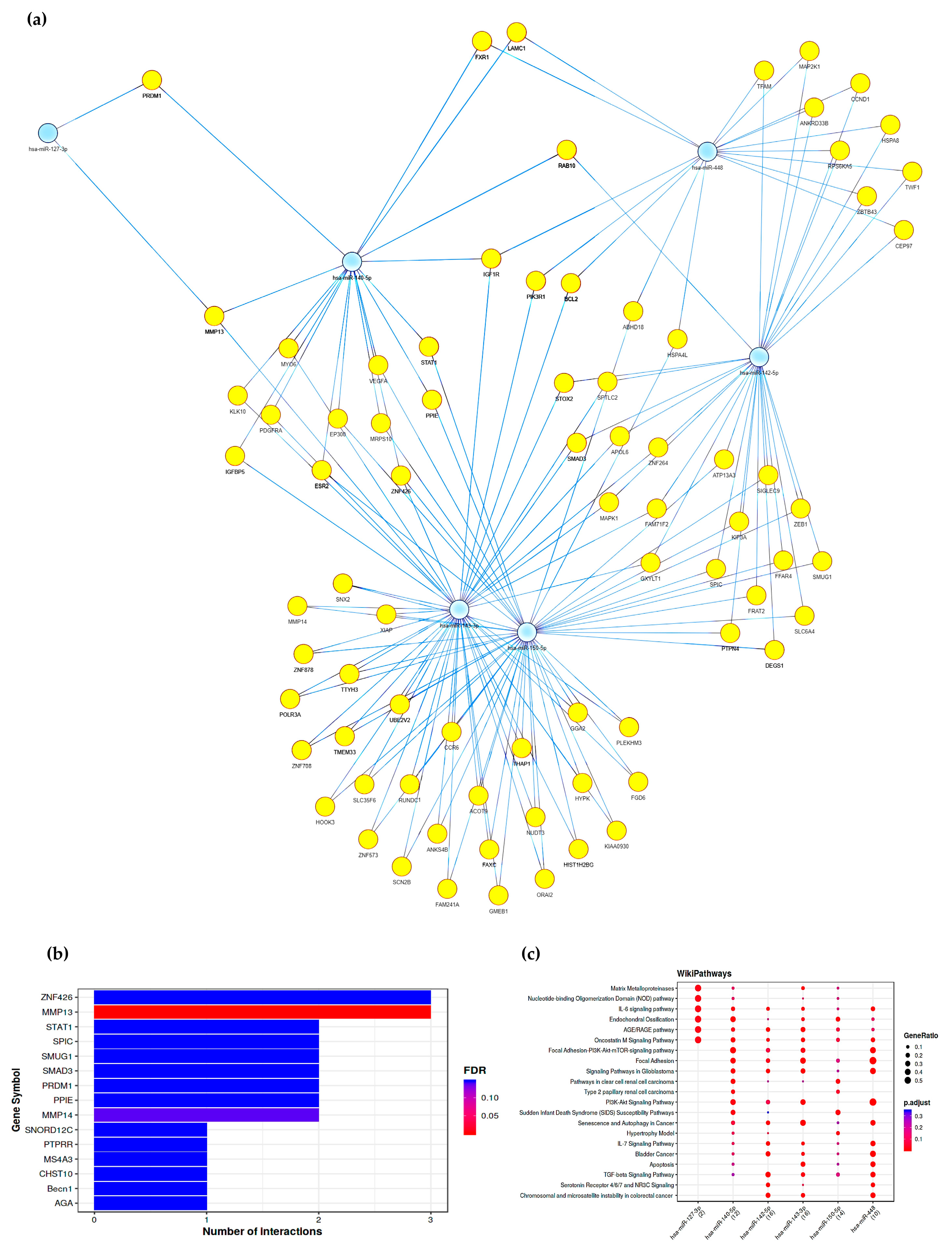
2.5. Gene Pathways Analysis
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Samples Collection
4.3. Cytokines Evaluation
4.4. DNA Extraction and Analyses of HHV-6A/B Presence
4.5. RNA Extraction and miRNA Analysis
4.6. Gene Pathways and Functional Enriched Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Estévez-López, F.; Mudie, K.; Wang-Steverding, X.; Bakken, I.J.; Ivanovs, A.; Castro-Marrero, J.; Nacul, L.; Alegre, J.; Zalewski, P.; Słomko, J.; et al. Systematic Review of the Epidemiological Burden of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Across Europe: Current Evidence and EUROMENE Research Recommendations for Epidemiology. J. Clin. Med. 2020, 9, 1557. [Google Scholar] [CrossRef]
- Fukuda, K.; Straus, S.E.; Hickie, I.; Sharpe, M.C.; Dobbins, J.G.; Komaroff, A. The Chronic Fatigue Syndrome: A Comprehensive Approach to Its Definition and Study. International Chronic Fatigue Syndrome Study Group. Ann. Intern. Med. 1994, 121, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Carruthers, B.M.; Jain, A.K.; De Meirleir, K.L.; Peterson, D.L.; Klimas, N.G.; Lemer, A.M.; Bested, A.C.; Flor-Henry, P.; Joshi, P.; Powles, A.C.P.; et al. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. J. Chronic Fatigue Syndr. 2011, 11, 7–115. [Google Scholar] [CrossRef]
- A Copy of the ‘London Criteria for M.E.’ as Revised in 2014|Archived Here for Reference Purposes|15 October 2016|The ME Association. Available online: https://meassociation.org.uk/2016/10/a-copy-of-the-london-criteria-as-revised-in-2014-archived-here-for-reference-purposes-15-october-2016/ (accessed on 20 January 2023).
- Carruthers, B.M.; Van de Sande, M.I.; De Meirleir, K.L.; Klimas, N.G.; Broderick, G.; Mitchell, T.; Staines, D.; Powles, A.C.P.; Speight, N.; Vallings, R.; et al. Myalgic Encephalomyelitis: International Consensus Criteria. J. Intern. Med. 2011, 270, 327–338. [Google Scholar] [CrossRef]
- Sunnquist, M.; Jason, L.A.; Nehrke, P.; Goudsmit, E.M. A Comparison of Case Definitions for Myalgic Encephalomyelitis and Chronic Fatigue Syndrome. J. Chronic Dis. Manag. 2017, 2, 1013. [Google Scholar] [PubMed]
- Ariza, M.E.; Sciortino, T.; Williams, M. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: The Human Herpesviruses Are Back! Biomolecules 2021, 11, 185. [Google Scholar] [CrossRef]
- Sotzny, F.; Blanco, J.; Capelli, E.; Castro-Marrero, J.; Steiner, S.; Murovska, M.; Scheibenbogen, C. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome—Evidence for an Autoimmune Disease. Autoimmun. Rev. 2018, 17, 601–609. [Google Scholar] [CrossRef]
- Rasa, S.; Nora-Krukle, Z.; Henning, N.; Eliassen, E.; Shikova, E.; Harrer, T.; Scheibenbogen, C.; Murovska, M.; Prusty, B.K. Chronic Viral Infections in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). J. Transl. Med. 2018, 16, 268. [Google Scholar] [CrossRef] [PubMed]
- Hickie, I.; Davenport, T.; Wakefield, D.; Vollmer-Conna, U.; Cameron, B.; Vernon, S.D.; Reeves, W.C.; Lloyd, A. Post-Infective and Chronic Fatigue Syndromes Precipitated by Viral and Non-Viral Pathogens: Prospective Cohort Study. BMJ 2006, 333, 575. [Google Scholar] [CrossRef]
- Chu, L.; Valencia, I.J.; Garvert, D.W.; Montoya, J.G. Onset Patterns and Course of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Front. Pediatr. 2019, 7, 12. [Google Scholar] [CrossRef]
- Chapenko, S.; Krumina, A.; Logina, I.; Rasa, S.; Chistjakovs, M.; Sultanova, A.; Viksna, L.; Murovska, M. Association of Active Human Herpesvirus-6, -7 and Parvovirus B19 Infection with Clinical Outcomes in Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Adv. Virol. 2012, 2012, 205085. [Google Scholar] [CrossRef] [PubMed]
- Rasa-Dzelzkaleja, S.; Krumina, A.; Capenko, S.; Nora-Krukle, Z.; Gravelsina, S.; Vilmane, A.; Ievina, L.; Shoenfeld, Y.; Murovska, M. The Persistent Viral Infections in the Development and Severity of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. J. Transl. Med. 2023, 21, 33. [Google Scholar] [CrossRef]
- Lee, J.S.; Lacerda, E.M.; Nacul, L.; Kingdon, C.C.; Norris, J.; O’Boyle, S.; Roberts, C.H.; Palla, L.; Riley, E.M.; Cliff, J.M. Salivary DNA Loads for Human Herpesviruses 6 and 7 Are Correlated With Disease Phenotype in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Front. Med. 2021, 8, 656692. [Google Scholar] [CrossRef] [PubMed]
- Kasimir, F.; Toomey, D.; Liu, Z.; Kaiping, A.C.; Ariza, M.E.; Prusty, B.K. Tissue Specific Signature of HHV-6 Infection in ME/CFS. Front. Mol. Biosci. 2022, 9, 1044964. [Google Scholar] [CrossRef]
- Schreiner, P.; Harrer, T.; Scheibenbogen, C.; Lamer, S.; Schlosser, A.; Naviaux, R.K.; Prusty, B.K. Human Herpesvirus-6 Reactivation, Mitochondrial Fragmentation, and the Coordination of Antiviral and Metabolic Phenotypes in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. ImmunoHorizons 2020, 4, 201–215. [Google Scholar] [CrossRef]
- Cox, B.S.; Alharshawi, K.; Mena-Palomo, I.; Lafuse, W.P.; Ariza, M.E. EBV/HHV-6A DUTPases Contribute to Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Pathophysiology by Enhancing TFH Cell Differentiation and Extrafollicular Activities. JCI Insight 2022, 7, e158193. [Google Scholar] [CrossRef]
- Kennedy, G.; Spence, V.A.; McLaren, M.; Hill, A.; Underwood, C.; Belch, J.J.F. Oxidative Stress Levels Are Raised in Chronic Fatigue Syndrome and Are Associated with Clinical Symptoms. Free Radic. Biol. Med. 2005, 39, 584–589. [Google Scholar] [CrossRef]
- Montoya, J.G.; Holmes, T.H.; Anderson, J.N.; Maecker, H.T.; Rosenberg-Hasson, Y.; Valencia, I.J.; Chu, L.; Younger, J.W.; Tato, C.M.; Davis, M.M. Cytokine Signature Associated with Disease Severity in Chronic Fatigue Syndrome Patients. Proc. Natl. Acad. Sci. USA 2017, 114, E7150–E7158. [Google Scholar] [CrossRef] [PubMed]
- Roerink, M.E.; van der Schaaf, M.E.; Hawinkels, L.J.A.C.; Raijmakers, R.P.H.; Knoop, H.; Joosten, L.A.B.; van der Meer, J.W.M. Pitfalls in Cytokine Measurements—Plasma TGF-Β1 in Chronic Fatigue Syndrome. Neth. J. Med. 2018, 76, 310–313. [Google Scholar]
- Scherbakov, N.; Szklarski, M.; Hartwig, J.; Sotzny, F.; Lorenz, S.; Meyer, A.; Grabowski, P.; Doehner, W.; Scheibenbogen, C. Peripheral Endothelial Dysfunction in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. ESC Heart Fail. 2020, 7, 1064–1071. [Google Scholar] [CrossRef]
- Gravelsina, S.; Vilmane, A.; Svirskis, S.; Rasa-Dzelzkaleja, S.; Nora-Krukle, Z.; Vecvagare, K.; Krumina, A.; Leineman, I.; Shoenfeld, Y.; Murovska, M. Biomarkers in the Diagnostic Algorithm of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Front. Immunol. 2022, 13, 5980. [Google Scholar] [CrossRef]
- Almenar-Pérez, E.; Sarría, L.; Nathanson, L.; Oltra, E. Assessing Diagnostic Value of MicroRNAs from Peripheral Blood Mononuclear Cells and Extracellular Vesicles in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Sci. Rep. 2020, 10, 2064. [Google Scholar] [CrossRef]
- Brenu, E.W.; Ashton, K.J.; Batovska, J.; Staines, D.R.; Marshall-Gradisnik, S.M. High-Throughput Sequencing of Plasma MicroRNA in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. PLoS ONE 2014, 9, e102783. [Google Scholar] [CrossRef] [PubMed]
- Nepotchatykh, E.; Elremaly, W.; Caraus, I.; Godbout, C.; Leveau, C.; Chalder, L.; Beaudin, C.; Kanamaru, E.; Kosovskaia, R.; Lauzon, S.; et al. Profile of Circulating MicroRNAs in Myalgic Encephalomyelitis and Their Relation to Symptom Severity, and Disease Pathophysiology. Sci. Rep. 2020, 10, 19620. [Google Scholar] [CrossRef]
- Jin, F.; Hu, H.; Xu, M.; Zhan, S.; Wang, Y.; Zhang, H.; Chen, X. Serum MicroRNA Profiles Serve as Novel Biomarkers for Autoimmune Diseases. Front. Immunol. 2018, 9, 2381. [Google Scholar] [CrossRef]
- Cheema, A.K.; Sarria, L.; Bekheit, M.; Collado, F.; Almenar-Pérez, E.; Martín-Martínez, E.; Alegre, J.; Castro-Marrero, J.; Fletcher, M.A.; Klimas, N.G.; et al. Unravelling Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS): Gender-Specific Changes in the MicroRNA Expression Profiling in ME/CFS. J. Cell. Mol. Med. 2020, 24, 5865–5877. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, E.D.; Veremeyko, T.; Barteneva, N.; Krichevsky, A.M.; Weiner, H.L. MicroRNA-124 Promotes Microglia Quiescence and Suppresses EAE by Deactivating Macrophages via the C/EBP-α-PU.1 Pathway. Nat. Med. 2011, 17, 64–70. [Google Scholar] [CrossRef]
- Nepotchatykh, E.; Caraus, I.; Elremaly, W.; Leveau, C.; Elbakry, M.; Godbout, C.; Rostami-Afshari, B.; Petre, D.; Khatami, N.; Franco, A.; et al. Circulating MicroRNA Expression Signatures Accurately Discriminate Myalgic Encephalomyelitis from Fibromyalgia and Comorbid Conditions. Sci. Rep. 2023, 13, 1896. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Liang, G.; Egger, G.; Friedman, J.M.; Chuang, J.C.; Coetzee, G.A.; Jones, P.A. Specific Activation of MicroRNA-127 with Downregulation of the Proto-Oncogene BCL6 by Chromatin-Modifying Drugs in Human Cancer Cells. Cancer Cell 2006, 9, 435–443. [Google Scholar] [CrossRef]
- Wei, G.; Tan, M.; Wang, C.; Liang, L. Decreased MiR-127 Promotes the Occurrence of Breast Cancer via Increasing the Expression of SPP1. Adv. Clin. Exp. Med. 2023, 32. Epub ahead of print. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Bahroudi, Z.; Shoorei, H.; Abak, A.; Ahin, M.; Taheri, M. MicroRNA-140: A MiRNA with Diverse Roles in Human Diseases. Biomed. Pharmacother. 2021, 135, 111256. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fu, M.; Li, Y. MiR-142-5p and MiR-212-5p Cooperatively Inhibit the Proliferation and Collagen Formation of Cardiac Fibroblasts by Regulating c-Myc/TP53INP1. Can. J. Physiol. Pharmacol. 2020, 98, 314–323. [Google Scholar] [CrossRef]
- Cheng, W.; Yan, K.; Xie, L.Y.; Chen, F.; Yu, H.C.; Huang, Y.X.; Dang, C.X. MiR-143-3p Controls TGF-Β1-Induced Cell Proliferation and Extracellular Matrix Production in Airway Smooth Muscle via Negative Regulation of the Nuclear Factor of Activated T Cells 1. Mol. Immunol. 2016, 78, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Ménoret, A.; Agliano, F.; Karginov, T.A.; Karlinsey, K.S.; Zhou, B.; Vella, A.T. Antigen-Specific Downregulation of MiR-150 in CD4 T Cells Promotes Cell Survival. Front. Immunol. 2023, 14, 1102403. [Google Scholar] [CrossRef]
- Liao, Z.B.; Tan, X.L.; Dong, K.S.; Zhang, H.W.; Chen, X.P.; Chu, L.; Zhang, B.X. miRNA-448 Inhibits Cell Growth by Targeting BCL-2 in Hepatocellular Carcinoma. Dig. Liver Dis. 2019, 51, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, L.; Han, W. Elevated Level of MiR-551b-5p Is Associated With Inflammation and Disease Progression in Patients With Severe Acute Pancreatitis. Ther. Apher. Dial. 2018, 22, 649–655. [Google Scholar] [CrossRef]
- Soffritti, I.; D’Accolti, M.; Ravegnini, G.; Arcangeletti, M.C.; Maccari, C.; De Conto, F.; Calderaro, A.; Caselli, E. Modulation of Micrornome by Human Cytomegalovirus and Human Herpesvirus 6 Infection in Human Dermal Fibroblasts: Possible Significance in the Induction of Fibrosis in Systemic Sclerosis. Cells 2021, 10, 1060. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Soffritti, I.; D’Accolti, M.; Bortolotti, D.; Di Luca, D.; Caselli, E. HHV-6A/6B Infection of NK Cells Modulates the Expression of MiRNAs and Transcription Factors Potentially Associated to Impaired NK Activity. Front. Microbiol. 2017, 8, 2143. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; D’Accolti, M.; Soffritti, I.; Zatelli, M.C.; Rossi, R.; Degli Uberti, E.; Di Luca, D. HHV-6A in vitro Infection of Thyrocytes and T Cells Alters the Expression of MiRNA Associated to Autoimmune Thyroiditis. Virol. J. 2017, 14, 3. [Google Scholar] [CrossRef]
- Minnock, P.; Ringnér, A.; Bresnihan, B.; Veale, D.; FitzGerald, O.; McKee, G. Perceptions of the Cause, Impact and Management of Persistent Fatigue in Patients with Rheumatoid Arthritis Following Tumour Necrosing Factor Inhibition Therapy. Musculoskelet. Care 2017, 15, 23–35. [Google Scholar] [CrossRef]
- Licursi, V.; Conte, F.; Fiscon, G.; Paci, P. MIENTURNET: An Interactive Web Tool for MicroRNA-Target Enrichment and Network-Based Analysis. BMC Bioinform. 2019, 20, 545. [Google Scholar] [CrossRef] [PubMed]
- Soffritti, I.; D’Accolti, M.; Maccari, C.; Bini, F.; Mazziga, E.; de Conto, F.; Calderaro, A.; Arcangeletti, M.C.; Caselli, E. Human Cytomegalovirus and Human Herpesvirus 6 Coinfection of Dermal Fibroblasts Enhances the Pro-Inflammatory Pathway Predisposing to Fibrosis: The Possible Impact on Systemic Sclerosis. Microorganisms 2022, 10, 1600. [Google Scholar] [CrossRef] [PubMed]
- Peterson, D.; Brenu, E.W.; Gottschalk, G.; Ramos, S.; Nguyen, T.; Staines, D.; Marshall-Gradisnik, S. Cytokines in the Cerebrospinal Fluids of Patients with Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. Mediat. Inflamm. 2015, 2015, 929720. [Google Scholar] [CrossRef]
- Tian, P.; Tao, L.; Wang, Y.; Han, X. MicroRNA-127 Inhibits the Progression of Melanoma by Downregulating Delta-Like Homologue 1. Biomed. Res. Int. 2020, 2020, 8523465. [Google Scholar] [CrossRef]
- Saito, Y.; Suzuki, H.; Tsugawa, H.; Imaeda, H.; Matsuzaki, J.; Hirata, K.; Hosoe, N.; Nakamura, M.; Mukai, M.; Saito, H.; et al. Overexpression of MiR-142-5p and MiR-155 in Gastric Mucosa-Associated Lymphoid Tissue (MALT) Lymphoma Resistant to Helicobacter Pylori Eradication. PLoS ONE 2012, 7, e47396. [Google Scholar] [CrossRef] [PubMed]
- Slezak, S.; Jin, P.; Caruccio, L.; Ren, J.; Bennett, M.; Zia, N.; Adams, S.; Wang, E.; Ascensao, J.; Schechter, G.; et al. Gene and MicroRNA Analysis of Neutrophils from Patients with Polycythemia Vera and Essential Thrombocytosis: Down-Regulation of Micro RNA-1 and -133a. J. Transl. Med. 2009, 7, 39. [Google Scholar] [CrossRef]
- Allantaz, F.; Cheng, D.T.; Bergauer, T.; Ravindran, P.; Rossier, M.F.; Ebeling, M.; Badi, L.; Reis, B.; Bitter, H.; D’Asaro, M.; et al. Expression Profiling of Human Immune Cell Subsets Identifies MiRNA-MRNA Regulatory Relationships Correlated with Cell Type Specific Expression. PLoS ONE 2012, 7, e29979. [Google Scholar] [CrossRef]
- Yu, B.; Yu, M.; Zhang, H.; Xie, D.; Nie, W.; Shi, K. Suppression of MiR-143-3p Contributes to the Anti-Fibrosis Effect of Atorvastatin on Myocardial Tissues via the Modulation of Smad2 Activity. Exp. Mol. Pathol. 2020, 112, 104346. [Google Scholar] [CrossRef]
- Xia, S.; Huang, J.; Yan, L.; Han, J.; Zhang, W.; Shao, H.; Shen, H.; Wang, J.; Wang, J.; Tao, C.; et al. MiR-150 Promotes Progressive T Cell Differentiation via Inhibiting FOXP1 and RC3H1. Hum. Immunol. 2022, 83, 778–788. [Google Scholar] [CrossRef]
- Hamada, S.; Masamune, A.; Kanno, A.; Shimosegawa, T. Comprehensive Analysis of Serum MicroRNAs in Autoimmune Pancreatitis. Digestion 2015, 91, 263–271. [Google Scholar] [CrossRef]
- Su, Y.; Li, Z.; Rang, X.; Wang, Y.; Fu, J. Integrated Analysis and Identification of CSF-Derived Risk MiRNAs and Pivotal Genes in Multiple Sclerosis. J. Mol. Neurosci. 2022, 72, 1916–1928. [Google Scholar] [CrossRef]
- Chen, J.Q.; Papp, G.; Póliska, S.; Szabó, K.; Tarr, T.; Bálint, B.L.; Szodoray, P.; Zeher, M. MicroRNA Expression Profiles Identify Disease-Specific Alterations in Systemic Lupus Erythematosus and Primary Sjögren’s Syndrome. PLoS ONE 2017, 12, e0174585. [Google Scholar] [CrossRef]
- Wu, R.; He, Q.; Chen, H.; Xu, M.; Zhao, N.; Xiao, Y.; Tu, Q.; Zhang, W.; Bi, X. MicroRNA-448 Promotes Multiple Sclerosis Development through Induction of Th17 Response through Targeting Protein Tyrosine Phosphatase Non-Receptor Type 2 (PTPN2). Biochem. Biophys. Res. Commun. 2017, 486, 759–766. [Google Scholar] [CrossRef]
- Yang, Z.; Wen, H.J.; Minhas, V.; Wood, C. The Zinc Finger DNA-Binding Domain of K-RBP Plays an Important Role in Regulating Kaposi’s Sarcoma-Associated Herpesvirus RTA-Mediated Gene Expression. Virology 2009, 391, 221–231. [Google Scholar] [CrossRef]
- Spiesz, E.M.; Thorpe, C.T.; Chaudhry, S.; Riley, G.P.; Birch, H.L.; Clegg, P.D.; Screen, H.R.C. Tendon Extracellular Matrix Damage, Degradation and Inflammation in Response to in Vitro Overload Exercise. J. Orthop. Res. 2015, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.B.; Li, Y.; Fung, D.T.; Majeska, R.J.; Schaffler, M.B.; Flatow, E.L. Coordinate Regulation of IL-1β and MMP-13 in Rat Tendons Following Subrupture Fatigue Damage. Clin. Orthop. Relat. Res. 2008, 466, 1555–1561. [Google Scholar] [CrossRef] [PubMed]
- Godwin, J.S.; Sexton, C.L.; Kontos, N.J.; Ruple, B.A.; Willoughby, D.S.; Young, K.C.; Mobley, C.B.; Roberts, M.D. Extracellular Matrix Content and Remodeling Markers Do Not Differ in College-Aged Men Classified as Higher and Lower Responders to Resistance Training. J. Appl. Physiol. 2023, 134, 731–741. [Google Scholar] [CrossRef]
- Rullman, E.; Norrbom, J.; Strömberg, A.; Wågsäter, D.; Rundqvist, H.; Haas, T.; Gustafsson, T. Endurance Exercise Activates Matrix Metalloproteinases in Human Skeletal Muscle. J. Appl. Physiol. 2009, 106, 804–812. [Google Scholar] [CrossRef]
- Zhuang, Y.; Yang, D.; Shi, S.; Wang, L.; Yu, M.; Meng, X.; Fan, Y.; Zhou, R.; Wang, F. MiR-375-3p Promotes Cardiac Fibrosis by Regulating the Ferroptosis Mediated by GPX4. Comput. Intell. Neurosci. 2022, 2022, 9629158. [Google Scholar] [CrossRef]
- Alphonse, N.; Wanford, J.J.; Voak, A.A.; Gay, J.; Venkhaya, S.; Burroughs, O.; Mathew, S.; Lee, T.; Evans, S.L.; Zhao, W.; et al. A Family of Conserved Bacterial Virulence Factors Dampens Interferon Responses by Blocking Calcium Signaling. Cell 2022, 185, 2354–2369.e17. [Google Scholar] [CrossRef] [PubMed]
- Ott, N.; Faletti, L.; Heeg, M.; Andreani, V.; Grimbacher, B. JAKs and STATs from a Clinical Perspective: Loss-of-Function Mutations, Gain-of-Function Mutations, and Their Multidimensional Consequences. J. Clin. Immunol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Rusin, A.; Seymour, C.; Mothersill, C. Chronic Fatigue and Immune Deficiency Syndrome (CFIDS), Cellular Metabolism, and Ionizing Radiation: A Review of Contemporary Scientific Literature and Suggested Directions for Future Research. Int. J. Radiat. Biol. 2018, 94, 212–228. [Google Scholar] [CrossRef]
- Okreglicka, K.; Iten, I.; Pohlmeier, L.; Onder, L.; Feng, Q.; Kurrer, M.; Ludewig, B.; Nielsen, P.; Schneider, C.; Kopf, M. PPARγ Is Essential for the Development of Bone Marrow Erythroblastic Island Macrophages and Splenic Red Pulp Macrophages. J. Exp. Med. 2021, 218, e20191314. [Google Scholar] [CrossRef]
- Saha, A.K.; Schmidt, B.R.; Wilhelmy, J.; Nguyen, V.; Abugherir, A.; Do, J.K.; Nemat-Gorgani, M.; Davis, R.W.; Ramasubramanian, A.K. Red Blood Cell Deformability Is Diminished in Patients with Chronic Fatigue Syndrome. Clin. Hemorheol. Microcirc. 2019, 71, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Solé, P.; Yamanouchi, J.; Garnica, J.; Uddin, M.M.; Clarke, R.; Moro, J.; Garabatos, N.; Thiessen, S.; Ortega, M.; Singha, S.; et al. A T Follicular Helper Cell Origin for T Regulatory Type 1 Cells. Cell. Mol. Immunol. 2023, 20, 489. [Google Scholar] [CrossRef]
- Yao Lan, H.; Ka, L.; Lan, H.Y.; Ka Shing, L. Diverse Roles of TGF-β/Smads in Renal Fibrosis and Inflammation. Int. J. Biol. Sci. 2011, 7, 1056–1067. [Google Scholar] [CrossRef] [PubMed]
- Rioux, K.L.; Delaney, S. Ionic Strength Modulates Excision of Uracil by SMUG1 from Nucleosome Core Particles. DNA Repair 2023, 125, 103482. [Google Scholar] [CrossRef]
- Mi, H.; Kops, O.; Zimmermann, E.; Jäschke, A.; Tropschug, M. A Nuclear RNA-Binding Cyclophilin in Human T Cells. FEBS Lett. 1996, 398, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yang, Z. Resistin’s, Obesity and Insulin Resistance: The Continuing Disconnect between Rodents and Humans. J. Endocrinol. Investig. 2016, 39, 607–615. [Google Scholar] [CrossRef]
- Patarca, R. Cytokines and Chronic Fatigue Syndrome. Ann. N. Y. Acad. Sci. 2001, 933, 185–200. [Google Scholar] [CrossRef]
- Hardcastle, S.L.; Brenu, E.W.; Johnston, S.; Nguyen, T.; Huth, T.; Ramos, S.; Staines, D.; Marshall-Gradisnik, S. Serum Immune Proteins in Moderate and Severe Chronic Fatigue Syndrome/Myalgic Encephalomyelitis Patients. Int. J. Med. Sci. 2015, 12, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Lisboa, F.A.; Bradley, M.J.; Hueman, M.T.; Schobel, S.A.; Gaucher, B.J.; Styrmisdottir, E.L.; Potter, B.K.; Forsberg, J.A.; Elster, E.A. Nonsteroidal Anti-Inflammatory Drugs May Affect Cytokine Response and Benefit Healing of Combat-Related Extremity Wounds. Surgery 2017, 161, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Fuss, I.J.; Kanof, M.E.; Smith, P.D.; Zola, H. Isolation of Whole Mononuclear Cells from Peripheral Blood and Cord Blood. Curr. Protoc. Immunol. 2009, 85, 7.1.1–7.1.8. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-Directed Enzymatic Amplification of DNA with a Thermostable DNA Polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef] [PubMed]
| Target miRNAs | Association and Potential Role in ME/CFS | References |
|---|---|---|
| miR-124-3p |
| |
| miR-127-3p |
| |
| miR-140-5p |
| |
| miR-142-5p |
| |
| miR-143-3p |
| |
| miR-150-5p |
| |
| miR-448 |
| |
| miR-551b-3p |
|
| Group | N | Age Mean ± SE 1 | Gender | ME/CFS Severity 2 | ||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ||||
| ME/CFS | 40 | 49.30 ± 2.234 | F: 31 (77.5%) M: 9 (22.5%) | 5 (12.5%) | 22 (55%) | 13 (32.5%) |
| Controls | 20 | 33.40 ± 2.634 | F: 16 (80%) M: 4 (40%) | - | - | - |
| p value | 0.0001 | 1.00 (n.s) | ||||
| Cytokine | Group (Subjects n°) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Controls (20) | ME/CFS (39) | p-Value 2 | ME/CFS Grade 1 (5) | p-Value 2 | ME/CFS Grade 2 (22) | p-Value 2 | ME/CFS Grade 3 (13) | p-Value 2 | |
| IFN-γ | 649.90 ± 61.33 | 507.7 ± 49.98 | 0.1560 | 675.8 ± 92.42 | 0.2890 | 429.2 ± 77.06 | 0.1634 | 572.3 ± 79.80 | 0.5089 |
| IL-17A | 103.80 ± 12.40 | 18.80 ± 5.54 | 0.0003 | 20.47 ± 20.43 | 0.1568 | 16.66 ± 5.55 | <0.0001 | 19.48 ± 11.76 | 0.0043 |
| IL-2 | 30.40 ± 3.99 | 4.52 ± 1.42 | 0.0001 | 7.26 ± 7.17 | 0.1989 | 4.28 ± 1.10 | <0.0001 | 4.40 ± 2.63 | 0.0004 |
| IL-21 | 43.97 ± 5.94 | 9.07 ± 2.13 | 0.0001 | 10.46 ± 9.20 | 0.1065 | 8.68 ± 2.18 | <0.0001 | 9.64 ± 3.99 | 0.0005 |
| IL-23 | 1.602 ± 235.00 | 407.5 ± 114.5 | 0.1065 | 527.5 ± 620.3 | 0.6728 | 403.2 ± 100.3 | 0.0013 | 377.4 ± 194.3 | 0.0156 |
| IL-6 | 16.34 ± 8.14 | 0.90 ± 2.13 | 0.0270 | 0.95 ± 0.23 | 0.0061 | 0.90 ± 2.96 | 0.0308 | 0.74 ± 4.31 | 0.0588 |
| TNF-α | 45.16 ± 4.06 | 12.35 ± 3.42 | 0.0052 | 13.44 ± 11.27 | 0.6397 | 12.35 ±4.04 | 0.0038 | 11.61 ± 6.89 | 0.0587 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soffritti, I.; Gravelsina, S.; D’Accolti, M.; Bini, F.; Mazziga, E.; Vilmane, A.; Rasa-Dzelzkaleja, S.; Nora-Krukle, Z.; Krumina, A.; Murovska, M.; et al. Circulating miRNAs Expression in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Int. J. Mol. Sci. 2023, 24, 10582. https://doi.org/10.3390/ijms241310582
Soffritti I, Gravelsina S, D’Accolti M, Bini F, Mazziga E, Vilmane A, Rasa-Dzelzkaleja S, Nora-Krukle Z, Krumina A, Murovska M, et al. Circulating miRNAs Expression in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. International Journal of Molecular Sciences. 2023; 24(13):10582. https://doi.org/10.3390/ijms241310582
Chicago/Turabian StyleSoffritti, Irene, Sabine Gravelsina, Maria D’Accolti, Francesca Bini, Eleonora Mazziga, Anda Vilmane, Santa Rasa-Dzelzkaleja, Zaiga Nora-Krukle, Angelika Krumina, Modra Murovska, and et al. 2023. "Circulating miRNAs Expression in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome" International Journal of Molecular Sciences 24, no. 13: 10582. https://doi.org/10.3390/ijms241310582
APA StyleSoffritti, I., Gravelsina, S., D’Accolti, M., Bini, F., Mazziga, E., Vilmane, A., Rasa-Dzelzkaleja, S., Nora-Krukle, Z., Krumina, A., Murovska, M., & Caselli, E. (2023). Circulating miRNAs Expression in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. International Journal of Molecular Sciences, 24(13), 10582. https://doi.org/10.3390/ijms241310582








