Abstract
Colorectal cancer (CRC) remains a major life-threatening malignancy, despite numerous therapeutic and screening attempts. Apoptosis and autophagy are two processes that share common signaling pathways, are linked by functional relationships and have similar protein components. During the development of cancer, the two processes can trigger simultaneously in the same cell, causing, in some cases, an inhibition of autophagy by apoptosis or apoptosis by autophagy. Malignant cells that have accumulated genetic alterations can take advantage of any alterations in the apoptotic process and as a result, progress easily in the cancerous transformation. Autophagy often plays a suppressive role during the initial stages of carcinogenicity, while in the later stages of cancer development it can play a promoting role. It is extremely important to determine the regulation of this duality of autophagy in the development of CRC and to identify the molecules involved, as well as the signals and the mechanisms behind it. All the reported experimental results indicate that, while the antagonistic effects of autophagy and apoptosis occur in an adverse environment characterized by deprivation of oxygen and nutrients, leading to the formation and development of CRC, the effects of promotion and collaboration usually involve an auxiliary role of autophagy compared to apoptosis. In this review, we elucidate the different roles of autophagy and apoptosis in human CRC development.
Keywords:
human colorectal cancer; apoptosis; autophagy; oxidative stress; non-coding-RNA; p53; NF-kB; SQSTM1/p62 1. Introduction
CRC remains a major life-threatening malignancy, despite numerous therapeutic and screening attempts [1]. During CRC development, programmed cell death plays a crucial role in the transformation process of benign polyps into adenomas and ultimately adenocarcinoma [2]. During the multistep process of human colorectal carcinogenesis, the key proteins regulating the programmed death of mucosal cells undergo fundamental expression changes. The cells of the normal mucosa evolve towards a neoplastic condition and acquire new characteristics and biological capabilities, among which the ability to escape cell death is one of the most important [3]. The multistep process of human carcinogenesis requires that cancer cells acquire the features enabling them to become tumorigenic and ultimately malignant. The apoptotic machinery is very complex and diversified and is composed of both upstream regulators and downstream effector components [4]. The existing balance between pro and anti-apoptotic factors belonging to the B-cell lymphoma-2 (Bcl-2 family) of regulatory proteins is the basis of the control of the “apoptotic trigger” that conveys signals between regulators and effectors. Moreover, several abnormality sensors that play key roles in tumor development have been identified, among which a DNA damage sensor that functions via the tumor suppressor p53 (TP53) is most notable [5]. Apoptosis and autophagy are two processes that share common signaling pathways, are linked by functional relationships and have similar protein components. During the development of cancer, the two processes can trigger simultaneously in the same cell, causing, in some cases, an inhibition of autophagy by apoptosis or apoptosis by autophagy [6]. In particular, several genes and proteins are part of the complex mechanism of autophagy. Recent evidence has demonstrated that autophagy plays a decisive role in all stages of human tumorigenesis, by leading initiation and contributing to progression and metastatic ability in tumors. The signal pathways that are activated in the autophagic process during carcinogenesis are not yet defined and appear to be dependent on the tumor microenvironment, as they can have both promotion and inhibition roles. Autophagy often plays a suppressive role during the initial stages of carcinogenicity, while in the later stages of cancer development, it can play a promoting role [6]. It is extremely important to determine the regulation of this duality of autophagy in the development of CRC and to identify the molecules involved, the signals and the mechanisms that allow autophagy to play a pro-malignant dominant role in some conditions and the opposite role in others. The most interesting research on autophagy in colorectal carcinogenesis has been focused on several molecules, mainly microtubule-associated protein 1A/1B-light chain 3 (LC3), sequestosome 1 (SQSTM1/p62) and nuclear factor-κB (NF-κB), and these studies have produced conflicting results. In this review, we try to elucidate the different roles of autophagy and apoptosis in human CRC development.
2. Apoptosis and Autophagy in Adenoma–Carcinoma Sequence
The crypts of the human colorectal mucosa exhibit a polarized structural organization. The reserve pool of the stem cells of the regenerative epithelium is very small and located at the base of the crypt. Stem cells move from the bottom of the crypt up, and on the apical margin of the mucosa about 10 million cells per day die from apoptosis, after which they fall into the lumen [7]. For this reason, adequate cell death is required to maintain the homeostasis of normal colorectal mucosa. However, defective signaling or unbalanced regulation of apoptosis is probably one of the causes of the onset and progression of an adenoma to the carcinoma sequence that results in CRC. It should be noted that the proteins most involved in apoptosis, such as Bcl-2 antagonist killer 1 (Bak) or Bcl-2, are not equally expressed in all anatomical regions of the colorectal mucosa, confirming the fact that there is a diversified regulation of death in the bowel [8,9]. Moreover, not only apoptosis, as a classical form of programmed cell death, has been shown to be operative and consistent in the colorectal glands, but also autophagy, a controlled process of cell self-digestion of great importance in situations of cellular stress, lack of nutrients, or energy deprivation (Figure 1) [10].
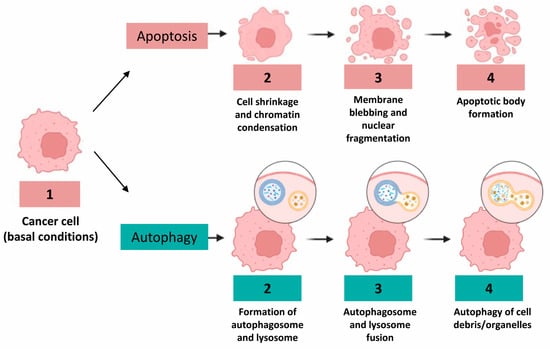
Figure 1.
Different morphological features of human colon cancer cells during apoptosis and autophagy considered as prototypic cell death processes, in particular, morphogenetic changes occurring in the nucleus during apoptosis, that is, fragmentation process or, conversely, vacuole formation during autophagy.
In healthy differentiated adult cells, autophagy delivers to the lysosome damaged cytosolic components and organelles, preventing ageing, and actively participates in intracellular quality control and renovation in order to avoid the formation of protein aggregates that could cause several diseases, including neurodegenerative ones [11]. In contrast to apoptosis, the intensity of autophagy cell expression decreases in the crypt from the bottom to the top. Autophagy is therefore confined to the lower part of the crypt [6]. Groulx et al. [12] suggested that, in the human colon, autophagy was physiologically present in the normal mucosa and is associated with a region containing the proliferative progenitor/stem cell populations, which are crucial for general metabolism and cellular turnover of the colonic epithelium. The integrity of the complex interaction between cell death and autophagy, a stress-reactive process, is necessary for the normal development and homeostasis of colorectal mucosa. Modifying the classic pathways of the cell death signal is often the trigger for the pathogenesis of various colorectal diseases, for example, chronic diseases and those sustained by a constant inflammation of the colorectal mucosa (Crohn’s disease as well as ulcerative colitis) [10]. Cancer cells evolve a variety of strategies to escape or circumvent the actions of the apoptotic machinery and program; moreover, autophagy appears to represent an additional barrier that must be overcome during cancer development and progression [13]. The development of CRC commonly requires three precisely connected steps: initiation, a process that modifies the molecular structure and phenotype of the normal cell; promotion, in which specific signal transduction cascades are activated; and progression, involving the activation of cells that have accumulated relevant gene mutations and phenotypic alterations (Figure 2). Changes in the expression pattern levels of different apoptotic proteins during the adenoma–carcinoma sequence highlights the crucial role of apoptosis during the progression of colon cancer. Moreover, both pro- and anti-apoptotic factors are upregulated in adenomas; therefore, tumor progression is probably due to a balance between these factors [14].
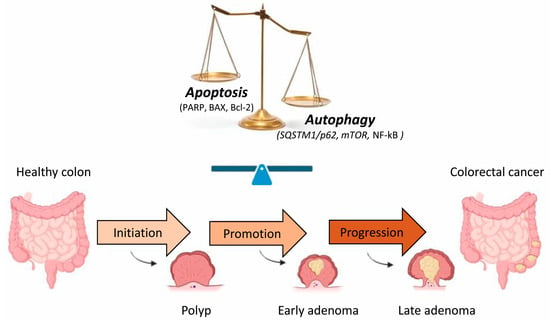
Figure 2.
Schematic illustration of the human CRC sequence and development. There are three stages in the development of colorectal carcinogenesis: initiation, promotion and progression. Changes in the expression of different apoptotic and autophagic proteins during the transformation of adenoma into CRC suggest the importance of the balance between these two events in the progression of CRC. [BAX, Bcl-2 associated X protein; Bcl-2, B-cell lymphoma-2; mTOR, mammalian target of rapamycin; NF-κB, nuclear factor-κB; PARP, poly (ADP-ribose) polymerase, SQSTM1/p62, sequestosome 1].
The tumor suppressor p53 function is frequently impaired in human cancers, and it is thought that p53 mutations play a pivotal role in the adenoma–carcinoma transition [15,16] and occur in 60% of reported CRCs [17]. For this reason, the transition from adenoma to carcinoma in the colorectal region is considered a mechanism in which the regulation of the apoptotic process, given its relationship to the p53 mutations, is essential [18]. As regards the autophagic process, its involvement in the initiation of the adenoma–carcinoma transition is still highly controversial. Studies of the relationship between autophagy and colorectal carcinogenesis have produced contradictory results: the conditional deletion of autophagy-related gene-7 (ATG7) in the intestinal epithelium does not increase the number of intestinal polyps [19], while the heterozygous deletion of autophagy-related gene-5 (ATG5)—an essential ATG protein in autophagy—in adenomatous polyposis coli (Apc) Min/+mice increases the number and size of adenomas through the activation of the epidermal growth factor receptor (EGFR)/extracellular signal-regulated kinase 1/2 (ERK1/2) and wingless and INT-1 (Wnt)/β-catenin pathways [20]. Li et al. [21] suggest that atractylenolide I, a natural substance that significantly decreased the viability of carcinoma cell lines through inducing intrinsic apoptosis, reduces intestinal adenoma formation by activating autophagy machinery to downregulate D-dopachrome tautomerase (D-DT). D-DT is a novel hypoxia-inducible gene and a direct target of hypoxia-inducible factor-1 alpha (HIF-1α) and hypoxia-inducible factor-2 alpha (HIF-2α) [22], and is highly expressed in two human CRC cell lines [21]. Studies, regarding the possible role of autophagy in the adenoma–carcinoma sequence initiation, indicate a possible activation of the immune system [23].
3. Regulatory Role of p53 in Apoptotic and Autophagic Processes
Over the years, the interest of researchers in determining different types of cell death has never diminished, since it remains an essential physiological process required to maintain tissue homeostasis. The recent recommendations of the Nomenclature Committee on Cell Death define autophagy-dependent cell death (ADCD) as regulated cell death that results from the autophagy machinery (i.e., pharmacological or genetic manipulations of autophagy genes block cell death), without involving alternative death pathways [24]. However, research attempts have not yet produced an accurate picture of cell death phenomena. The fate of a cell, whether it lives or dies, is crucial and can be the determining factor in the progression of tumors [25]. Incorrect regulation of the cell death signaling cascades is considered fundamental for the initiation and progression of CRC. Many in vitro studies have elucidated that the interplay between apoptosis and autophagy is highly complex and their influence in cancer can be modified by several factors, such as the types of tissue and cells involved, tumor stage and malignancy grading, type of oncogenic mutation, the entity of damage or cellular stress, intra-tumoral oxygen levels or the presence of nutrient deprivation [26]. The tumor suppressor protein, p53, presents a plethora of anticancer functions [27]. p53 is activated in the presence of cellular stress, including DNA damage, hypoxia, senescence and DNA repair, inducing cell cycle arrest to allow cell repair or apoptosis (mainly via the intrinsic pathway) [28]. The p53 gene is mutated in a high percentage of colorectal tumors and can be found in both adenomas and malignant cells, resulting in the disruption of tumor suppressor functions [24]. There is evidence that p53 mRNA expression may represent a useful survival predictor in patients with stage III CRC or rectal carcinoma [29]. On the other hand, Sakanashi F in one morphological study showed that autophagy and apoptosis are overrepresented in CRC and that p53 mutation may lead to the upregulation of autophagy [30]. p53 mutation may alter the ability to reduce autophagy, which implies that the p53 mutant increases autophagy, leading to more aggressive CRCs. Recently, researchers demonstrated that, after pharmacological or genetic protein kinase B (AKT) inhibition, p53 interacted with sirtuin 6 (SIRT6) and poly (ADP-ribose) polymerase1 (PARP1) directly to activate it, and promoted the formation of PAR polymer in human colon cancer cell lines. Consequently, the PAR polymer, after being transported to the outer membrane of the mitochondria, resulted in the release and transfer of apoptosis-inducing factor (AIF) into the nucleus and the promotion of cell death (Figure 3). These processes were inhibited by p53 deletion or mutation and protective autophagy was increased by the inhibition of AKT [31].
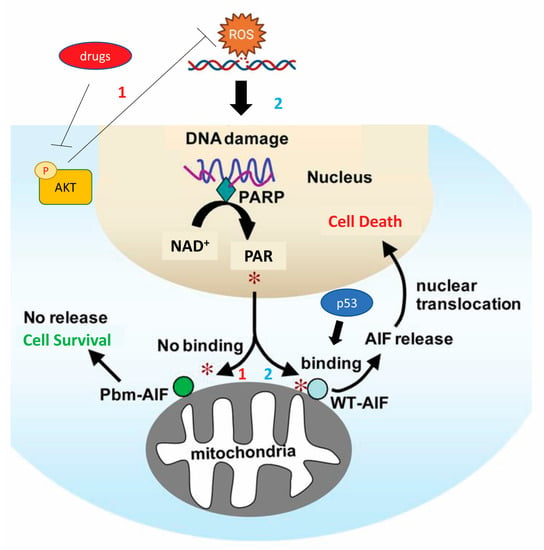
Figure 3.
Mechanism by which p53 promotes the apoptosis of human colon cancer cell lines by releasing and transferring AIF into the nucleus after the formation of the PAR polymer. This death signaling pathway is inhibited by p53 deletion or mutation and protective autophagy is increased by inhibition of AKT. *1, No binding between PAR and AIF; *2, binding between PAR and AIF [AIF, apoptosis-inducing factor; AKT, protein kinase B; NAD, Nicotinamide adenine dinucleotide; p53, protein-53; PAR, poly (ADP-ribose); PARP, poly (ADP-ribose) polymerase].
Another interesting paper showed that apoptotic cell death induced by N1, N11-diethylnorspermine (DENSpm) is boosted when autophagy is inhibited by 3-mase9pethyladenine (3-MA), an autophagy inhibitor, or beclin-1 siRNA through polyamine depletion or polyamine catabolic activation in colon cancer cells, regardless p53 mutation status. In conclusion, the researchers stated that p53 deficiency does not alter the response of colon cancer cells to apoptotic death induced by DENSpm in autophagy suppression conditions [32]. p53 is linked to the regulation of autophagy by affecting the availability of nutrients. The loss of p53 leads to the overproduction of LC3, a standard marker for autophagosomes, and forces cells to maintain elevated levels of autophagy. A study including HCT116 colorectal cells highlighted that p53 promotes selective downregulation of LC3 mRNA and protein in conditions of protracted nutrient deprivation. Initially, autophagy has a protective effect against cellular damage, avoiding nutrient starvation through the recycling of cellular substrate as proteins, protein complexes, lipids, ions and whole organelles. However, after complete nutrient depletion, this form of self-cannibalism is stalled, leading to the accumulation of aberrant metabolic intermediates that activate signaling promoting the execution of programmed cell death, such as apoptosis [33].
4. SQSTM1/p62 Is Related to Autophagy and Apoptosis in CRC Cells
SQSTM1/p62 is an autophagy receptor involved in optimizing the autophagosome formation process. After the fusion of autophagosomes with lysosomes, the autophagosome content, as well as SQSTM1/p62, is degraded [34]. SQSTM1/p62 plays specific and indispensable roles in selective autophagy and SQSTM1/p62 protein levels have been used as an indicator of autophagic flux [35]. The pivotal roles of SQSTM1/p62 in ingested protein catabolism through autophagy and lysosomal targeting have been well elucidated in the literature [34,35,36]. Although the role of SQSTM1/p62 in solid tumors is still debated, evidence has shown that SQSTM1/p62 is upregulated in different cancers and promotes tumor growth. Moreover, it has been assessed that SQSTM1/p62, as a tumor oncogene, is frequently abnormally upregulated and engaged in the acquired malignancy of gastrointestinal tumors, such as CRC [37]. However, the molecular mechanisms that regulate SQSTM1/p62 expression in CRC are still widely debated. Recently, Mukherjee et al. stated that, in HCT116 colorectal cells, SQSTM1/p62 is a mutp53 interactor, which associates selectively with the DNA contact mutant p53R273H but not with the structural mutant p53R175H. They further assess that the interaction with SQSTM1/p62 is needed for the ability of p53R273H to trigger cancer cell migration and invasion. The authors assumed that this acquired ability is due to the involvement of the mutp53-p62 axis in directing the ubiquitin-dependent proteasome degradation of connexin 43 and cell junction proteins [38]. Nevertheless, the autophagic process can be repressed by SQSTM1/p62 inhibition, leading to the arrest of cancer cell growth and the inhibition of cancer development [39]. On the other hand, literature has reported that SQSTM1/p62 expression in CRC cells was suppressed by the β-catenin/transcription factor (TCF)4 complex, which blocked phagocytic ingestion, but under stress conditions induced by nutrient depletion, SQSTM1/p62 was markedly raised, as β-catenin was immediately degraded by LC3 binding to the LC3-interacting region (LIR) of β-catenin [39]. Another interesting paper showed that poly C binding protein 1 (PCBP1), a novel tumor suppressor gene, withdraws or balances the basal cell autophagy necessary to retain cell homeostasis in CRC cells [40]. The authors stated that PCBP1 can affect various autophagy-related genes expression at the different autophagic stages, including the autophagosome formation, LC3B, the autophagosome-lysosome fusion and SQSTM1/p62. Moreover, the PCBP1–SQSTM1/p62 signaling axis may play a repressive role in the early phases of tumorigenesis. PCBP-1 acts by inhibiting both autophagy and proteasome-mediated SQSTM1/p62 degradation and stabilizing SQSTM1/p62 m-RNA. Moreover, considering that SQSTM1/p62 associates with caspase-8 and the subsequent apoptotic pathway [41], PCBP1 downregulation or depletion would be a relevant strategy for tumor cell survival via downregulation of SQSTM1/p62 and caspase-8, suggesting the concept that autophagy inhibition could be an efficient strategy to activate the apoptotic cell death mechanism in colorectal tumor cells [40]. In contrast, Zhang et al. [42] demonstrated that SQSTM1/p62 acts as an oncogene in CRC, both in vivo and in vitro, and that the overexpression of SQSTM1/p62 significantly inhibited apoptosis. The authors demonstrated that SQSTM1/p62 binds the vitamin D receptor (VDR) and may target the nuclear factor-erythroid-2-related factor 2 (NRF2)-quinone oxidoreductase-1 (NQO1) axis via VDR in human CRC cells, promoting CRC cell proliferation and invasion in vivo. Therefore, SQSTM1/p62 may act as a protective agent in tumor cell viability by triggering autophagy and blocking apoptosis under drug-induced stress. Interestingly, escin, a triterpene saponin, inhibited cell viability and colony formation in CRC cells. The treatment of escin induced DNA damage, leading to phospho ataxia-telangiectasia mutated (p-ATM) and histone γh2ax upregulation. In particular, the escin process increased the expression of p62, which had a protective effect against DNA escin-induced damage: knockdown of P62 increased DNA escin-induced damage, while overexpression of p62 decreased the rate of apoptosis, inducing DNA damage repair in escin-treated cells. Moreover, treatment with escin induced apoptosis in a manner directly proportional to concentration and time. Similarly, knockdown of SQSTM1/p62 produced an evident anti-cancer effect in vivo [43]. This result can be reported at the genetic level, since CRC cells treated with escin activated autophagy and overexpressed SQSTM1/p62 played an important protective role against escin-induced apoptosis and DNA damage, as SQSTM1/p62 suppresses the ataxia-telangiectasia mutated/phosphorylated histone family member X pathway [44]. Finally, SQSTM1/p62 may act as a shielded factor in cancer cell survival by triggering autophagy and blocking apoptosis under drug-induced stress; otherwise, it may act as a driving force to determine the adverse fate of tumor cells via mechanisms that remain partially unclear.
5. NF-κB as a Potential Matchmaker between Autophagy and Apoptosis
Studies on CRC have reported that the NF-kB signaling pathway promotes tumor initiation and contributes to cancer cell metastasis formation and epithelial to mesenchymal transition [45,46,47]. In the literature, there is a plethora of papers that confirm the role of NF-kB in regulating the apoptotic process. In fact, the molecular mechanisms by which NF-kB exerts its regulatory action are well known, for instance: NF-κB signaling prevents apoptosis by up-regulating anti-apoptotic genes expression such as B-cell lymphoma-extra-large (Bcl-xL), the Bcl-2-related gene (A1/BFL1), cellular inhibitors of apoptosis proteins (cIAPs) and caspase-8/FAS-associated death domain-like IL-1beta-converting enzyme inhibitory protein (c-FLIP) [46]. Over the past few years, some very interesting studies have highlighted a correlation between NF-kB, autophagy and apoptotic processes. A study showed that metformin had an antiproliferative effect related to changes in the expression of nuclear factor E2-related factor (NRF-2)/NF-κB pathways on human colon cancer cells (HT-29) in a dose- and time-dependent manner, and exerted growth inhibitory effects by increasing both apoptosis and autophagy [48]. Recently, the autophagy-regulated cytotoxic effect of green synthesized silver nanoparticles (Brassica Ag-NPs) against human epithelial colorectal adenocarcinoma cells was demonstrated. The authors found that Brassica Ag-NPs induced NF-κB mediated autophagy in Caco-2 cells. The decreased expression of NFκB was associated with an increased expression of inhibitor of NF-κB (IκB)-kinase, which is involved in autophagic process initiation. Moreover, the activity of p53 and light chain 3 (LC3) II greatly accelerated autophagosome formation, and the inhibition of Akt and mammalian target of rapamycin (mTOR) was evident. In conclusion, the authors stated that, in the event of excessive activation of the autophagic process, after the complete depletion of nutrients, this process stalled, leading to the accumulation of aberrant metabolic intermediates that can trigger apoptotic cell death, which subsequently resulted in necrosis [49]. A study examining the relationship between long non-coding RNA cancer susceptibility candidate 2 (lncRNA CASC2) and microRNA19 (miR19) in colon cancer reported that lncRNA CASC2 suppressed expressions of LC3 and SQSTM1/p62 and significantly reduced cell viability of colon cancer cells, promoting apoptosis. Overexpressed miR19a reduced the expression of lncRNA CASC2 and increased cell viability. NF-kB was upregulated in colon cancer cell lines; otherwise, after treatment by N4-(4-phenoxyphenethyl) quinazoline-4,6-diamine (QNZ), a NF-kB inhibitor, the expression of Bcl2 was downregulated and Bax expression was upregulated. As for autophagy, expressions of LC3 and SQSTM1/p62 were reduced by inhibition of NF-kB. Therefore, inhibition of NF-kB reversed the role of lncRNA CASC2 and enhanced the effect of miR19a. The authors deduced that lncRNA CASC2 and miR19a might have a close correlation during the progression of colon cancer cells and that NF-kB was the signaling pathway, which could regulate the correlation between lncRNA CASC2 and miR19a [50]. Researchers discovered that bupivacaine, a local anesthetic, inhibited colorectal adenocarcinoma cell proliferation and migration, which are related to increased endoplasmic reticulum stress [51]. It seems that bupivacaine significantly inhibited CRC cell vitality, promoted the apoptosis rate and expression of apoptosis genes such as Bax and caspase-3, inhibited Bcl-2 expression, prevented cancer cell migration, increased autophagy-related protein LC3B II/LC3B I ratio and Beclin-1 expression and hindered p62 expression. Finally, bupivacaine could influence the expression of inflammatory factors such as interleukin 1 beta (IL-1 beta), interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α), increasing them and inhibiting phosphorylation of proteins I kappa B kinase (IKK), IκB and NF-κB, related to the signaling pathway of NF-κB in SW480 and SW620 cells. In in vivo experiments, bupivacaine prevented tumor volume increase, as well as NF-κB expression [52].
6. Oxidative Stress and Modulation of Autophagy and Apoptosis
Oxidative stress has been implicated in the pathophysiology of cancer: high levels of intra-cytoplasmatic reactive oxygen species (ROS), generated by accelerate aerobic glycolysis followed by “selfish” metabolic reprogramming (the Warburg effect), increase oncogene activity, the activation of growth factor dependent pathways or the presence of an increase in the pool of oxidizing enzymes, leads to genetic instability [53,54]. Aerobic glycolysis or the Warburg effect is a hallmark of metabolic phenotypes of cancer; in fact, cancer cells, including CRC cells, present altered glucose metabolism and are characterized by an enhanced uptake of glucose and an increased conversion of glucose to lactate [55]. In the past few years, many studies have focused on the importance of the role played by ROS in the crosstalk between autophagy and apoptosis of colon cancer cells. ROS has a negative feedback action on the autophagic process: ROS can promote autophagy, and at the same time, autophagy reduces ROS production by removing damaged mitochondria, endoplasmic reticulum and other materials that contribute to ROS generation [56,57]. On the other hand, ROS can trigger DNA damage, initiating the apoptotic process pathway in colon cancer cells [58,59]. The most common method by which ROS delete transformed cells is the activation of programmed cell death, which is completed by an extrinsic or an intrinsic pathway; both pathways culminate in caspase-induced final cell demise with the formation of apoptotic bodies that are removed by adjacent phagocytes (Figure 4) [60].
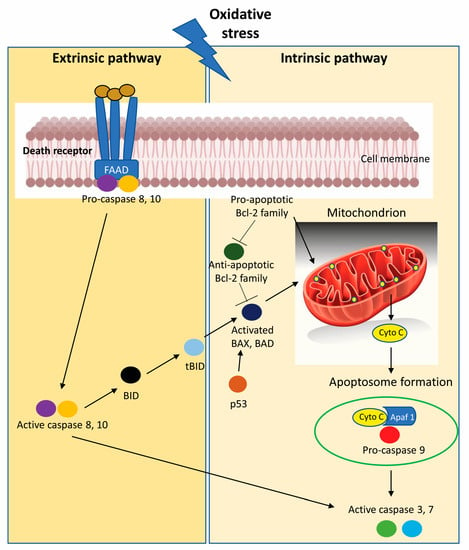
Figure 4.
Oxidative stress promotes the activation of programmed cell death initiated by intrinsic apoptotic signaling in the mitochondria or extrinsic apoptotic signaling by death receptor pathways in CRC cells. [Apaf 1, Apoptotic protease activating factor 1; BAD, Bcl-2-Associated agonist of cell death; BAX, Bcl-2-associated X protein; Bcl-2, B-cell lymphoma-2; BID, BH3-interacting domain death agonist; cyto c, cytochrome c; tBID, C-terminal fragment termed truncated Bid].
In the past few years, several interesting articles have been published concerning the inducing effect on both apoptosis and autophagy of drugs, mostly of natural origin, through regulatory pathways mediated by oxidative stress [61,62,63,64,65]. In this regard, a study investigated how β-elemene, extracted from the genus Curcuma, affected cells in vitro and in vivo. The results reported that after β-elemene treatment, CRC cells exhibited apoptotic bodies and increased levels of cleaved-caspase-3/9 and PARP proteins. The in vivo experiments conducted on mouse-xenograft models inoculated with mouse colon carcinoma HT-29 cells presented the same expression pattern of apoptotic markers. Moreover, β-elemene promoted the formation of intracellular acidic vesicle organelles (autophagy lysosomes) and boosted the expression of autophagic markers such as LC3B and SQSTM1/p62 in human CRC cells. In addition, β-elemene increased the expression of LC3B-II in tumor tissues, indicating that the autophagosomes–lysosomes fusion is very active. Moreover, the authors stated that β-elemene increased ROS intracytoplasmic levels, enhanced the phosphorylation of activated AMP-activated protein kinase (AMPK) and reduced the phosphorylation of mTOR in human CRC cells; hence, β-elemene leads to autophagy and apoptosis via the activation of the ROS/AMPK/mTOR pathway in human CRC cells [66]. Meanwhile, other authors demonstrated that polysaccharide from Dendrobium officinale (DOP) indirectly targeted mitochondria and induced excess ROS production, which subsequently caused mitochondrial dysfunction and inhibited ATP synthesis. Then, the low ratio of ATP/AMP activated AMPK/mTOR signaling, which in turn increased autophagy in CT26 cells. Finally, excessive activation of the autophagic process induced cancer cell death [67]. Similarly, polyphyllin I (PPI) induces ROS production and down-regulates the AKT/mTOR pathway, which in turn triggers the autophagic cell death and apoptosis of SW480 colon cancer cells [68]. Intriguingly, other articles emphasize the protective role played by the autophagic process on the survival of colon cancer cells treated with anticancer drugs. In fact, Antrodia salmonea (AS), a traditional Chinese medicinal fungus with anti-cancer proprieties, reduced the phosphorylation of AKT and mTOR in SW620 cells, inhibiting AKT/mTOR signaling cascades. Moreover, AS decreased cell viability and induced apoptosis and cytoprotective autophagy via intra-cytoplasmatic ROS buildup and interruption of the signaling pathways for ERK [69]. In the literature, it was reported that selenium exhibited chemo-preventive and chemotherapeutic effects against CRCs [70]. As regards this topic, Yu et al. identified a signaling pathway involved in selenite-induced protective autophagy in CRC cells undergoing apoptosis. Selenite leads to ROS production and promotes phosphorylation of AMPK, which binds and activates forkhead box O3 (FoxO3a). The phosphorylated FoxO3a binds to γ-aminobutyric acid receptor-associated protein-like 1 (GABARAPL-1) promoter and upregulates the transcription level of GABARAPL-1, activating an autophagic process that serves as a survival signal against apoptosis in both CRC cells and colon xenograft models [71].
7. Role of Non-Coding RNA in Enhancing or Inhibiting Autophagy and Apoptosis
With the deepening of the knowledge about the interaction between autophagy and apoptosis in CRC, researchers gradually begin to pay attention to the role of non-coding-RNA in this interplay. microRNAs (miRNAs) are a group of evolutionarily conserved small RNAs, endogenous, non-coding, single-stranded RNAs that regulate the expression of their target genes through mRNA degradation and serve a negative role at post-transcriptional levels. miRNAs have been found to influence many cellular biological processes including apoptosis, cell cycle distribution and autophagy in CRC [72,73,74,75] (Table 1 and Table 2). In detail, as regards autophagy, miRNAs may play a dual, and therefore conflicting, role in tumor cell proliferation through modulation of the autophagic process. While miR-211 promote CRC proliferation by inhibiting autophagy and targeting tumor protein 53-induced nuclear protein 1 (TP53INP1) [76], on the other hand, miR-30d suppresses cell proliferation in CRC by inhibiting autophagy. The authors demonstrated that miR-30d inhibited autophagy and promoted apoptosis of colon cancer cells by directly targeting ATG5, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta (PIK3CB) and beclin 1 (BECN1); the inhibition of proliferation was due to negative regulation of cell autophagy and promotion of cell death [77]. More recently, a study focused on the effect of miR-126 on the autophagy and apoptosis of CRC cells in vitro and in vivo highlighted the synergistic role of the two processes in the inhibition of CRC cell proliferation by modulating the mTOR pathway [78]. Otherwise, overexpressing miR-142-3p facilitated proliferation and inhibited apoptosis of CRC cells. miR-142-3p targeted and decreased TP53INP2 levels. On the other hand, TP53INP2 overexpression suppressed the HEDGEHOG signaling pathway and induced the activation of CRC cell autophagy. In conclusion, miR-142-3p hindered tumor cell autophagy and promoted CRC progression via targeting TP53INP2 [79]. Moreover, a very intriguing study of Zhang et al. has shown the interplay between long non-coding RNA growth arrest specific 5 (GAS5) and miR-34a, and their regulative role on macroautophagy and apoptosis in CRC patients [80]. GAS5 is a newly discovered lncRNA that exerts inhibitory effects through the modulation of several target microRNAs in cancer [81], and miR-34a acts as a tumor-suppressor gene in human CRC by blocking autophagy through sirtuin 1 (SIRT1) regulating [82]. Finally, this study demonstrated that miR-34a participates in regulating GAS5-suppressed CRC cell macroautophagy and leads to apoptosis through the mTOR/SIRT1 pathway. GAS5-mediated regulation of macroautophagy retains CRC cells in an equilibrium state that protects against apoptosis. Consistent with this notion, miR-483 promotes the development of CRC by inhibiting the expression level of etoposide-induced 2.4 (EI24), an autophagy associated transmembrane protein [83], and the knockdown of homeobox transcript antisense intergenic RNA (HOTAIR), a well-known lncRNA, stimulates cell apoptosis and suppresses cell autophagy by upregulating microRNA-93 and downregulating autophagy-related 12 (ATG12) in CRC [84]. Circular RNA (circRNA) is a kind of high abundance non-coding covalent closed RNA formed by exon sequence and intron sequence, and many studies have shown that a variety of circRNA is abnormally expressed in CRC, and its abnormal expression is related to the proliferation, apoptosis and autophagy of CRC cells [85]. Despite the relationship between circRNAs and apoptosis in CRC having been confirmed by some studies, their regulation of autophagy molecules is gradually being revealed. In fact, silencing of circular homeodomain-interacting protein kinase (circ HIPK3) and hsa_circ_0007534 induces apoptosis in HCT116 and HT-29 CRC cells [86,87]. In the past few years, many circular RNAs have been identified, the downregulation of which induces the activation of the apoptotic process in CRC cells, i.e., hsa_circ_0020397, hsa_circ_0000523 [88,89]. Recently, Chen et al. showed that silencing circular RNA ubiquitin-associated protein 2 (circUBAP2) decreased the formation of total autophagosomes and autolysosomes in CRC cells, and when circUBAP2 is knocked down, a decrement of LC3B-II expression, a downregulation of LC3B-I/II conversion and the degradation of autophagy-related protein 6 (beclin1), autophagy-related protein 7 (ATG7) and forkhead box O1 (FOXO1) proteins were observed. In conclusion, overexpression of circUBAP2 can effectively activate autophagy in CRC cells by targeting the miR-582-5p/FOXO1 axis [90]. Authors found that circCCDC66 in CRC under hypoxic conditions promoted the malignant behaviors of CRC cells by regulating the miR-3140/autophagy pathway [91]. Moreover, circular autophagy-related protein 4B (circATG4B) encodes a novel functional protein (circATG4B-222aa) that promoted autophagy in CRC cells both in vitro and in vivo. Therefore, circATG4B-222aa interacts competitively with transmembrane p24 trafficking protein 10 (TMED10) and functions as a decoy that prevents TMED10 from binding to ATG4B, leading to increased autophagy [92].

Table 1.
Non-coding-RNA associated with apoptosis in CRC.

Table 2.
Non-coding-RNA associated with autophagy in CRC.
8. Pathways Implicated in Regulation of Apoptosis and Autophagy and Possible Therapeutic Targets
The most important pathway involved in the induction and regulation of apoptosis and autophagy in CRC is the Wnt pathway and its component components, such as β-catenin, disheveled (Dvl), adenomatous polyposis coli (APC) and axin, that are activated along with the Wnt signaling cascade during cancer development [93]. Wnt signal transduction is typically divided into classical and non-classical pathways. The classical pathway is involved in cell survival, proliferation, apoptosis and autophagy, while the non-classical pathway regulates cell polarity and migration. The Wnt/β-catenin signaling and autophagy pathways play important roles in tissue homeostasis and tumorigenesis. A variety of studies, reporting experimental design of the genomic manipulation of β-catenin expression levels in vitro and in vivo, show that β-catenin inhibits autophagosome formation and directly blocks SQSTM1/p62 via T cell-specific DNA-binding protein (TCF4) [94]. During starvation, β-catenin is selectively degraded through the formation of β-catenin-LC3 complexes, which avoid β-catenin/T cell-specific DNA-binding protein (TCF) mediated transcription and proliferation in order to better adapt to metabolic stress conditions. The formation of the β-catenin-LC3 complex is mediated by the W/YXXI/L motif and the LC3 interaction region (LIR) in β-catenin. Therefore, as previously described, Wnt/β-catenin inhibits autophagy and SQSTM1/p62 expression, while β-catenin itself is targeted for autophagic degradation [95]. Autophagy negatively controls Wnt signaling by promoting Dvl degradation. Von Hippel–Lindau protein-mediated ubiquitination is necessary for the binding of Dvl2 to SQSTM1/p62, leading to the formation of Dvl2 aggregates under conditions of nutrient deprivation and LC3-mediated autophagosome recruitment. Finally, the ubiquitylated Dvl2 aggregates are degraded through the autophagy-lysosome pathway [96]. Interestingly, an inverse correlation between Dvl expression and autophagy was found in late stages of CRC development, supporting the hypothesis that autophagy may contribute to the aberrant promotion of Wnt signaling during tumorigenesis [93]. In the literature, many reports indicate that Wnt/β-catenin signaling plays a pivotal role in tumor initiation and development, and its function of inhibiting cell apoptosis may result in tumor chemoresistance [97]. As regard autophagy, APC, a modulator of the Wnt signaling pathway, can interact with other members of the Wnt cascade to regulate the development of bowel diseases by affecting autophagy [98]. Recently, researchers showed that Na/H exchange 3 regulator 1 (NHERF1), which is normally lost or down-regulated in early dysplastic adenomas, triggers nuclear β-catenin activity. The depletion of NHERF1 is responsible for the exacerbation of the transformed phenotype in vitro and in vivo in CRC cells. The genetic or pharmacologic manipulation of β-catenin caused an autophagy-to-apoptosis switch’ that implied the activation of caspase-3, the cleavage of PARP and the decrement of phosphorylated ERK1/2, beclin-1 and ras-related in brain 7 (Rab7) autophagy proteins levels, which in turn strikingly impacted the fate of CRC cells. Finally, the researchers hypothesized that doubleβ-catenin/NHERF1-inhibitory strategy may be fruitful to augment the induction of apoptotic death in CRC cells refractory to Wnt/β-catenin-targeted therapeutics [99]. Casein kinase 1 α (CK1α) is an enzyme that simultaneously regulates both Wnt/β-catenin and AKT. Despite the link of the AKT and Wnt pathways to autophagy in RAS-mutated CRC cells not being well defined, a pharmacological study demonstrated that CK1α is involved in regulation of autophagy, Wnt/β-catenin and AKT pathways in rat sarcoma (RAS)-mutant CRC cell lines. In particular, CK1α inhibition significantly reduced the AKT/phospho-β-catenin axis, and at the same time blocked the autophagy flux. In addition, CK1α inhibition induces the activation of apoptotic machinery [100]. According to recently published literature, achaete-scute homolog 2 (Ascl2), as the target molecule of the Wnt signaling pathway, is an important marker of colon cancer stem/precursor cells [101]. Wang et al. [102] showed that the suppression of Ascl2 expression exerts a tumor suppressor function in CRC by inducing excessive autophagy. Apoptosis, which is triggered by si-Ascl2 (small/short interference), could be counteracted by treatment with autophagy inhibitors, such as 3-methyladenine (3-MA) and chloroquine (CQ), indicating that Ascl2 targeted therapy may represent a new strategy for the treatment of CRC. Studies published thus far established that SLC6A14, a Na+/Cl− coupled neutral and cationic amino acid transporter, is up-regulated in CRC tissues and in colon cancer cell lines. Recently, researchers have paid attention to the evaluation of the impact of SLC6A14 deletion on colon cancer in multiple preclinical model systems, comparing the effects with pharmacologic SLC6A14 blockade. Treatment of colon cancer cells with α-methyltryptophan (α-MT), a blocker of SLC6A14, decreases mTOR activity, triggers autophagy and promotes apoptosis. Moreover, in xenograft and syngeneic mouse tumor models, silencing of SLC6A14 or suppressing its action by α-MT reduces the size of tumor via targeting of β-catenin [103]. Tumor necrosis factor-α-inducing protein 8-like 2 (TIPE2) is a novel potential therapeutic target for advanced and recurrent human rectal adenocarcinoma [104]. TIPE2 overexpression modulates apoptosis through the Wnt/β-Catenin signaling by down-regulating the expression levels of Wnt3a, phospho(p)-β-Catenin and p-glycogen synthase kinase-3β. Family with sequence similarity 134, member B (FAM134B), in CRC, act as a cancer suppressor and inhibits autophagy by regulating the autophagic turnover of endoplasmic reticulum [105]. A fairly complex experimental study has demonstrated that exogenous inhibition of FAM134B leads to upregulation of EB1, which could in turn promote the WNT/β-catenin pathway by inactivating the tumor suppressor gene APC and consequently activating β-catenin in colon cancer cells. FAM134B is probably involved in the activation and regulation of earlier events/steps in the adenoma–carcinoma sequence of colorectal tumorigenesis by affecting autophagy via the WNT/β-catenin pathway [106].
9. Discussion
It is necessary to revisit, update and understand the main pathways and components that control cell death and autophagy, the connection between the pathways associated with CRC and the possible interaction between these pathways to obtain a better understanding of the biology of cell death in CRC context. Apoptosis plays an important role in many biological events, including the elimination of harmful cells. Malignant cells that have accumulated genetic alterations can take advantage of any alterations in the apoptotic process and, as a result, progress easily in the cancerous transformation [6]. A dysregulation of autophagic flux leads to an intracytoplasmic accumulation of organelles, protein aggregates and metabolic intermediates. These accumulations may trigger the over-production of reactive oxygen species and cause metabolic insufficiency. Especially in stressful situations and in conditions of nutrient deprivation, impairment of autophagic flux can promote tumorigenesis [107]. By contrast, autophagy is essential for the survival of cancer cells and in malignant cells usually a high level of autophagy is evident. However, autophagy induction promotes cancer cells survival under conditions of hypoxia and metabolic and energy depletion. As regards CRC, autophagosome formation is most prominent in tumors cells growing in a hypoxic environment [108]. Importantly, there is a wide overlap of the apoptosis and autophagy signaling networks and providing mechanistic insight their relationships is very difficult. Most prominently, Bcl-2 proteins are at the crossroad between the two processes; they are inhibitors of both apoptosis and autophagy by binding pro-autophagic beclin1 [109]. Therefore, it has been shown that dual specificity protein phosphatase 4 (DUSP4) silencing blocked the protein interaction between Bcl-2 and beclin1 or Bax in HCT116 cells. Moreover, the survival of HCT116 cells inhibited by DUSP4 silencing was prevented by autophagy impairment with spautin-1. DUSP4 can promote the viability and function of CRC cells by inhibiting Bcl-2 phosphorylation-dependent autophagic cell death and apoptosis [110]. Colitis and colon cancer initiation are prevented by the activation of intestinal epithelial autophagy; however, colon cancer growth and progression are aided by this process [47]. The relationship between autophagy and apoptosis is complex in CRC because normally the two processes are active in the same cells, but not simultaneously: autophagy always precedes the initiation of apoptosis [9]. At the same time, a variety of signal transduction pathways and regulators are involved and autophagy may induce or inhibit apoptosis depending on the cell type involved and the nature and duration of the stimulus/stress [10,111,112]. For these reasons, depending on the type of interaction between autophagy and apoptosis, an antagonistic or synergistic effect may occur in CRC cells. As described previously, excessive autophagy induces cancer cell death through the PI3K/AKT/mTOR pathway, which plays different roles in autophagy and apoptosis of CRC [68]. Autophagy and apoptosis can comparably induce cancer cell death in CRC. Another instance of cooperation between the two processes is represented by the induction of apoptosis in HT-29 and Caco-2 human cells by Rhus coriaria extract (RCE). RCE significantly impaired the viability of colon cancer cells through the caspase-7-dependent pathway, but it does not represent the main mechanism of cell death, which is triggered through programmed cell death type II (PCD-II), probably due to excessive autophagy activation [113].
10. Conclusions
All the reported experimental results essentially indicate that, while the antagonistic effects of autophagy and apoptosis occur in an adverse environment characterized by deprivation of oxygen and nutrients leading to the formation and development of CRC, the effects of promotion and collaboration usually involve an auxiliary role of autophagy compared to apoptosis in colon cancer cells. In an overwhelming majority of cases, studies examining the mechanistic role of autophagy and apoptosis on CRC tumorigenesis used xenograft mice models and CRC cell lines. Moreover, many of these studies have targeted autophagy and apoptosis transiently through genetic manipulations or the use of drugs, which can influence the study in various ways. From our point of view, it would be necessary for preclinical research to focus on the improvement of gene-editing techniques, which are necessary to delineate the mechanistic role of autophagy and apoptosis in CRC. The results obtained at this stage would be of great use in clinical trials in patients to test the response to immune-checkpoint inhibitors. In addition, it is necessary to focus attention on ongoing clinical trials that are using apoptosis or autophagy inhibitors in combination with chemotherapeutics or drugs. These studies aim to capture the positive results of using autophagy as a process of inducing programmed cell death in cancer cells. Such autophagic manipulation can lead to promising effects in the treatment of CRC if it will attempt to solve the autophagy paradox that migrates from an anti-cancer to a pro-cancer mechanism, so treatments need to be highly specific for the given setting.
We believe that it will also be a major challenge to address the interaction pattern between autophagy and apoptosis, as well as the regulatory signaling network and cooperating factors involved, which could play a pivotal role in the management of CRC. In this perspective, autophagy and apoptosis can be used conveniently in the specific context of pharmacological interventions as new targets to provide a basis for the development of drugs in the field of CRC.
Author Contributions
Conceptualization, P.S., G.O. and L.R.; writing—original draft preparation, writing P.S., G.O. and G.C.; writing—review and editing, P.S. and L.R.; visualization, G.O. and G.C.; supervision, P.S.; funding acquisition, L.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by ARTI (Associazione Ricerca Tumori Intestinali), Modena and Piano di sviluppo dipartimentale (FAR 2022).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created.
Acknowledgments
Some of the figures were created partly using Biorender.com (accessed on 15 April 2023).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Leslie, A.; Carey, F.A.; Pratt, N.R.; Steele, R.J.C. The Colorectal Adenoma–Carcinoma Sequence. Br. J. Surg. 2002, 89, 845–860. [Google Scholar] [CrossRef] [PubMed]
- Lee-Six, H.; Olafsson, S.; Ellis, P.; Osborne, R.J.; Sanders, M.A.; Moore, L.; Georgakopoulos, N.; Torrente, F.; Noorani, A.; Goddard, M.; et al. The Landscape of Somatic Mutation in Normal Colorectal Epithelial Cells. Nature 2019, 574, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lai, Y.; Hua, Z.-C. Apoptosis and Apoptotic Body: Disease Message and Therapeutic Target Potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef] [PubMed]
- Strasser, A.; Vaux, D.L. Cell Death in the Origin and Treatment of Cancer. Mol. Cell 2020, 78, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- D’Arcy, M.S. Cell Death: A Review of the Major Forms of Apoptosis, Necrosis and Autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, N.; Manieri, E.; Storm, E.E.; Saadatpour, A.; Luoma, A.M.; Kapoor, V.N.; Madha, S.; Gaynor, L.T.; Cox, C.; Keerthivasan, S.; et al. Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell 2020, 26, 391–402.e5. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, C.A.; Pritchard, D.M. Suppression of Apoptosis, Crypt Hyperplasia, and Altered Differentiation in the Colonic Epithelia of Bak-Null Mice. Gastroenterology 2009, 136, 943–952. [Google Scholar] [CrossRef]
- Qiao, L.; Wong, B.C.Y. Targeting Apoptosis as an Approach for Gastrointestinal Cancer Therapy. Drug Resist. Updates 2009, 12, 55–64. [Google Scholar] [CrossRef]
- Wu, Y.; Yao, J.; Xie, J.; Liu, Z.; Zhou, Y.; Pan, H.; Han, W. The Role of Autophagy in Colitis-Associated Colorectal Cancer. Signal Transduct. Target. Ther. 2018, 3, 31. [Google Scholar] [CrossRef]
- Finkbeiner, S. The Autophagy Lysosomal Pathway and Neurodegeneration. Cold Spring Harb. Perspect. Biol. 2020, 12, a033993. [Google Scholar] [CrossRef] [PubMed]
- Groulx, J.-F.; Khalfaoui, T.; Benoit, Y.D.; Bernatchez, G.; Carrier, J.C.; Basora, N.; Beaulieu, J.-F. Autophagy Is Active in Normal Colon Mucosa. Autophagy 2012, 8, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Koustas, E.; Sarantis, P.; Kyriakopoulou, G.; Papavassiliou, A.G.; Karamouzis, M.V. The Interplay of Autophagy and Tumor Microenvironment in Colorectal Cancer—Ways of Enhancing Immunotherapy Action. Cancers 2019, 11, 533. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Ceteci, F.; Gupta, J.; Pesic, M.; Böttger, T.W.; Nicolas, A.M.; Kennel, K.B.; Engel, E.; Schewe, M.; Callak Kirisözü, A.; et al. Colon Tumour Cell Death Causes MTOR Dependence by Paracrine P2X4 Stimulation. Nature 2022, 612, 347–353. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational Landscape and Significance across 12 Major Cancer Types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Hasbullah, H.H.; Musa, M. Gene Therapy Targeting P53 and KRAS for Colorectal Cancer Treatment: A Myth or the Way Forward? Int. J. Mol. Sci. 2021, 22, 11941. [Google Scholar] [CrossRef]
- Nakayama, M.; Oshima, M. Mutant P53 in Colon Cancer. J. Mol. Cell Biol. 2019, 11, 267–276. [Google Scholar] [CrossRef]
- Malki, A.; ElRuz, R.A.; Gupta, I.; Allouch, A.; Vranic, S.; Al Moustafa, A.-E. Molecular Mechanisms of Colon Cancer Progression and Metastasis: Recent Insights and Advancements. Int. J. Mol. Sci. 2020, 22, 130. [Google Scholar] [CrossRef]
- Nishiumi, S.; Fujishima, Y.; Inoue, J.; Masuda, A.; Azuma, T.; Yoshida, M. Autophagy in the Intestinal Epithelium Is Not Involved in the Pathogenesis of Intestinal Tumors. Biochem. Biophys. Res. Commun. 2012, 421, 768–772. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Lu, Y.; Zhang, Q.; Qu, X. Heterozygous Deletion of ATG5 in Apc Min/+ Mice Promotes Intestinal Adenoma Growth and Enhances the Antitumor Efficacy of Interferon-Gamma. Cancer Biol. Ther. 2015, 16, 383–391. [Google Scholar] [CrossRef]
- Li, L.; Jing, L.; Wang, J.; Xu, W.; Gong, X.; Zhao, Y.; Ma, Y.; Yao, X.; Sun, X. Autophagic Flux Is Essential for the Downregulation of D-Dopachrome Tautomerase by Atractylenolide I to Ameliorate Intestinal Adenoma Formation. J. Cell Commun. Signal. 2018, 12, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Xiao, Z.; Dekker, F.J.; Poelarends, G.J.; Melgert, B.N. Macrophage Migration Inhibitory Factor Family Proteins Are Multitasking Cytokines in Tissue Injury. Cell. Mol. Life Sci. 2022, 79, 105. [Google Scholar] [CrossRef] [PubMed]
- Lévy, J.; Cacheux, W.; Bara, M.A.; L’Hermitte, A.; Lepage, P.; Fraudeau, M.; Trentesaux, C.; Lemarchand, J.; Durand, A.; Crain, A.-M.; et al. Intestinal Inhibition of Atg7 Prevents Tumour Initiation through a Microbiome-Influenced Immune Response and Suppresses Tumour Growth. Nat. Cell Biol. 2015, 17, 1062–1073. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular Mechanisms of Cell Death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Hu, B.; Yin, Y.; Li, S.; Guo, X. Insights on Ferroptosis and Colorectal Cancer: Progress and Updates. Molecules 2022, 28, 243. [Google Scholar] [CrossRef]
- Dong, Y.; Wu, Y.; Zhao, G.-L.; Ye, Z.-Y.; Xing, C.-G.; Yang, X.-D. Inhibition of Autophagy by 3-MA Promotes Hypoxia-Induced Apoptosis in Human Colorectal Cancer Cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1047–1054. [Google Scholar] [CrossRef]
- Haronikova, L.; Olivares-Illana, V.; Wang, L.; Karakostis, K.; Chen, S.; Fåhraeus, R. The P53 MRNA: An Integral Part of the Cellular Stress Response. Nucleic Acids Res. 2019, 47, 3257–3271. [Google Scholar] [CrossRef]
- Yamada, K.; Yoshida, K. Mechanical Insights into the Regulation of Programmed Cell Death by P53 via Mitochondria. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2019, 1866, 839–848. [Google Scholar] [CrossRef]
- Shajani-Yi, Z.; de Abreu, F.B.; Peterson, J.D.; Tsongalis, G.J. Frequency of Somatic TP53 Mutations in Combination with Known Pathogenic Mutations in Colon Adenocarcinoma, Non–Small Cell Lung Carcinoma, and Gliomas as Identified by Next-Generation Sequencing. Neoplasia 2018, 20, 256–262. [Google Scholar] [CrossRef]
- Sakanashi, F.; Shintani, M.; Tsuneyoshi, M.; Ohsaki, H.; Kamoshida, S. Apoptosis, Necroptosis and Autophagy in Colorectal Cancer: Associations with Tumor Aggressiveness and P53 Status. Pathol. Res. Pract. 2019, 215, 152425. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, C.; Li, J.; Jiang, M.; Guo, S.; Yang, G.; Zhang, L.; Wang, F.; Yi, S.; Wang, J.; et al. Inhibition of AKT Induces P53/SIRT6/PARP1-Dependent Parthanatos to Suppress Tumor Growth. Cell Commun. Signal. 2022, 20, 93. [Google Scholar] [CrossRef] [PubMed]
- Gurkan, A.C.; Arisan, E.D.; Yerlikaya, P.O.; Ilhan, H.; Unsal, N.P. Inhibition of Autophagy Enhances DENSpm-Induced Apoptosis in Human Colon Cancer Cells in a P53 Independent Manner. Cell. Oncol. 2018, 41, 297–317. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.J.; Indra, R.; Wohak, L.E.; Sozeri, O.; Feser, K.; Mrizova, I.; Phillips, D.H.; Stiborova, M.; Arlt, V.M. The Impact of Chemotherapeutic Drugs on the CYP1A1-Catalysed Metabolism of the Environmental Carcinogen Benzo[a]Pyrene: Effects in Human Colorectal HCT116 TP53(+/+), TP53(+/−) and TP53(−/−) Cells. Toxicology 2018, 398–399, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of Selective Autophagy: The P62/SQSTM1 Paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Salazar, G.; Cullen, A.; Huang, J.; Zhao, Y.; Serino, A.; Hilenski, L.; Patrushev, N.; Forouzandeh, F.; Hwang, H.S. SQSTM1/P62 and PPARGC1A/PGC-1alpha at the Interface of Autophagy and Vascular Senescence. Autophagy 2020, 16, 1092–1110. [Google Scholar] [CrossRef]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Kosumi, K.; Masugi, Y.; Yang, J.; Qian, Z.R.; Kim, S.A.; Li, W.; Shi, Y.; da Silva, A.; Hamada, T.; Liu, L.; et al. Tumor SQSTM1 (P62) Expression and T Cells in Colorectal Cancer. OncoImmunology 2017, 6, e1284720. [Google Scholar] [CrossRef]
- Mukherjee, S.; Maddalena, M.; Lü, Y.; Martinez, S.; Nataraj, N.B.; Noronha, A.; Sinha, S.; Teng, K.; Cohen-Kaplan, V.; Ziv, T.; et al. Cross-Talk between Mutant P53 and P62/SQSTM1 Augments Cancer Cell Migration by Promoting the Degradation of Cell Adhesion Proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2119644119. [Google Scholar] [CrossRef]
- Tang, J.; Li, Y.; Xia, S.; Li, J.; Yang, Q.; Ding, K.; Zhang, H. Sequestosome 1/P62: A Multitasker in the Regulation of Malignant Tumor Aggression (Review). Int. J. Oncol. 2021, 59, 77. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, S.; Guan, W.; Huang, Z.; Kong, J.; Huang, C.; Wang, H.; Yang, S. Poly C Binding Protein 1 Regulates P62/SQSTM1 MRNA Stability and Autophagic Degradation to Repress Tumor Progression. Front. Genet. 2020, 11, 930. [Google Scholar] [CrossRef]
- Yan, X.; Zhong, X.; Yu, S.; Zhang, L.; Liu, Y.; Zhang, Y.; Sun, L.; Su, J. P62 Aggregates Mediated Caspase 8 Activation Is Responsible for Progression of Ovarian Cancer. J. Cell. Mol. Med. 2019, 23, 4030–4042. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, S.; Xu, B.; Wang, T.; Zheng, Y.; Liu, F.; Ren, F.; Jiang, J.; Shi, H.; Zou, B.; et al. P62 Functions as an Oncogene in Colorectal Cancer through Inhibiting Apoptosis and Promoting Cell Proliferation by Interacting with the Vitamin D Receptor. Cell Prolif. 2019, 52, e12585. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, Q.; Li, B.; Xie, J.; Yang, X.; Zhao, K.; Wu, Y.; Ye, Z.; Chen, Z.; Qin, Z.; et al. Escin-Induced DNA Damage Promotes Escin-Induced Apoptosis in Human Colorectal Cancer Cells via P62 Regulation of the ATM/ΓH2AX Pathway. Acta Pharm. Sin. 2018, 39, 1645–1660. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, Z.; Xie, J.; Wang, G.; Qian, L.; Guan, X.; Shen, X.; Qin, Z.; Shen, G.; Li, X.; et al. TIGAR Knockdown Enhanced the Anticancer Effect of Aescin via Regulating Autophagy and Apoptosis in Colorectal Cancer Cells. Acta Pharm. Sin. 2019, 40, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Plewka, D.; Plewka, A.; Miskiewicz, A.; Morek, M.; Bogunia, E. Nuclear Factor-Kappa B as Potential Therapeutic Target in Human Colon Cancer. J. Can. Res. Ther. 2018, 14, 516. [Google Scholar] [CrossRef]
- Soleimani, A.; Rahmani, F.; Ferns, G.A.; Ryzhikov, M.; Avan, A.; Hassanian, S.M. Role of the NF-ΚB Signaling Pathway in the Pathogenesis of Colorectal Cancer. Gene 2020, 726, 144132. [Google Scholar] [CrossRef]
- Xin, J. Critical Signaling Pathways Governing Colitis-Associated Colorectal Cancer: Signaling, Therapeutic Implications, and Challenges. Dig. Liver Dis. 2023, 55, 169–177. [Google Scholar] [CrossRef]
- Sena, P.; Mancini, S.; Benincasa, M.; Mariani, F.; Palumbo, C.; Roncucci, L. Metformin Induces Apoptosis and Alters Cellular Responses to Oxidative Stress in Ht29 Colon Cancer Cells: Preliminary Findings. Int. J. Mol. Sci. 2018, 19, 1478. [Google Scholar] [CrossRef]
- Akter, M.; Atique Ullah, A.K.M.; Banik, S.; Sikder, M.T.; Hosokawa, T.; Saito, T.; Kurasaki, M. Green Synthesized Silver Nanoparticles-Mediated Cytotoxic Effect in Colorectal Cancer Cells: NF-ΚB Signal Induced Apoptosis Through Autophagy. Biol. Trace Elem. Res. 2021, 199, 3272–3286. [Google Scholar] [CrossRef]
- Zhang, P.; Pan, Y.; Sun, J.; Pan, G. Aberrant Expression of LncRNA CASC2 Mediated the Cell Viability, Apoptosis and Autophagy of Colon Cancer Cells by Sponging MiR-19a via NF-κB Signaling Pathway. Int. J. Exp. Path 2021, 102, 163–171. [Google Scholar] [CrossRef]
- Li, T.; Chen, L.; Zhao, H.; Wu, L.; Masters, J.; Han, C.; Hirota, K.; Ma, D. Both Bupivacaine and Levobupivacaine Inhibit Colon Cancer Cell Growth but Not Melanoma Cells in Vitro. J. Anesth. 2019, 33, 17–25. [Google Scholar] [CrossRef]
- Liu, B.; Yan, X.; Hou, Z.; Zhang, L.; Zhang, D. Impact of Bupivacaine on Malignant Proliferation, Apoptosis and Autophagy of Human Colorectal Cancer SW480 Cells through Regulating NF-ΚB Signaling Path. Bioengineered 2021, 12, 2723–2733. [Google Scholar] [CrossRef]
- Vaupel, P.; Multhoff, G. Revisiting the Warburg Effect: Historical Dogma versus Current Understanding. J. Physiol. 2021, 599, 1745–1757. [Google Scholar] [CrossRef]
- Keller, F.; Bruch, R.; Schneider, R.; Meier-Hubberten, J.; Hafner, M.; Rudolf, R. A Scaffold-Free 3-D Co-Culture Mimics the Major Features of the Reverse Warburg Effect In Vitro. Cells 2020, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Kasprzak, A. Insulin-Like Growth Factor 1 (IGF-1) Signaling in Glucose Metabolism in Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 6434. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, Mitochondria and Oxidative Stress: Cross-Talk and Redox Signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Yepes, J.; Burns, M.; Anandhan, A.; Khalimonchuk, O.; del Razo, L.M.; Quintanilla-Vega, B.; Pappa, A.; Panayiotidis, M.I.; Franco, R. Oxidative Stress, Redox Signaling, and Autophagy: Cell Death Versus Survival. Antioxid. Redox Signal. 2014, 21, 66–85. [Google Scholar] [CrossRef]
- Yu, G.; Luo, H.; Zhang, N.; Wang, Y.; Li, Y.; Huang, H.; Liu, Y.; Hu, Y.; Liu, H.; Zhang, J.; et al. Loss of P53 Sensitizes Cells to Palmitic Acid-Induced Apoptosis by Reactive Oxygen Species Accumulation. Int. J. Mol. Sci. 2019, 20, 6268. [Google Scholar] [CrossRef]
- da Silva, E.L.; Mesquita, F.P.; Ramos, I.N.d.F.; Gomes, C.B.d.S.M.R.; Moreira, C.d.S.; Ferreira, V.F.; da Rocha, D.R.; Bahia, M.d.O.; Moreira-Nunes, C.A.; de Souza, C.R.T.; et al. Antitumoral Effect of Novel Synthetic 8-Hydroxy-2-((4-Nitrophenyl)Thio)Naphthalene-1,4-Dione (CNN16) via ROS-Mediated DNA Damage, Apoptosis and Anti-Migratory Effect in Colon Cancer Cell Line. Toxicol. Appl. Pharmacol. 2022, 456, 116256. [Google Scholar] [CrossRef]
- Gao, L.; Loveless, J.; Shay, C.; Teng, Y. Targeting ROS-Mediated Crosstalk Between Autophagy and Apoptosis in Cancer. In Reviews on New Drug Targets in Age-Related Disorders; Guest, P.C., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2020; Volume 1260, pp. 1–12. ISBN 978-3-030-42666-8. [Google Scholar]
- Lee, S.-J.; Lee, D.-E.; Choi, S.-Y.; Kwon, O.-S. OSMI-1 Enhances TRAIL-Induced Apoptosis through ER Stress and NF-ΚB Signaling in Colon Cancer Cells. Int. J. Mol. Sci. 2021, 22, 11073. [Google Scholar] [CrossRef]
- Shi, W.; Men, L.; Pi, X.; Jiang, T.; Peng, D.; Huo, S.; Luo, P.; Wang, M.; Guo, J.; Jiang, Y.; et al. Shikonin Suppresses Colon Cancer Cell Growth and Exerts Synergistic Effects by Regulating ADAM17 and the IL-6/STAT3 Signaling Pathway. Int. J. Oncol. 2021, 59, 99. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Ning, X.; Ye, M.; Yin, Y. Antrodia Camphorata Extract (ACE)-Induced Apoptosis Is Associated with BMP4 Expression and P53-Dependent ROS Generation in Human Colon Cancer Cells. J. Ethnopharmacol. 2021, 268, 113570. [Google Scholar] [CrossRef]
- Hong, H.; Cao, W.; Wang, Q.; Liu, C.; Huang, C. Synergistic Antitumor Effect of Andrographolide and Cisplatin through ROS-Mediated ER Stress and STAT3 Inhibition in Colon Cancer. Med. Oncol. 2022, 39, 101. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lee, J.; Ha, S.H.; Lee, H.K.; Lim, H.M.; Yu, S.; Lee, C.M.; Nam, M.J.; Yang, Y.; Park, K.; et al. 6, 8-Diprenylorobol Induces Apoptosis in Human Colon Cancer Cells via Activation of Intracellular Reactive Oxygen Species and P53. Environ. Toxicol. 2021, 36, 914–925. [Google Scholar] [CrossRef]
- Wang, G.-Y.; Zhang, L.; Geng, Y.-D.; Wang, B.; Feng, X.-J.; Chen, Z.-L.; Wei, W.; Jiang, L. β-Elemene Induces Apoptosis and Autophagy in Colorectal Cancer Cells through Regulating the ROS/AMPK/MTOR Pathway. Chin. J. Nat. Med. 2022, 20, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhou, X.; Wang, J.; Zhou, Y.; Qi, W.; Chen, H.; Nie, S.; Xie, M. Dendrobium Officinale Polysaccharide Triggers Mitochondrial Disorder to Induce Colon Cancer Cell Death via ROS-AMPK-Autophagy Pathway. Carbohydr. Polym. 2021, 264, 118018. [Google Scholar] [CrossRef]
- Luo, Q.; Jia, L.; Huang, C.; Qi, Q.; Jahangir, A.; Xia, Y.; Liu, W.; Shi, R.; Tang, L.; Chen, Z. Polyphyllin I Promotes Autophagic Cell Death and Apoptosis of Colon Cancer Cells via the ROS-Inhibited AKT/MTOR Pathway. Int. J. Mol. Sci. 2022, 23, 9368. [Google Scholar] [CrossRef]
- Yang, H.-L.; Liu, H.-W.; Shrestha, S.; Thiyagarajan, V.; Huang, H.-C.; Hseu, Y.-C. Antrodia Salmonea Induces Apoptosis and Enhances Cytoprotective Autophagy in Colon Cancer Cells. Aging 2021, 13, 15964–15989. [Google Scholar] [CrossRef]
- Carlisle, A.E.; Lee, N.; Matthew-Onabanjo, A.N.; Spears, M.E.; Park, S.J.; Youkana, D.; Doshi, M.B.; Peppers, A.; Li, R.; Joseph, A.B.; et al. Selenium Detoxification Is Required for Cancer-Cell Survival. Nat. Metab. 2020, 2, 603–611. [Google Scholar] [CrossRef]
- Yu, H.; Huang, Y.; Ge, Y.; Hong, X.; Lin, X.; Tang, K.; Wang, Q.; Yang, Y.; Sun, W.; Huang, Y.; et al. Selenite-Induced ROS/AMPK/FoxO3a/GABARAPL-1 Signaling Pathway Modulates Autophagy That Antagonize Apoptosis in Colorectal Cancer Cells. Discov. Oncol. 2021, 12, 35. [Google Scholar] [CrossRef]
- Zhang, N.; Hu, X.; Du, Y.; Du, J. The Role of MiRNAs in Colorectal Cancer Progression and Chemoradiotherapy. Biomed. Pharmacother. 2021, 134, 111099. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, W.; Xiao, Y.; Shan, L. MiR-125b Inhibits Cell Proliferation and Induces Apoptosis in HumanColon Cancer SW480 Cells via Targeting STAT3. Recent Pat. Anti-Cancer Drug Discov. 2022, 17, 187–194. [Google Scholar] [CrossRef]
- Gao, J.; Fei, L.; Wu, X.; Li, H. MiR-766-3p Suppresses Malignant Behaviors and Stimulates Apoptosis of Colon Cancer Cells via Targeting TGFBI. Can. J. Gastroenterol. Hepatol. 2022, 2022, 7234704. [Google Scholar] [CrossRef]
- Chen, S.; Wang, Y.; Xu, M.; Zhang, L.; Su, Y.; Wang, B.; Zhang, X. MiR-1184 Regulates the Proliferation and Apoptosis of Colon Cancer Cells via Targeting CSNK2A1. Mol. Cell. Probes 2020, 53, 101625. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Li, T.; Ye, C.; Zeng, L.; Li, H.; Pu, X.; Ding, C.; He, Z.; Huang, G. MiR-221 Inhibits Autophagy and Targets TP53INP1 in Colorectal Cancer Cells. Exp. Ther. Med. 2017, 15, 1712–1717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, J.; Zhao, J.; Bai, J. Mir-30d Suppresses Cell Proliferation of Colon Cancer Cells by Inhibiting Cell Autophagy and Promoting Cell Apoptosis. Tumour Biol. 2017, 39, 101042831770398. [Google Scholar] [CrossRef]
- Wei, L.; Chen, Z.; Cheng, N.; Li, X.; Chen, J.; Wu, D.; Dong, M.; Wu, X. MicroRNA-126 Inhibit Viability of Colorectal Cancer Cell by Repressing MTOR Induced Apoptosis and Autophagy. OncoTargets Ther. 2020, 13, 2459–2468. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Cheng, C.; Xu, J.; Gao, P.; Wang, J.; Chen, L. MiR-142-3p Regulates Tumor Cell Autophagy and Promotes Colon Cancer Progression by Targeting TP53INP2. Chemotherapy 2022, 67, 57–66. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, F.; Wang, Y.; Zhao, Z.; Qiao, P. LncRNA GAS5 Inhibits Malignant Progression by Regulating Macroautophagy and Forms a Negative Feedback Regulatory Loop with the MiR-34a/MTOR/SIRT1 Pathway in Colorectal Cancer. Oncol. Rep. 2020, 45, 202–216. [Google Scholar] [CrossRef]
- Yang, X.; Xie, Z.; Lei, X.; Gan, R. Long Non-coding RNA GAS5 in Human Cancer (Review). Oncol. Lett. 2020, 20, 2587–2594. [Google Scholar] [CrossRef]
- Qiao, P.; Yao, L.; Zeng, Z. Catalpol-mediated MicroRNA-34a Suppresses Autophagy and Malignancy by Regulating SIRT1 in Colorectal Cancer. Oncol. Rep. 2020, 43, 1053–1066. [Google Scholar] [CrossRef]
- Zhou, W.; Yang, W.; Yang, J.; Zhu, H.; Duan, L.; Wang, X.; Li, Y.; Niu, L.; Xiao, S.; Zhang, R.; et al. MiR-483 Promotes the Development of Colorectal Cancer by Inhibiting the Expression Level of EI24. Mol. Med. Rep. 2021, 24, 567. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Chen, X.; Liu, J.; Gu, H.; Fan, R.; Ge, H. Long Non-Coding RNA HOTAIR Knockdown Enhances Radiosensitivity through Regulating MicroRNA-93/ATG12 Axis in Colorectal Cancer. Cell Death Dis. 2020, 11, 175. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Kong, S.; Wang, F.; Ju, S. CircRNAs: Biogenesis, Functions, and Role in Drug-Resistant Tumours. Mol. Cancer 2020, 19, 119. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. CircHIPK3 Promotes Colorectal Cancer Growth and Metastasis by Sponging MiR-7. Cell Death Dis. 2018, 9, 417. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, J.; Zhao, J.; Wang, X. Silencing of Hsa_circ_0007534 Suppresses Proliferation and Induces Apoptosis in Colorectal Cancer Cells. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 118–126. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, L.; Wang, F. Hsa_circ_0020397 Regulates Colorectal Cancer Cell Viability, Apoptosis and Invasion by Promoting the Expression of the MiR-138 Targets TERT and PD-L1: Circ_0020397 Regulates Growth in Colorectal Cancer Cells by Sequestering MiR-138. Cell Biol. Int. 2017, 41, 1056–1064. [Google Scholar] [CrossRef]
- Jin, Y.; Yu, L.L.; Zhang, B.; Liu, C.F.; Chen, Y. Circular RNA Hsa_circ_0000523 Regulates the Proliferation and Apoptosis of Colorectal Cancer Cells as MiRNA Sponge. Braz. J. Med. Biol. Res. 2018, 51, e7811. [Google Scholar] [CrossRef]
- Chen, F.; Guo, L.; Di, J.; Li, M.; Dong, D.; Pei, D. Circular RNA Ubiquitin-Associated Protein 2 Enhances Autophagy and Promotes Colorectal Cancer Progression and Metastasis via MiR-582-5p/FOXO1 Signaling. J. Genet. Genom. 2021, 48, 1091–1103. [Google Scholar] [CrossRef]
- Feng, J.; Li, Z.; Li, L.; Xie, H.; Lu, Q.; He, X. Hypoxia-induced CircCCDC66 Promotes the Tumorigenesis of Colorectal Cancer via the MiR-3140/Autophagy Pathway. Int. J. Mol. Med. 2020, 46, 1973–1982. [Google Scholar] [CrossRef]
- Pan, Z.; Zheng, J.; Zhang, J.; Lin, J.; Lai, J.; Lyu, Z.; Feng, H.; Wang, J.; Wu, D.; Li, Y. A Novel Protein Encoded by Exosomal CircATG4B Induces Oxaliplatin Resistance in Colorectal Cancer by Promoting Autophagy. Adv. Sci. 2022, 9, 2204513. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt Signaling in Colorectal Cancer: Pathogenic Role and Therapeutic Target. Mol. Cancer 2022, 21, 144. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-Catenin Signalling: Function, Biological Mechanisms, and Therapeutic Opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Petherick, K.J.; Williams, A.C.; Lane, J.D.; Ordóñez-Morán, P.; Huelsken, J.; Collard, T.J.; Smartt, H.J.; Batson, J.; Malik, K.; Paraskeva, C.; et al. Autolysosomal β-Catenin Degradation Regulates Wnt-Autophagy-P62 Crosstalk. EMBO J. 2013, 32, 1903–1916. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Cao, W.; Bao, L.; Zuo, W.; Xie, G.; Cai, T.; Fu, W.; Zhang, J.; Wu, W.; Zhang, X.; et al. Autophagy Negatively Regulates Wnt Signalling by Promoting Dishevelled Degradation. Nat. Cell. Biol. 2010, 12, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Tao, F.; Zhang, X.; Zhang, Y.; Sun, X.; Wu, D. Role of Wnt/ β -Catenin Signaling in the Chemoresistance Modulation of Colorectal Cancer. BioMed Res. Int. 2020, 2020, 9390878. [Google Scholar] [CrossRef] [PubMed]
- Foerster, E.G.; Mukherjee, T.; Cabral-Fernandes, L.; Rocha, J.D.B.; Girardin, S.E.; Philpott, D.J. How Autophagy Controls the Intestinal Epithelial Barrier. Autophagy 2022, 18, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, C.; Sergio, S.; Coluccia, A.; De Luca, M.; La Regina, G.; Mologni, L.; Famiglini, V.; Naccarato, V.; Bonetti, D.; Gautier, C.; et al. β-Catenin Knockdown Promotes NHERF1-Mediated Survival of Colorectal Cancer Cells: Implications for a Double-Targeted Therapy. Oncogene 2018, 37, 3301–3316. [Google Scholar] [CrossRef]
- Behrouj, H.; Seghatoleslam, A.; Mokarram, P.; Ghavami, S. Effect of Casein Kinase 1α Inhibition on Autophagy Flux and the AKT/Phospho-β-Catenin (S552) Axis in HCT116, a RAS-Mutated Colorectal Cancer Cell Line. Can. J. Physiol. Pharmacol. 2021, 99, 284–293. [Google Scholar] [CrossRef]
- Murata, K.; Jadhav, U.; Madha, S.; Van Es, J.; Dean, J.; Cavazza, A.; Wucherpfennig, K.; Michor, F.; Clevers, H.; Shivdasani, R.A. Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells. Cell Stem Cell 2020, 26, 377–390.e6. [Google Scholar] [CrossRef]
- Wang, H.; Ye, T.; Cai, Y.; Chen, W.; Xie, H.; Ke, C. Downregulation of Ascl2 Promotes Cell Apoptosis by Enhancing Autophagy in Colorectal Cancer Cells. J. Gastrointest. Oncol. 2021, 12, 630–638. [Google Scholar] [CrossRef]
- Sikder, M.O.F.; Sivaprakasam, S.; Brown, T.P.; Thangaraju, M.; Bhutia, Y.D.; Ganapathy, V. SLC6A14, a Na+/Cl−-Coupled Amino Acid Transporter, Functions as a Tumor Promoter in Colon and Is a Target for Wnt Signaling. Biochem. J. 2020, 477, 1409–1425. [Google Scholar] [CrossRef]
- Wu, D.-D.; Liu, S.-Y.; Gao, Y.-R.; Lu, D.; Hong, Y.; Chen, Y.-G.; Dong, P.-Z.; Wang, D.-Y.; Li, T.; Li, H.-M.; et al. Tumour Necrosis Factor-α-Induced Protein 8-like 2 Is a Novel Regulator of Proliferation, Migration, and Invasion in Human Rectal Adenocarcinoma Cells. J. Cell. Mol. Med. 2019, 23, 1698–1713. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Gopalan, V.; Wahab, R.; Smith, R.A.; Qiao, B.; Lam, A.K.-Y. Stage Dependent Expression and Tumor Suppressive Function of FAM134B (JK1) in Colon Cancer: TUMOR SUPPRESSOR FUNCTION OF FAM134B. Mol. Carcinog. 2017, 56, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Chaousis, S.; Wahab, R.; Gopalan, V.; Lam, A.K.-Y. Protein Interactions of FAM134B with EB1 and APC/Beta-catenin in Vitro in Colon Carcinoma. Mol. Carcinog. 2018, 57, 1480–1491. [Google Scholar] [CrossRef] [PubMed]
- Sana-Eldine, A.O.; Abdelgawad, H.M.; Kotb, N.S.; Shehata, N.I. The Potential Effect of Schisandrin-B Combination with Panitumumab in Wild-type and Mutant Colorectal Cancer Cell Lines: Role of Apoptosis and Autophagy. J. Biochem. Mol. Toxicol. 2023, 37, e23324. [Google Scholar] [CrossRef]
- Qureshi-Baig, K.; Kuhn, D.; Viry, E.; Pozdeev, V.I.; Schmitz, M.; Rodriguez, F.; Ullmann, P.; Koncina, E.; Nurmik, M.; Frasquilho, S.; et al. Hypoxia-Induced Autophagy Drives Colorectal Cancer Initiation and Progression by Activating the PRKC/PKC-EZR (Ezrin) Pathway. Autophagy 2020, 16, 1436–1452. [Google Scholar] [CrossRef]
- Xu, H.-D.; Qin, Z.-H. Beclin 1, Bcl-2 and Autophagy. In Autophagy: Biology and Diseases; Qin, Z.-H., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2019; Volume 1206, pp. 109–126. ISBN 9789811506017. [Google Scholar]
- Xu, W.; Nie, C.; Chen, X. DUSP4 Inhibits Autophagic Cell Death and Apoptosis in Colorectal Cancer by Regulating BCL2-Beclin1/Bax Signaling. Mol. Biol. Rep. 2023, 50, 3229–3239. [Google Scholar] [CrossRef]
- Tang, H.; Wu, D.; Yang, H.; Yang, J.; Zhang, Y.; Li, M.; Liu, H.; Li, Q. Inhibition of autophagy enhances apoptosis induced by doxorubicin hydrochloride in human colon cancer cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2022, 38, 237–243. [Google Scholar]
- Athamneh, K.; Alneyadi, A.; Alsamri, H.; Alrashedi, A.; Palakott, A.; El-Tarabily, K.A.; Eid, A.H.; Al Dhaheri, Y.; Iratni, R. Origanum Majorana Essential Oil Triggers P38 MAPK-Mediated Protective Autophagy, Apoptosis, and Caspase-Dependent Cleavage of P70S6K in Colorectal Cancer Cells. Biomolecules 2020, 10, 412. [Google Scholar] [CrossRef]
- Athamneh, K.; Hasasna, H.E.; Samri, H.A.; Attoub, S.; Arafat, K.; Benhalilou, N.; Rashedi, A.A.; Dhaheri, Y.A.; AbuQamar, S.; Eid, A.; et al. Rhus Coriaria Increases Protein Ubiquitination, Proteasomal Degradation and Triggers Non-Canonical Beclin-1-Independent Autophagy and Apoptotic Cell Death in Colon Cancer Cells. Sci. Rep. 2017, 7, 11633. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).