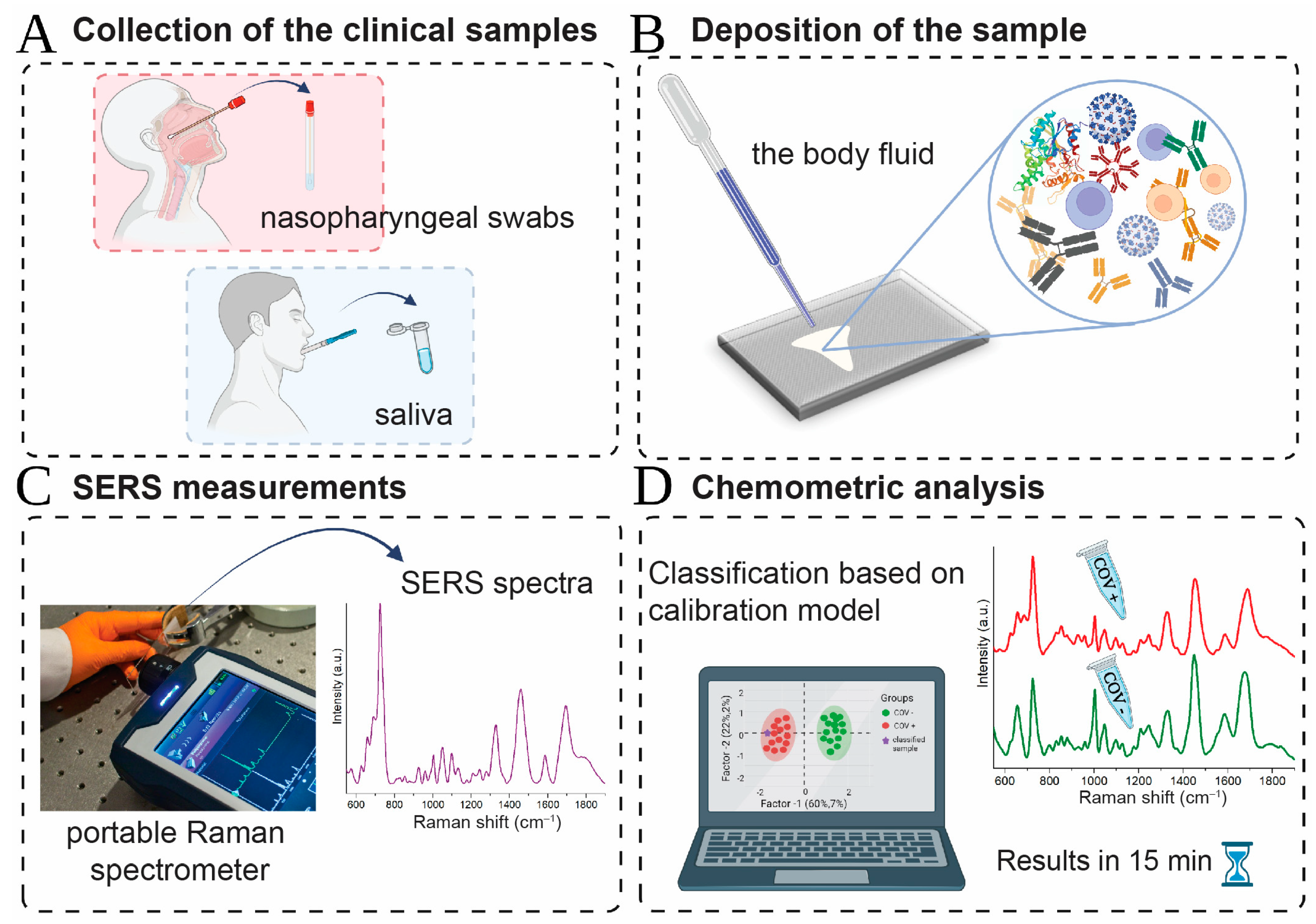
SERS Signature of SARS-CoV-2 in Saliva and Nasopharyngeal Swabs: Towards Perspective COVID-19 Point-of-Care Diagnostics
Abstract
1. Introduction
2. Results and Discussion
2.1. Salivary SERS Fingerprint of COVID-19
2.1.1. The Classification and Prediction Methods for Diagnosis COVID-19 in Saliva Samples
2.1.2. Prediction by Means of PLS-DA Analysis
2.1.3. Classification Results of PCA-LDA and SVMC Analysis
2.2. Nasopharyngeal SERS Fingerprint of COVID-19
2.2.1. Prediction by Means of PLS-DA Analysis
2.2.2. Classification Results of PCA-LDA and SVMC Analysis
3. Materials and Methods
3.1. Viral RNA Extraction
3.2. SARS-CoV-2 RNA Detection Using Quantitative Reverse Transcriptase Real-Time Polymerase Chain Reaction (qRT-PCR)
3.3. SERS Platform Preparation
3.4. SERS Measurements
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 25 February 2022).
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial Undocumented Infection Facilitates the Rapid Dissemination of Novel Coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.J.; Lu, X.X.; Deng, Y.; Tang, Y.J.; Lu, J.C. COVID-19: Asymptomatic Carrier Transmission Is an Underestimated Problem. Epidemiol. Infect. 2020, 148, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Day, M. Covid-19: Four Fifths of Cases Are Asymptomatic, China Figures Indicate. BMJ 2020, 369, m1375. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, Y.-R.; Luo, F.-M.; Tu, W.-W.; Zhan, D.-C.; Yu, G.; Zhou, Z.-H. COVID-19 Asymptomatic Infection Estimation. medRxiv 2020. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, Y.; Sun, C.; Wang, X.; Guo, Y.; Qiu, S.; Ma, K. A Systematic Review of Asymptomatic Infections with COVID-19. J. Microbiol. Immunol. Infect. 2021, 54, 12–16. [Google Scholar] [CrossRef]
- Pathan, R.K.; Biswas, M.; Khandaker, M.U. Time Series Prediction of COVID-19 by Mutation Rate Analysis Using Recurrent Neural Network-Based LSTM Model. Chaos Solitons Fractals 2020, 138, 110018. [Google Scholar] [CrossRef]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 Mutation Hot Spots Include a Novel RNA-Dependent-RNA Polymerase Variant. J. Transl. Med. 2020, 18, 179. [Google Scholar] [CrossRef]
- Chand, G.B.; Banerjee, A.; Azad, G.K. Identification of Novel Mutations in RNA-Dependent RNA Polymerases of SARS-CoV-2 and Their Implications on Its Protein Structure. PeerJ 2020, 8, e9492. [Google Scholar] [CrossRef]
- Zahradník, J.; Marciano, S.; Shemesh, M.; Zoler, E.; Chiaravalli, J.; Meyer, B.; Dym, O.; Elad, N.; Schreiber, G. SARS-CoV-2 RBD in Vitro Evolution Follows Contagious Mutation Spread, yet Generates an Able Infection Inhibitor. bioRxiv 2021. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Q.; Wei, P.; Chen, Z.; Aviszus, K.; Yang, J.; Downing, W.; Jiang, C.; Liang, B.; Reynoso, L.; et al. The Basis of a More Contagious 501Y.V1 Variant of SARS-CoV-2. Cell Res. 2021, 31, 720–722. [Google Scholar] [CrossRef]
- Priesemann, V.; Balling, R.; Brinkmann, M.M.; Ciesek, S.; Czypionka, T.; Eckerle, I.; Giordano, G.; Hanson, C.; Hel, Z.; Hotulainen, P.; et al. An Action Plan for Pan-European Defence against New SARS-CoV-2 Variants. Lancet 2021, 397, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.R.; Townsend, J.P.; Pandey, A.; Moghadas, S.M.; Krieger, G.; Singer, B.; McDonald, R.H.; Fitzpatrick, M.C.; Galvani, A.P. Optimal COVID-19 Quarantine and Testing Strategies. Nat. Commun. 2021, 12, 356. [Google Scholar] [CrossRef] [PubMed]
- Grassly, N.C.; Pons-Salort, M.; Parker, E.P.K.; White, P.J.; Ferguson, N.M.; Ainslie, K.; Baguelin, M.; Bhatt, S.; Boonyasiri, A.; Brazeau, N.; et al. Comparison of Molecular Testing Strategies for COVID-19 Control: A Mathematical Modelling Study. Lancet Infect. Dis. 2020, 20, 1381–1389. [Google Scholar] [CrossRef]
- Mina, B.M.J.; Andersen, K.G. COVID-19 Testing: One Size Does Not Fit All. Science 2021, 37, 126–128. [Google Scholar] [CrossRef]
- Kamel Boulos, M.N.; Geraghty, E.M. Geographical Tracking and Mapping of Coronavirus Disease COVID-19/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Epidemic and Associated Events around the World: How 21st Century GIS Technologies Are Supporting the Global Fight against Outbr. Int. J. Health Geogr. 2020, 19, 8. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.E.; Caliendo, A.M.; Arias, C.A.; Hayden, M.K.; Englund, J.A.; Lee, M.J.; Loeb, M.; Patel, R.; El Alayli, A.; Altayar, O.; et al. The Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19: Molecular Diagnostic Testing. Clin. Infect. Dis. 2021, ciab557. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A. Molecular Diagnostic Technologies for COVID-19: Limitations and Challenges. J. Adv. Res. 2020, 26, 149–159. [Google Scholar] [CrossRef]
- Touma, M. COVID-19: Molecular Diagnostics Overview. J. Mol. Med. 2020, 98, 947–954. [Google Scholar] [CrossRef]
- Weissleder, R.; Lee, H.; Ko, J.; Pittet, M.J. COVID-19 Diagnostics in Context. Sci. Transl. Med. 2020, 12, eabc1931. [Google Scholar] [CrossRef]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic Accuracy of Serological Tests for Covid-19: Systematic Review and Meta-Analysis. BMJ 2020, 370, m2516. [Google Scholar] [CrossRef]
- Espejo, A.P.; Akgun, Y.; Al Mana, A.F.; Tjendra, Y.; Millan, N.C.; Gomez-Fernandez, C.; Cray, C. Review of Current Advances in Serologic Testing for COVID-19. Am. J. Clin. Pathol. 2020, 154, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Simon, V. Serology Assays to Manage COVID-19. Science 2020, 368, 1060–1061. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, L.; Deng, Q.; Zhang, G.; Wu, K.; Ni, L.; Yang, Y.; Liu, B.; Wang, W.; Wei, C.; et al. The Presence of SARS-CoV-2 RNA in the Feces of COVID-19 Patients. J. Med. Virol. 2020, 92, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Ardebili, A. Real-Time RT-PCR in COVID-19 Detection: Issues Affecting the Results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Xu, D.; Ye, G.; Xia, C.; Wang, S.; Li, Y.; Xu, H. Positive RT-PCR Test Results in Patients Recovered from COVID-19. JAMA—J. Am. Med. Assoc. 2020, 323, 1502–1503. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.T.; Tong, Y.X.; Zhang, S. False Negative of RT-PCR and Prolonged Nucleic Acid Conversion in COVID-19: Rather than Recurrence. J. Med. Virol. 2020, 92, 1755–1756. [Google Scholar] [CrossRef]
- van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.E.M.; Meijer, A. Comparison of Seven Commercial RT-PCR Diagnostic Kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Zheng, J.; He, L. Surface-Enhanced Raman Spectroscopy for the Chemical Analysis of Food. Compr. Rev. Food Sci. Food Saf. 2014, 13, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Schlücker, S. Surface-Enhanced Raman Spectroscopy: Concepts and Chemical Applications. Angew. Chemie—Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef] [PubMed]
- Moskovits, M. Surface-Enhanced Spectroscopy. Rev. Mod. Phys. 1985, 57, 783. [Google Scholar] [CrossRef]
- Otto, A. Surface-Enhanced Raman Scattering of Adsorbates. J. Raman Spectrosc. 1991, 22, 743–752. [Google Scholar] [CrossRef]
- Kamińska, A.; Witkowska, E.; Winkler, K.; Dzięcielewski, I.; Weyher, J.L.; Waluk, J. Detection of Hepatitis B Virus Antigen from Human Blood: SERS Immunoassay in a Microfluidic System. Biosens. Bioelectron. 2015, 66, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Chrimes, A.F.; Khoshmanesh, K.; Tang, S.-Y.; Wood, B.R.; Stoddart, P.R.; Collins, S.S.E.; Mitchell, A.; Kalantar-zadeh, K. In Situ SERS Probing of Nano-Silver Coated Individual Yeast Cells. Biosens. Bioelectron. 2013, 49, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Premasiri, W.R.; Lee, J.C.; Sauer-Budge, A.; Théberge, R.; Costello, C.E.; Ziegler, L.D. The Biochemical Origins of the Surface-Enhanced Raman Spectra of Bacteria: A Metabolomics Profiling by SERS. Anal. Bioanal. Chem. 2016, 408, 4631–4647. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, X.; Ma, R.; Deng, S.; Wang, X.; Wang, X.; Zhang, X.; Huang, X.; Liu, Y.; Li, G.; et al. Ultra-Fast and Onsite Interrogation of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Waters via Surface Enhanced Raman Scattering (SERS). Water Res. 2021, 200, 117243. [Google Scholar] [CrossRef]
- Lee, S.; Chon, H.; Lee, J.; Ko, J.; Chung, B.H.; Lim, D.W.; Choo, J. Rapid and Sensitive Phenotypic Marker Detection on Breast Cancer Cells Using Surface-Enhanced Raman Scattering (SERS) Imaging. Biosens. Bioelectron. 2014, 51, 238–243. [Google Scholar] [CrossRef]
- Kadam, U.S.; Schulz, B.; Lrudayaraj, J. Detection and Quantification of Alternative Splice Sites in Arabidopsis Genes AtDCL2 and AtPTB2 with Highly Sensitive Surface Enhanced Raman Spectroscopy (SERS) and Gold Nanoprobes. FEBS Lett. 2014, 588, 1637–1643. [Google Scholar] [CrossRef]
- Kadam, U.S.; Schulz, B.; Irudayaraj, J.M.K. Multiplex Single-Cell Quantification of Rare RNA Transcripts from Protoplasts in a Model Plant System. Plant J. 2017, 90, 1187–1195. [Google Scholar] [CrossRef]
- Kadam, U.; Moeller, C.A.; Irudayaraj, J.; Schulz, B. Effect of T-DNA Insertions on MRNA Transcript Copy Numbers Upstream and Downstream of the Insertion Site in Arabidopsis Thaliana Explored by Surface Enhanced Raman Spectroscopy. Plant Biotechnol. J. 2014, 12, 568–577. [Google Scholar] [CrossRef]
- Carlomagno, C.; Bertazioli, D.; Gualerzi, A.; Picciolini, S.; Banfi, P.I.; Lax, A.; Messina, E.; Navarro, J.; Bianchi, L.; Caronni, A.; et al. COVID-19 Salivary Raman Fingerprint: Innovative Approach for the Detection of Current and Past SARS-CoV-2 Infections. Sci. Rep. 2021, 11, 4943. [Google Scholar] [CrossRef]
- Ember, K.J.; Daoust, F.; Mahfoud, M.; Dallaire, F.; Zamani, E.Z.; Tran, T.; Plante, A.; Diop, M.-K.; Nguyen, T.; St-Georges-Robillard, A.; et al. Saliva-Based Detection of COVID-19 Infection in a Real-World Setting Using Reagent-Free Raman Spectroscopy and Machine Learning. J. Biomed. Opt. 2022, 27, 025002. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, V.; Joseph, M.M.; Yadev, I.; Sharma, H.; Shamna, K.; Saurav, S.; Sreejith, R.P.; Anand, V.; Beegum, R.; Regi David, S.; et al. A Non-Invasive Ultrasensitive Diagnostic Approach for COVID-19 Infection Using Salivary Label-Free SERS Fingerprinting and Artificial Intelligence. J. Photochem. Photobiol. B Biol. 2022, 234, 112545. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, H.; Jones, L.; Murray, J.; Haverstick, J.; Naikare, H.K.; Mosley, Y.Y.C.; Tripp, R.A.; Ai, B.; Zhao, Y. Rapid Detection of SARS-CoV-2 RNA in Human Nasopharyngeal Specimens Using Surface-Enhanced Raman Spectroscopy and Deep Learning Algorithms. ACS Sens. 2023, 8, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R.; Kochan, K.; Bedolla, D.E.; Salazar-Quiroz, N.; Grimley, S.L.; Perez-Guaita, D.; Baker, M.J.; Vongsvivut, J.; Tobin, M.J.; Bambery, K.R.; et al. Infrared Based Saliva Screening Test for COVID-19. Angew. Chemie 2021, 133, 17239–17244. [Google Scholar] [CrossRef]
- Sharma, C.P.; Sharma, S.; Sharma, V.; Singh, R. Rapid and Non-Destructive Identification of Claws Using ATR-FTIR Spectroscopy–A Novel Approach in Wildlife Forensics. Sci. Justice 2019, 59, 622–629. [Google Scholar] [CrossRef]
- Bordoloi, D.J.; Tiwari, R. Optimum Multi-Fault Classification of Gears with Integration of Evolutionary and SVM Algorithms. Mech. Mach. Theory 2014, 73, 49–60. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Szymborski, T.; Stepanenko, Y.; Niciński, K.; Piecyk, P.; Berus, S.M.; Adamczyk-Popławska, M.; Kamińska, A. Ultrasensitive SERS Platform Made via Femtosecond Laser Micromachining for Biomedical Applications. J. Mater. Res. Technol. 2021, 12, 1496–1507. [Google Scholar] [CrossRef]
- Cao, G.; Chen, M.; Chen, Y.; Huang, Z.; Lin, J.; Lin, J.; Xu, Z.; Wu, S.; Huang, W.; Weng, G.; et al. A Potential Method for Non-Invasive Acute Myocardial Infarction Detection Based on Saliva Raman Spectroscopy and Multivariate Analysis. Laser Phys. Lett. 2015, 12, 125702. [Google Scholar] [CrossRef]
- Baghizadeh Fini, M. Oral Saliva and COVID-19. Oral Oncol. 2020, 108, 104821. [Google Scholar] [CrossRef]
- Isho, B.; Abe, K.T.; Zuo, M.; Jamal, A.J.; Rathod, B.; Wang, J.H.; Li, Z.; Chao, G.; Rojas, O.L.; Bang, Y.M.; et al. Persistence of Serum and Saliva Antibody Responses to SARS-CoV-2 Spike Antigens in COVID-19 Patients. Sci. Immunol. 2020, 5, eabe5511. [Google Scholar] [CrossRef] [PubMed]
- Torreggiani, A.; Barata-Vallejo, S.; Chatgilialoglu, C. Combined Raman and IR Spectroscopic Study on the Radical-Based Modifications of Methionine. Anal. Bioanal. Chem. 2011, 401, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Pérez, P.; Kato, T.; Mikami, Y.; Okuda, K.; Gilmorre, R.C.; Conde, C.D.; Gasmi, B.; Stein, S.; Beach, M.; et al. SARS-CoV-2 Infection of the Oral Cavity and Saliva. Nat. Med. 2021, 27, 892–903. [Google Scholar] [CrossRef] [PubMed]
- Koziorowska, J.; Mazurowa, N.; Tautt, J. Methionine Dependence of Virus-Infected Cells. Exp. Cell Res. 1990, 190, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, N.; Thiruvengadam, C.; Ramani, P.; Premkumar, P.; Natesan, A.; Sherlin, H.J. Salivary Ferritin as a Predictive Marker of Iron Deficiency Anemia in Children. J. Clin. Pediatr. Dent. 2012, 37, 25–30. [Google Scholar] [CrossRef]
- Franco-Martínez, L.; Cerón, J.J.; Vicente-Romero, M.R.; Bernal, E.; Cantero, A.T.; Tecles, F.; Resalt, C.S.; Martínez, M.; Tvarijonaviciute, A.; Martínez-Subiela, S. Salivary Ferritin Changes in Patients with COVID-19. Int. J. Environ. Res. Public Health 2021, 19, 41. [Google Scholar] [CrossRef]
- Giuca, M.R.; Pasini, M.; Tecco, S.; Giuca, G.; Marzo, G. Levels of Salivary Immunoglobulins and Periodontal Evaluation in Smoking Patients. BMC Immunol. 2014, 15, 5. [Google Scholar] [CrossRef]
- Ashton, L.; Brewster, V.L.; Correa, E.; Goodacre, R. Detection of Glycosylation and Iron-Binding Protein Modifications Using Raman Spectroscopy. Analyst 2017, 142, 808–814. [Google Scholar] [CrossRef]
- Ettah, I.; Ashton, L. Engaging with Raman Spectroscopy to Investigate Antibody Aggregation. Antibodies 2018, 7, 24. [Google Scholar] [CrossRef]
- Lin, X.; Lin, D.; Ge, X.; Qiu, S.; Feng, S.; Chen, R. Noninvasive Detection of Nasopharyngeal Carcinoma Based on Saliva Proteins Using Surface-Enhanced Raman Spectroscopy. J. Biomed. Opt. 2017, 22, 105004. [Google Scholar] [CrossRef]
- Li, X.; Yang, T.; Lin, J. Spectral Analysis of Human Saliva for Detection of Lung Cancer Using Surface-Enhanced Raman Spectroscopy. J. Biomed. Opt. 2012, 17, 037003. [Google Scholar] [CrossRef] [PubMed]
- Talari, A.C.S.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2015, 50, 46–111. [Google Scholar] [CrossRef]
- Austin, L.A.; Osseiran, S.; Evans, C.L. Raman Technologies in Cancer Diagnostics. Analyst 2016, 141, 476–503. [Google Scholar] [CrossRef] [PubMed]
- Muro, C.K.; Doty, K.C.; de Souza Fernandes, L.; Lednev, I.K. Forensic Body Fluid Identification and Differentiation by Raman Spectroscopy. Forensic Chem. 2016, 1, 31–38. [Google Scholar] [CrossRef]
- Oliveira, E.M.; Rogero, M.; Ferreira, E.C.; Gomes Neto, J.A. Simultaneous Determination of Phosphite and Phosphate in Fertilizers by Raman Spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 246, 119025. [Google Scholar] [CrossRef]
- Virkler, K.; Lednev, I.K. Raman Spectroscopy Offers Great Potential for the Nondestructive Confirmatory Identification of Body Fluids. Forensic Sci. Int. 2008, 181, e1–e5. [Google Scholar] [CrossRef]
- Hu, P.; Zheng, X.S.; Zong, C.; Li, M.H.; Zhang, L.Y.; Li, W.; Ren, B. Drop-Coating Deposition and Surface-Enhanced Raman Spectroscopies (DCDRS and SERS) Provide Complementary Information of Whole Human Tears. J. Raman Spectrosc. 2014, 45, 565–573. [Google Scholar] [CrossRef]
- Virkler, K.; Lednev, I.K. Forensic Body Fluid Identification: The Raman Spectroscopic Signature of Saliva. Analyst 2010, 135, 512–517. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; Li, M.; Xiong, L.; Xu, X.; Ji, N.; Dai, L.; Sun, Q. The Formation of a Protein Corona and the Interaction with α-Amylase by Chitin Nanowhiskers in Simulated Saliva Fluid. Food Hydrocoll. 2020, 102, 105615. [Google Scholar] [CrossRef]
- Zamora-Mendoza, B.N.; Espinosa-Tanguma, R.; Ramírez-Elías, M.G.; Cabrera-Alonso, R.; Montero-Moran, G.; Portales-Pérez, D.; Rosales-Romo, J.A.; Gonzalez, J.F.; Gonzalez, C. Surface-Enhanced Raman Spectroscopy: A Non Invasive Alternative Procedure for Early Detection in Childhood Asthma Biomarkers in Saliva. Photodiagnosis Photodyn. Ther. 2019, 27, 85–91. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Q.; Hu, J.; Zhou, M.; Yu, M.Q.; Li, K.Y.; Xu, D.; Xiao, Y.; Yang, J.Y.; Lu, Y.J.; et al. Nasopharyngeal Swabs Are More Sensitive Than Oropharyngeal Swabs for COVID-19 Diagnosis and Monitoring the SARS-CoV-2 Load. Front. Med. 2020, 7, 334. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, B.A.; Higgins, T.S.; Han, J.K. Bacterial Pathogens in the Nasopharynx, Nasal Cavity, and Osteomeatal Complex during Wellness and Viral Infection. Am. J. Rhinol. Allergy 2013, 27, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Sam, S.S.; Caliendo, A.M.; Ingersoll, J.; Abdul-Ali, D.; Hill, C.E.; Kraft, C.S. Evaluation of Performance Characteristics of Panther Fusion Assays for Detection of Respiratory Viruses from Nasopharyngeal and Lower Respiratory Tract Specimens. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef]
- Silva, M.J.A.; Ribeiro, L.R.; Gouveia, M.I.M.; Marcelino, B.D.R.; Santos, C.S.D.; Lima, K.V.B.; Lima, L.N.G.C. Hyperinflammatory Response in COVID-19: A Systematic Review. Viruses 2023, 15, 553. [Google Scholar] [CrossRef] [PubMed]
- Gudowska-Sawczuk, M.; Mroczko, B. What Is Currently Known about the Role of CXCL10 in SARS-CoV-2 Infection? Int. J. Mol. Sci. 2022, 23, 3673. [Google Scholar] [CrossRef] [PubMed]
- Hailemichael, W.; Kiros, M.; Akelew, Y.; Getu, S.; Andualem, H. Neopterin: A Promising Candidate Biomarker for Severe COVID-19. J. Inflamm. Res. 2021, 14, 245. [Google Scholar] [CrossRef] [PubMed]
- Kamińska, A.; Witkowska, E.; Kowalska, A.; Skoczyńska, A.; Gawryszewska, I.; Guziewicz, E.; Snigurenko, D.; Waluk, J. Highly Efficient SERS-Based Detection of Cerebrospinal Fluid Neopterin as a Diagnostic Marker of Bacterial Infection. Anal. Bioanal. Chem. 2016, 408, 4319–4327. [Google Scholar] [CrossRef]
- Chan, C.P.Y.; Choi, J.W.Y.; Cao, K.Y.; Wang, M.; Gao, Y.; Zhou, D.H.; Di, B.; Xu, H.F.; Leung, M.F.; Bergmann, A.; et al. Detection of Serum Neopterin for Early Assessment of Dengue Virus Infection. J. Infect. 2006, 53, 152–158. [Google Scholar] [CrossRef]
- Fuchs, D.; Avanzas, P.; Arroyo-Espliguero, R.; Jenny, M.; Consuegra-Sanchez, L.; Kaski, J. The Role of Neopterin in Atherogenesis and Cardiovascular Risk Assessment. Curr. Med. Chem. 2009, 16, 4644–4653. [Google Scholar] [CrossRef]
- Murr, C.; Fuith, L.C.; Widner, B.; Wirleitner, B.; Baier-Bitterlich, G.; Fuchs, D. Increased Neopterin Concentrations in Patients with Cancer: Indicator of Oxidative Stress? Anticancer Res. 1999, 19, 1721–1728. [Google Scholar]
- Yadav, A.K.; Sharma, V.; Jha, V. Association between Serum Neopterin and Inflammatory Activation in Chronic Kidney Disease. Mediat. Inflamm. 2012, 2012, 6. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Cao, K.Y.; Chan, C.P.Y.; Choi, J.W.Y.; Leung, W.; Leung, M.; Duan, Z.H.; Gao, Y.; Wang, M.; Di, B.; et al. Serum Neopterin for Early Assessment of Severity of Severe Acute Respiratory Syndrome. Clin. Immunol. 2005, 116, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Chauvin, M.; Larsen, M.; Quirant, B.; Quentric, P.; Dorgham, K.; Royer, L.; Vallet, H.; Guihot, A.; Combadière, B.; Combadière, C.; et al. Elevated Neopterin Levels Predict Fatal Outcome in SARS-CoV-2-Infected Patients. Front. Cell. Infect. Microbiol. 2021, 11, 764. [Google Scholar] [CrossRef] [PubMed]
- Bellmann-Weiler, R.; Lanser, L.; Burkert, F.; Seiwald, S.; Fritsche, G.; Wildner, S.; Schroll, A.; Koppelstätter, S.; Kurz, K.; Griesmacher, A.; et al. Neopterin Predicts Disease Severity in Hospitalized Patients With COVID-19. Open Forum Infect Dis. 2020, 8, 1–5. [Google Scholar] [CrossRef]
- Bevilacqua, M.; Marini, F. Local Classification: Locally Weighted–Partial Least Squares-Discriminant Analysis (LW–PLS-DA). Anal. Chim. Acta 2014, 838, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Höskuldsson, A. PLS Regression Methods. J. Chemom. 1988, 2, 211–228. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.Y.; Jemain, A.A. Partial Least Squares-Discriminant Analysis (PLS-DA) for Classification of High-Dimensional (HD) Data: A Review of Contemporary Practice Strategies and Knowledge Gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Peerbhay, K.Y.; Mutanga, O.; Ismail, R. Commercial Tree Species Discrimination Using Airborne AISA Eagle Hyperspectral Imagery and Partial Least Squares Discriminant Analysis (PLS-DA) in KwaZulu–Natal, South Africa. ISPRS J. Photogramm. Remote Sens. 2013, 79, 19–28. [Google Scholar] [CrossRef]
- Suhandy, D.; Suhandy, D.; Yulia, M. Luwak Coffee Classification Using UV-Vis Spectroscopy Data: Comparison of Linear Discriminant Analysis and Support Vector Machine Methods. Aceh Int. J. Sci. Technol. 2018, 7, 115–121. [Google Scholar] [CrossRef]
- Terouzi, W.; Chem, M.J.; Rizki, H.; Kzaiber, F.; Hanine, H.; Nabloussi, A.; Oussama, A. Characterization and Rapid Detection of Adulterations in Sesame Oil Using FT-MIR and PCA-LDA. Moroccan J. Chem. 2016, 4, 1052–1060. [Google Scholar] [CrossRef]
- Yu, H.; Yu, H.; Yang, J. A Direct LDA Algorithm for High-Dimensional Data–with Application to Face Recognition. J. Pattern Recognitio Soc. 2001, 34, 2067–2070. [Google Scholar] [CrossRef]
- Lu, J.; Plataniotis, K.N.; Venetsanopoulos, A.N. Face Recognition Using LDA-Based Algorithms. IEEE Trans. Neural Netw. 2003, 14, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Mohan, V. Liver Disease Prediction Using SVM and Naïve Bayes Algorithms Privacy Preserving Data Mining View Project. Int. J. Sci. Eng. Technol. Res. 2015, 4, 816–820. [Google Scholar]
- Yao, Y.; Liu, Y.; Yu, Y.; Xu, H.; Lv, W.; Li, Z.; Chen, X. K-SVM: An Effective SVM Algorithm Based on K-Means Clustering. J. Comput. 2013, 8, 2632–2639. [Google Scholar] [CrossRef]
- Battineni, G.; Chintalapudi, N.; Amenta, F. Machine Learning in Medicine: Performance Calculation of Dementia Prediction by Support Vector Machines (SVM). Informatics Med. Unlocked 2019, 16, 100200. [Google Scholar] [CrossRef]
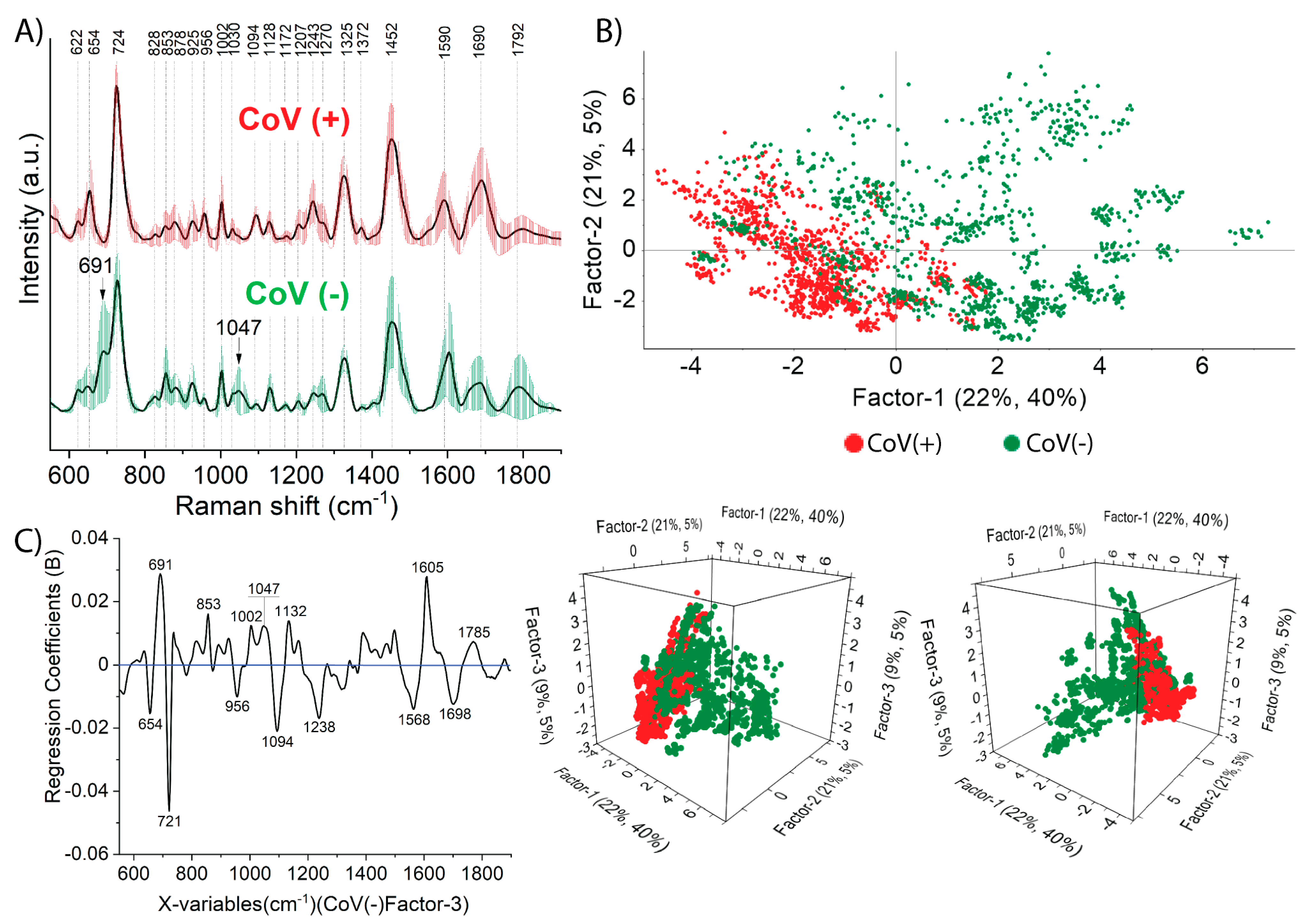
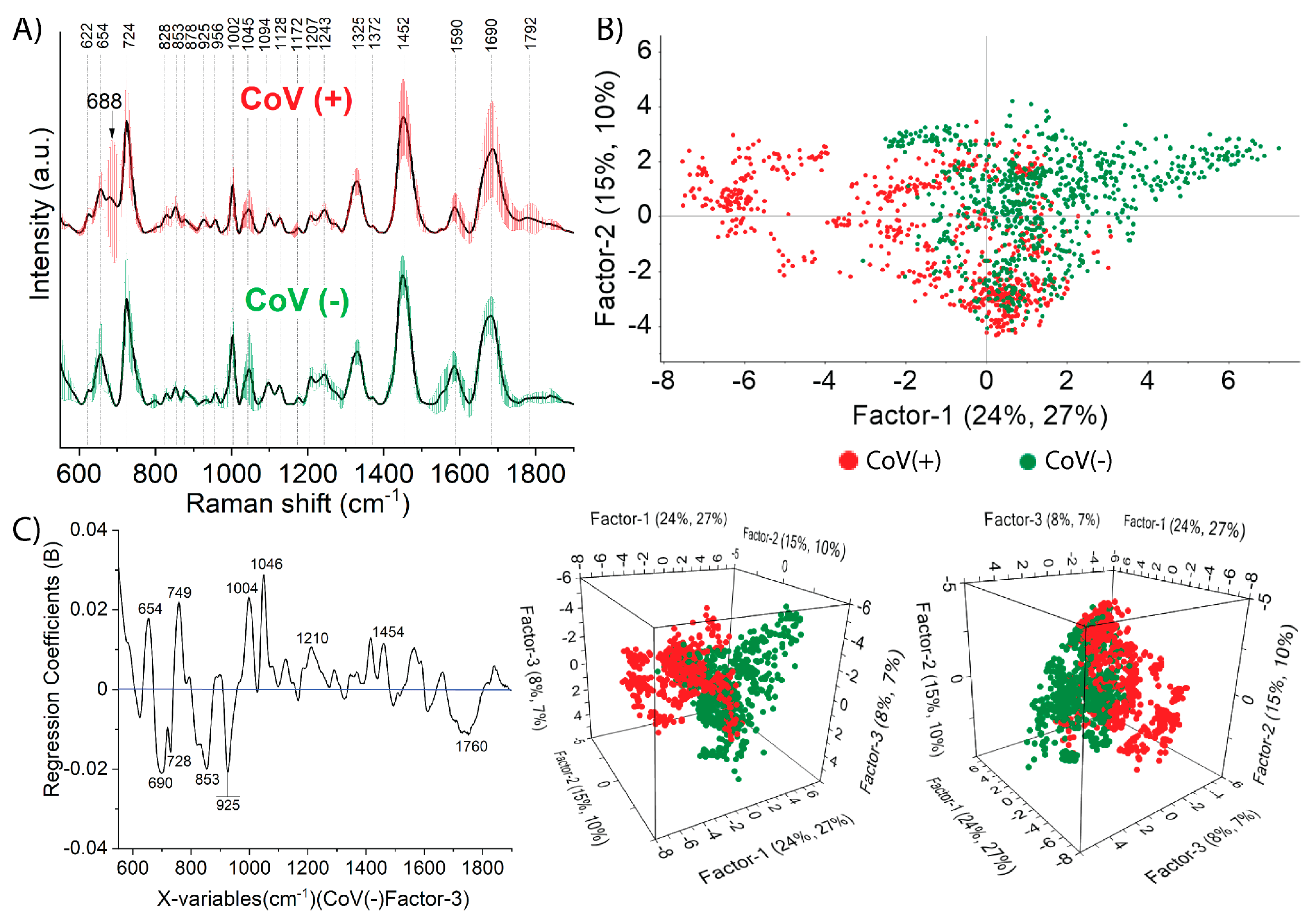
| Sample | Assignment | |||
|---|---|---|---|---|
| Saliva | Nasopharyngeal Swabs | |||
| CoV(−) | CoV(+) | CoV(−) | CoV(+) | |
| 622 | 622 | 623 | 623 | adenine, C-C twisting mode of phenylalanine (protein) |
| 649 | 654 | 654 | 654 | C-S stretching vibration in methionine C-C twisting mode of tyrosine |
| - | - | - | 679 | Ring breathing modes in the DNA bases, G (ring breathing modes in the DNA bases) neopterin |
| 691 | - | - | - | δ(O–C=O) Creatinine, cytosine |
| 724 | 724 | 724 | 724 | O-O stretching vibration in oxygenated proteins, glycoproteins such as mucines, ring breathing mode of tryptophan (protein assignment), C-N head group choline (H3C)3N+ (lipid assignment) |
| 828 | 828 | 828 | 828 | Ring breathing mode of tyrosine, Transferrin (Tyrosine, H-bonding) |
| 853 | 853 | 853 | 853 | Ring breathing mode of tyrosine, Transferrin (Tyrosine, H-bonding) |
| 878 | 878 | 878 | 878 | Proline, valine, glycine, tryptophan, glutamate or ν (C─C) Hydro-oxyproline, Transferrin (Tryptophan, H-bonding) or νsP(OH)2 of phosphate |
| 925 | 925 | 925 | 925 | C-C stretching proline ring, carboxylates including glucose and glycogen |
| 956 | 956 | 956 | 956 | hydroxyapatite, xanthine proline, valine |
| 1002 | 1002 | 1002 | 1002 | aromatic ring breathing of phenylalanine phenylalanine in Lysozyme, lactoferrin, albumin, Transferrin (Phenylalanine) |
| 1030 | 1030 | - | - | C-H in-plane bending mode of phenylalanine Phenylalanine in Lysozyme, lactoferrin, albumin |
| 1047 | - | 1046 | 1046 | C-O and C-N stretching in proteins, Glycogen C–CH3 vibration |
| 1094 | 1094 | 1094 | 1094 | Symmetric PO2− stretching vibration of the DNA backbone T cells |
| 1128 | 1128 | 1128 | 1128 | C-O stretching (carbohydrates), C-N stretching (proteins) |
| 1172 | 1172 | 1172 | 1172 | bending C-H tyrosine, Transferrin (Tyrosine, CH3) |
| 1207 | 1207 | 1207 | 1207 | tryptophan and phenylalanine v(C-C6H5) mode, Hydroxyproline, tyrosine Tryptophan in Lysozyme, lactoferrin, albumin |
| 1243 | 1243 | 1243 | 1243 | phosphodiester group associate with nucleic acid B-sheet (the most common secondary structures in proteins, e.g., alfa amylase) |
| 1270 | 1270 | 1270 | 1270 | Stretching C-N, bending N-H—amide III band in proteins Transferrin (Tyrosine/α-helix) α-helix (the most common secondary structures in proteins, e.g., alfa amylase) |
| 1325 | 1325 | 1325 | 1325 | amide III band in proteins CH3CH2 wagging mode in purine bases of nucleic acids T cells |
| 1372 | 1372 | 1372 | 1372 | Lipids, proteins (tryptophan) T, A, G (ring breathing modes of the DNA/RNA bases) T cells |
| 1402 | - | - | - | Bending of methyl groups in proteins |
| 1452 | 1452 | 1452 | 1452 | the C-H stretching of glycoproteins including mucines or Hydrocarbon chain of lipid, Triglycerides CH3 Deformation of lipids CH2, CH3 bend of tryptophan Tryptophan in Lysozyme, lactoferrin, albumin T cells |
| 1550 | 1550 | 1553 | 1553 | ʋ(CN) and δ(NH) amide II ν (C=C) tryptophan |
| 1604 | 1590 | 1585 | 1585 | phenylalanine, tryptophan, hydroxyproline, hypoxanthine C=C in-plane bending mode of phenylalanine and tyrosine Cytosine (NH2) |
| 1690 | 1690 | 1680 | 1680 | Amide I of proteins (Lysozyme, lactoferrin, albumin) |
| Type of Sample | Numerical Analysis | Sensitivity (%) | Specificity (%) | Accuracy (%) | Number of Samples |
|---|---|---|---|---|---|
| Saliva | PLS-DA | C: 97.0 | 90.0 | 93.0 | 149 |
| V: 90.0 | 70.0 | 80.0 | 20 | ||
| PCA-LDA | C: 97.0 | 79.0 | 88.0 | 149 | |
| V: 100.0 | 60.0 | 80.0 | 20 | ||
| SVMC | C: 100.0 | 100.0 | 100.0 | 149 | |
| V: 100.0 | 80.0 | 90.0 | 20 | ||
| Nasopharyngeal swabs | PLS-DA | C: 100.0 | 96.0 | 98.0 | 104 |
| V: 63.0 | 75.0 | 69.0 | 16 | ||
| PCA-LDA | C: 96.0 | 83.0 | 89.0 | 104 | |
| V: 63.0 | 75.0 | 69.0 | 16 | ||
| SVMC | C: 100.0 | 100.0 | 100.0 | 104 | |
| V: 88.0 | 63.0 | 75.0 | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berus, S.M.; Nowicka, A.B.; Wieruszewska, J.; Niciński, K.; Kowalska, A.A.; Szymborski, T.R.; Dróżdż, I.; Borowiec, M.; Waluk, J.; Kamińska, A. SERS Signature of SARS-CoV-2 in Saliva and Nasopharyngeal Swabs: Towards Perspective COVID-19 Point-of-Care Diagnostics. Int. J. Mol. Sci. 2023, 24, 9706. https://doi.org/10.3390/ijms24119706
Berus SM, Nowicka AB, Wieruszewska J, Niciński K, Kowalska AA, Szymborski TR, Dróżdż I, Borowiec M, Waluk J, Kamińska A. SERS Signature of SARS-CoV-2 in Saliva and Nasopharyngeal Swabs: Towards Perspective COVID-19 Point-of-Care Diagnostics. International Journal of Molecular Sciences. 2023; 24(11):9706. https://doi.org/10.3390/ijms24119706
Chicago/Turabian StyleBerus, Sylwia M., Ariadna B. Nowicka, Julia Wieruszewska, Krzysztof Niciński, Aneta A. Kowalska, Tomasz R. Szymborski, Izabela Dróżdż, Maciej Borowiec, Jacek Waluk, and Agnieszka Kamińska. 2023. "SERS Signature of SARS-CoV-2 in Saliva and Nasopharyngeal Swabs: Towards Perspective COVID-19 Point-of-Care Diagnostics" International Journal of Molecular Sciences 24, no. 11: 9706. https://doi.org/10.3390/ijms24119706
APA StyleBerus, S. M., Nowicka, A. B., Wieruszewska, J., Niciński, K., Kowalska, A. A., Szymborski, T. R., Dróżdż, I., Borowiec, M., Waluk, J., & Kamińska, A. (2023). SERS Signature of SARS-CoV-2 in Saliva and Nasopharyngeal Swabs: Towards Perspective COVID-19 Point-of-Care Diagnostics. International Journal of Molecular Sciences, 24(11), 9706. https://doi.org/10.3390/ijms24119706







