The Exosome-like Vesicles of Giardia Assemblages A, B, and E Are Involved in the Delivering of Distinct Small RNA from Parasite to Parasite
Abstract
1. Introduction
2. Results
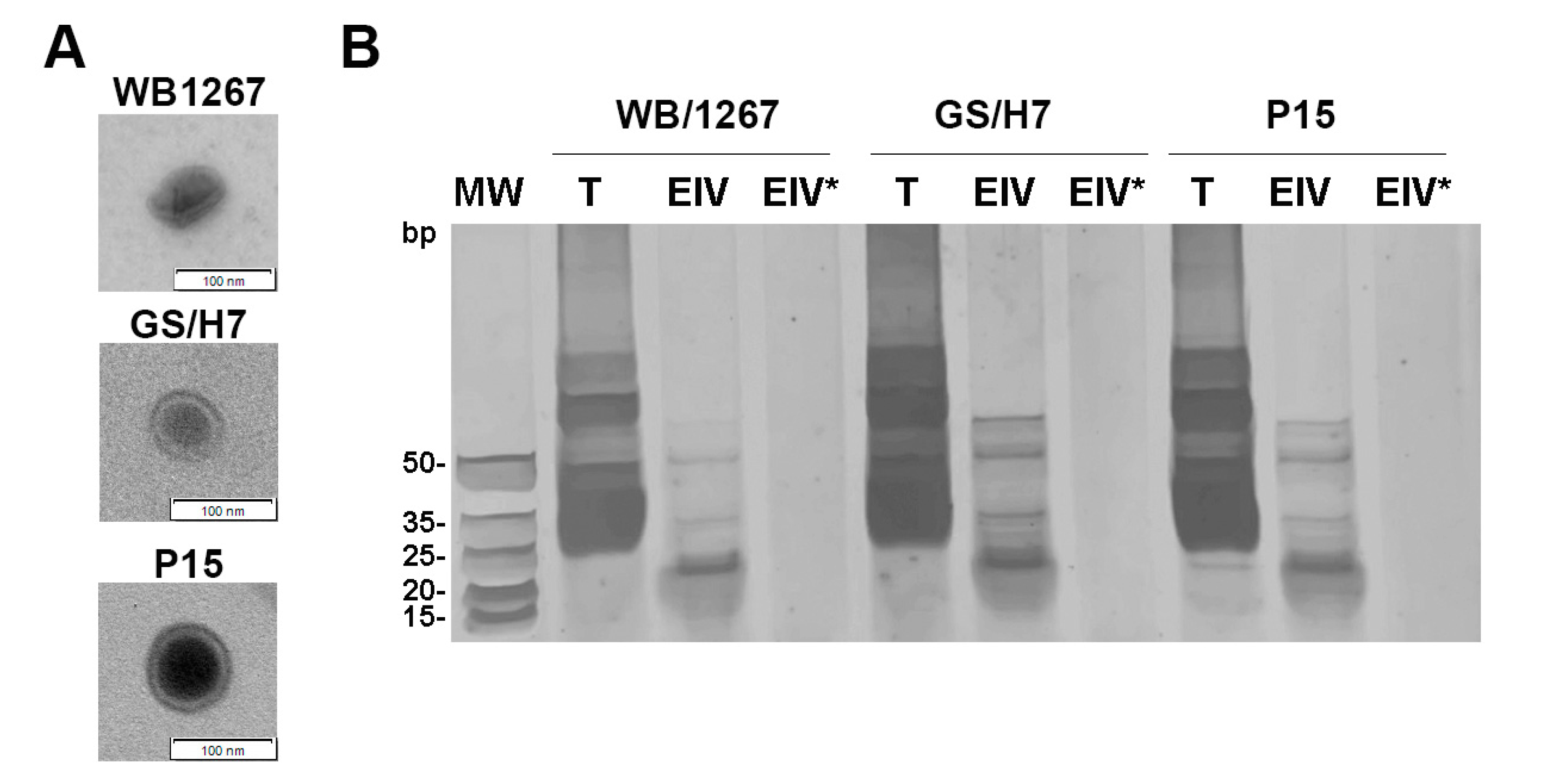
2.1. The Assemblages A, B, and E of Giardia Release Exosome-like Vesicles Containing Small RNAs
2.2. RNA-Seq of Giardia ElVs Revealed the Presence of Different sRNA Biotypes
2.3. The ElVs of the Assemblages A, B, and E of Giardia Contain Unique and Shared Small RNAs
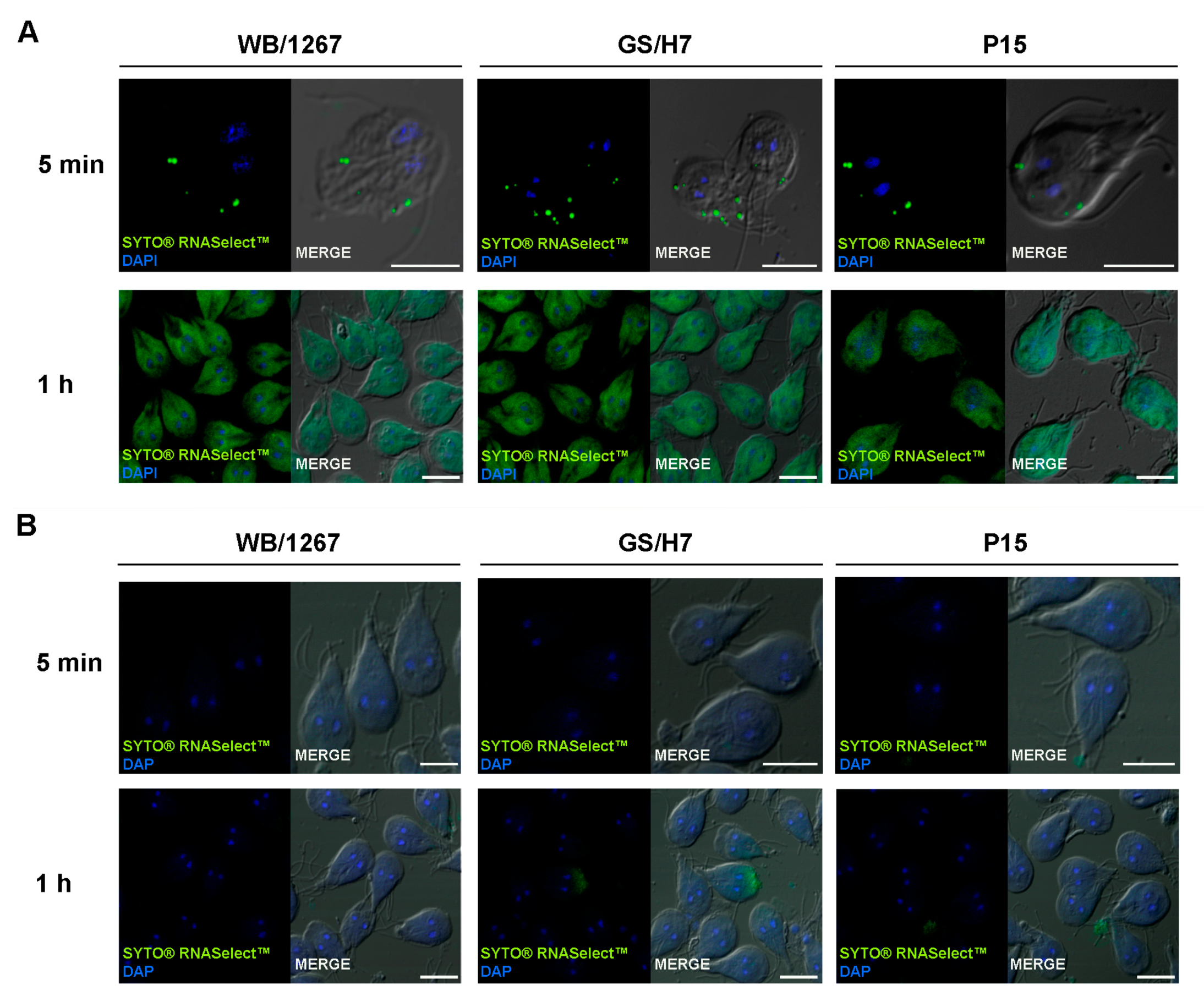
2.4. The Extracellular Vesicle RNA Cargo Is Internalized by the Trophozoites
3. Discussion
4. Materials and Methods
4.1. Cell Culture and EV Isolation
4.2. Exosome-like Vesicles (ElVs) Size and Electron-Microscopy Images
4.3. ElVs RNA Profiling
4.4. Small RNA Libraries and RNA-Seq
4.5. Bioinformatic Analysis
4.6. PCR Detection and Quantification of Small RNAs
4.7. Labelling the RNA from EVs
4.8. Cell Uptake Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sprong, H.; Caccio, S.M.; van der Giessen, J.W. Identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl. Trop. Dis. 2009, 3, e558. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.D. Biology of Giardia lamblia. Clin. Microbiol. Rev. 2001, 14, 447–475. [Google Scholar] [CrossRef] [PubMed]
- Monis, P.T.; Caccio, S.M.; Thompson, R.C. Variation in Giardia: Towards a taxonomic revision of the genus. Trends Parasitol. 2009, 25, 93–100. [Google Scholar] [CrossRef]
- Steuart, R.F.; O’Handley, R.; Lipscombe, R.J.; Lock, R.A.; Thompson, R.C. Alpha 2 giardin is an assemblage A-specific protein of human infective Giardia duodenalis. Parasitology 2008, 135, 1621–1627. [Google Scholar] [CrossRef]
- Andrews, R.H.; Chilton, N.B.; Mayrhofer, G. Selection of specific genotypes of Giardia intestinalis by growth in vitro and in vivo. Parasitology 1992, 105 Pt 3, 375–386. [Google Scholar] [CrossRef]
- Thompson, R.C.; Lymbery, A.J. Genetic variability in parasites and host-parasite interactions. Parasitology 1996, 112, S7–S22. [Google Scholar] [CrossRef]
- Homan, W.L.; Mank, T.G. Human giardiasis: Genotype linked differences in clinical symptomatology. Int. J. Parasitol. 2001, 31, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Sahagun, J.; Clavel, A.; Goni, P.; Seral, C.; Llorente, M.T.; Castillo, F.J.; Capilla, S.; Arias, A.; Gomez-Lus, R. Correlation between the presence of symptoms and the Giardia duodenalis genotype. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2008, 27, 81–83. [Google Scholar] [CrossRef]
- Haque, R.; Roy, S.; Kabir, M.; Stroup, S.E.; Mondal, D.; Houpt, E.R. Giardia assemblage A infection and diarrhea in Bangladesh. J. Infect. Dis. 2005, 192, 2171–2173. [Google Scholar] [CrossRef]
- Dubourg, A.; Xia, D.; Winpenny, J.P.; Al Naimi, S.; Bouzid, M.; Sexton, D.W.; Wastling, J.M.; Hunter, P.R.; Tyler, K.M. Giardia secretome highlights secreted tenascins as a key component of pathogenesis. GigaScience 2018, 7, 1–13. [Google Scholar] [CrossRef]
- Ma’ayeh, S.Y.; Liu, J.; Peirasmaki, D.; Hornaeus, K.; Bergstrom Lind, S.; Grabherr, M.; Bergquist, J.; Svard, S.G. Characterization of the Giardia intestinalis secretome during interaction with human intestinal epithelial cells: The impact on host cells. PLoS Negl. Trop. Dis. 2017, 11, e0006120. [Google Scholar] [CrossRef]
- Grajeda, B.I.; De Chatterjee, A.; Villalobos, C.M.; Pence, B.C.; Ellis, C.C.; Enriquez, V.; Roy, S.; Roychowdhury, S.; Neumann, A.K.; Almeida, I.C.; et al. Giardial lipid rafts share virulence factors with secreted vesicles and participate in parasitic infection in mice. Front. Cell. Infect. Microbiol. 2022, 12, 974200. [Google Scholar] [CrossRef]
- Pu, X.; Li, X.; Cao, L.; Yue, K.; Zhao, P.; Wang, X.; Li, J.; Zhang, X.; Zhang, N.; Zhao, Z.; et al. Giardia duodenalis Induces Proinflammatory Cytokine Production in Mouse Macrophages via TLR9-Mediated p38 and ERK Signaling Pathways. Front. Cell Dev. Biol. 2021, 9, 694675. [Google Scholar] [CrossRef] [PubMed]
- Moyano, S.; Musso, J.; Feliziani, C.; Zamponi, N.; Frontera, L.S.; Ropolo, A.S.; Lanfredi-Rangel, A.; Lalle, M.; Touz, M. Exosome Biogenesis in the Protozoa Parasite Giardia lamblia: A Model of Reduced Interorganellar Crosstalk. Cells 2019, 8, 1600. [Google Scholar] [CrossRef]
- Gruenberg, J.; Stenmark, H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004, 5, 317–323. [Google Scholar] [CrossRef]
- Hurley, J.H. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 2008, 20, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Montaner, S.; Galiano, A.; Trelis, M.; Martin-Jaular, L.; Del Portillo, H.A.; Bernal, D.; Marcilla, A. The Role of Extracellular Vesicles in Modulating the Host Immune Response during Parasitic Infections. Front. Immunol. 2014, 5, 433. [Google Scholar] [CrossRef] [PubMed]
- Coakley, G.; Maizels, R.M.; Buck, A.H. Exosomes and Other Extracellular Vesicles: The New Communicators in Parasite Infections. Trends Parasitol. 2015, 31, 477–489. [Google Scholar] [CrossRef]
- Marti, M.; Johnson, P.J. Emerging roles for extracellular vesicles in parasitic infections. Curr. Opin. Microbiol. 2016, 32, 66–70. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luo, J.; Zhou, H.; Liao, J.Y.; Ma, L.M.; Chen, Y.Q.; Qu, L.H. Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008, 36, 6048–6055. [Google Scholar] [CrossRef]
- Saraiya, A.A.; Wang, C.C. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008, 4, e1000224. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Gutierrez-Vazquez, C.; Sanchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin Cancer Biol 2014, 28, 3–13. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Madrid, F.; Mittelbrunn, M. Analysis of microRNA and protein transfer by exosomes during an immune synapse. Methods Mol. Biol. 2013, 1024, 41–51. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Ma’ayeh, S.Y.; Knorr, L.; Skold, K.; Garnham, A.; Ansell, B.R.E.; Jex, A.R.; Svard, S.G. Responses of the Differentiated Intestinal Epithelial Cell Line Caco-2 to Infection With the Giardia intestinalis GS Isolate. Front. Cell. Infect. Microbiol. 2018, 8, 244. [Google Scholar] [CrossRef] [PubMed]
- Gavinho, B.; Sabatke, B.; Feijoli, V.; Rossi, I.V.; da Silva, J.M.; Evans-Osses, I.; Palmisano, G.; Lange, S.; Ramirez, M.I. Peptidylarginine Deiminase Inhibition Abolishes the Production of Large Extracellular Vesicles From Giardia intestinalis, Affecting Host-Pathogen Interactions by Hindering Adhesion to Host Cells. Front. Cell. Infect. Microbiol. 2020, 10, 417. [Google Scholar] [CrossRef]
- Axtell, M.J. ShortStack: Comprehensive annotation and quantification of small RNA genes. Rna 2013, 19, 740–751. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- An, J.; Lai, J.; Lehman, M.L.; Nelson, C.C. miRDeep*: An integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2013, 41, 727–737. [Google Scholar] [CrossRef]
- Abels, E.R.; Maas, S.L.N.; Nieland, L.; Wei, Z.; Cheah, P.S.; Tai, E.; Kolsteeg, C.J.; Dusoswa, S.A.; Ting, D.T.; Hickman, S.; et al. Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular miR-21. Cell Rep. 2019, 28, 3105–3119.e3107. [Google Scholar] [CrossRef]
- Nicola, A.M.; Frases, S.; Casadevall, A. Lipophilic dye staining of Cryptococcus neoformans extracellular vesicles and capsule. Eukaryot. Cell 2009, 8, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Twu, O.; de Miguel, N.; Lustig, G.; Stevens, G.C.; Vashisht, A.A.; Wohlschlegel, J.A.; Johnson, P.J. Trichomonas vaginalis exosomes deliver cargo to host cells and mediate hostratioparasite interactions. PLoS Pathog. 2013, 9, e1003482. [Google Scholar] [CrossRef]
- Olmos-Ortiz, L.M.; Barajas-Mendiola, M.A.; Barrios-Rodiles, M.; Castellano, L.E.; Arias-Negrete, S.; Avila, E.E.; Cuellar-Mata, P. Trichomonas vaginalis exosome-like vesicles modify the cytokine profile and reduce inflammation in parasite-infected mice. Parasite Immunol. 2017, 39, e12426. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, F.M.C.; Dupin, T.V.; Toledo, M.D.S.; Reis, N.; Ribeiro, K.; Cronemberger-Andrade, A.; Rugani, J.N.; De Lorenzo, B.H.P.; Novaes, E.B.R.R.; Soares, R.P.; et al. Extracellular Vesicles Released by Leishmania (Leishmania) amazonensis Promote Disease Progression and Induce the Production of Different Cytokines in Macrophages and B-1 Cells. Front. Microbiol. 2018, 9, 3056. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, P.M.; de Menezes-Neto, A.; Borges, V.M.; Descoteaux, A.; Torrecilhas, A.C.; Xander, P.; Revach, O.Y.; Regev-Rudzki, N.; Soares, R.P. Immunomodulatory Properties of Leishmania Extracellular Vesicles During Host-Parasite Interaction: Differential Activation of TLRs and NF-kappaB Translocation by Dermotropic and Viscerotropic Species. Front. Cell. Infect. Microbiol. 2020, 10, 380. [Google Scholar] [CrossRef]
- Nogueira, P.M.; Ribeiro, K.; Silveira, A.C.; Campos, J.H.; Martins-Filho, O.A.; Bela, S.R.; Campos, M.A.; Pessoa, N.L.; Colli, W.; Alves, M.J.; et al. Vesicles from different Trypanosoma cruzi strains trigger differential innate and chronic immune responses. J. Extracell. Vesicles 2015, 4, 28734. [Google Scholar] [CrossRef]
- Cronemberger-Andrade, A.; Xander, P.; Soares, R.P.; Pessoa, N.L.; Campos, M.A.; Ellis, C.C.; Grajeda, B.; Ofir-Birin, Y.; Almeida, I.C.; Regev-Rudzki, N.; et al. Trypanosoma cruzi-Infected Human Macrophages Shed Proinflammatory Extracellular Vesicles That Enhance Host-Cell Invasion via Toll-Like Receptor 2. Front. Cell. Infect. Microbiol. 2020, 10, 99. [Google Scholar] [CrossRef]
- Diaz-Godinez, C.; Rios-Valencia, D.G.; Garcia-Aguirre, S.; Martinez-Calvillo, S.; Carrero, J.C. Immunomodulatory effect of extracellular vesicles from Entamoeba histolytica trophozoites: Regulation of NETs and respiratory burst during confrontation with human neutrophils. Front. Cell. Infect. Microbiol. 2022, 12, 1018314. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Xiu, F.; Wang, J.; Cong, H.; He, S.; Shi, Y.; Wang, X.; Li, X.; Zhou, H. Characterization of exosomes derived from Toxoplasma gondii and their functions in modulating immune responses. Int. J. Nanomed. 2018, 13, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.E.; Leonard, J.N. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J. Extracell. Vesicles 2016, 5, 31027. [Google Scholar] [CrossRef]
- Solaymani-Mohammadi, S.; Singer, S.M. Giardia duodenalis: The double-edged sword of immune responses in giardiasis. Exp. Parasitol. 2010, 126, 292–297. [Google Scholar] [CrossRef]
- Singer, S.M.; Nash, T.E. T-cell-dependent control of acute Giardia lamblia infections in mice. Infect. Immun. 2000, 68, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Einarsson, E.; Ma’ayeh, S.; Svard, S.G. An up-date on Giardia and giardiasis. Curr. Opin. Microbiol. 2016, 34, 47–52. [Google Scholar] [CrossRef]
- Einarsson, E.; Troell, K.; Hoeppner, M.P.; Grabherr, M.; Ribacke, U.; Svard, S.G. Coordinated Changes in Gene Expression Throughout Encystation of Giardia intestinalis. PLoS Negl. Trop. Dis. 2016, 10, e0004571. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.; Cremer, T.; Gallo, R.C.; Margolis, L.B. Extracellular vesicles and viruses: Are they close relatives? Proc. Natl. Acad. Sci. USA 2016, 113, 9155–9161. [Google Scholar] [CrossRef]
- Lefebvre, F.A.; Benoit Bouvrette, L.P.; Perras, L.; Blanchet-Cohen, A.; Garnier, D.; Rak, J.; Lecuyer, E. Comparative transcriptomic analysis of human and Drosophila extracellular vesicles. Sci. Rep. 2016, 6, 27680. [Google Scholar] [CrossRef]
- Fiskaa, T.; Knutsen, E.; Nikolaisen, M.A.; Jorgensen, T.E.; Johansen, S.D.; Perander, M.; Seternes, O.M. Distinct Small RNA Signatures in Extracellular Vesicles Derived from Breast Cancer Cell Lines. PLoS ONE 2016, 11, e0161824. [Google Scholar] [CrossRef]
- Hewson, C.; Capraro, D.; Burdach, J.; Whitaker, N.; Morris, K.V. Extracellular vesicle associated long non-coding RNAs functionally enhance cell viability. Noncoding RNA Res 2016, 1, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhou, B.; Zhang, F.; Tu, Y.; Hu, Y.; Zhang, B.; Zhai, Q. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PLoS ONE 2013, 8, e56842. [Google Scholar] [CrossRef] [PubMed]
- Keam, S.P.; Hutvagner, G. tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression. Life 2015, 5, 1638–1651. [Google Scholar] [CrossRef] [PubMed]
- Lambertz, U.; Oviedo Ovando, M.E.; Vasconcelos, E.J.; Unrau, P.J.; Myler, P.J.; Reiner, N.E. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Genom. 2015, 16, 151. [Google Scholar] [CrossRef]
- Dou, S.; Wang, Y.; Lu, J. Metazoan tsRNAs: Biogenesis, Evolution and Regulatory Functions. Noncoding RNA 2019, 5, 18. [Google Scholar] [CrossRef]
- Di Fazio, A.; Schlackow, M.; Pong, S.K.; Alagia, A.; Gullerova, M. Dicer dependent tRNA derived small RNAs promote nascent RNA silencing. Nucleic Acids Res. 2022, 50, 1734–1752. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.Y.; Guo, Y.H.; Zheng, L.L.; Li, Y.; Xu, W.L.; Zhang, Y.C.; Zhou, H.; Lun, Z.R.; Ayala, F.J.; Qu, L.H. Both endo-siRNAs and tRNA-derived small RNAs are involved in the differentiation of primitive eukaryote Giardia lamblia. Proc. Natl. Acad. Sci. USA 2014, 111, 14159–14164. [Google Scholar] [CrossRef]
- Garcia-Silva, M.R.; Sanguinetti, J.; Cabrera-Cabrera, F.; Franzen, O.; Cayota, A. A particular set of small non-coding RNAs is bound to the distinctive Argonaute protein of Trypanosoma cruzi: Insights from RNA-interference deficient organisms. Gene 2014, 538, 379–384. [Google Scholar] [CrossRef]
- Fernandez-Calero, T.; Garcia-Silva, R.; Pena, A.; Robello, C.; Persson, H.; Rovira, C.; Naya, H.; Cayota, A. Profiling of small RNA cargo of extracellular vesicles shed by Trypanosoma cruzi reveals a specific extracellular signature. Mol. Biochem. Parasitol. 2015, 199, 19–28. [Google Scholar] [CrossRef]
- Bayer-Santos, E.; Lima, F.M.; Ruiz, J.C.; Almeida, I.C.; da Silveira, J.F. Characterization of the small RNA content of Trypanosoma cruzi extracellular vesicles. Mol. Biochem. Parasitol. 2014, 193, 71–74. [Google Scholar] [CrossRef]
- Artuyants, A.; Campos, T.L.; Rai, A.K.; Johnson, P.J.; Dauros-Singorenko, P.; Phillips, A.; Simoes-Barbosa, A. Extracellular vesicles produced by the protozoan parasite Trichomonas vaginalis contain a preferential cargo of tRNA-derived small RNAs. Int. J. Parasitol. 2020, 50, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.P.; Kim, E.Y.; Badr, C.E.; Weissleder, R.; Mempel, T.R.; Tannous, B.A.; Breakefield, X.O. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015, 6, 7029. [Google Scholar] [CrossRef] [PubMed]
- Zomer, A.; Maynard, C.; Verweij, F.J.; Kamermans, A.; Schafer, R.; Beerling, E.; Schiffelers, R.M.; de Wit, E.; Berenguer, J.; Ellenbroek, S.I.J.; et al. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015, 161, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- de la Cuesta, F.; Passalacqua, I.; Rodor, J.; Bhushan, R.; Denby, L.; Baker, A.H. Extracellular vesicle cross-talk between pulmonary artery smooth muscle cells and endothelium during excessive TGF-beta signalling: Implications for PAH vascular remodelling. Cell Commun Signal 2019, 17, 143. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Saraiya, A.A.; Wang, C.C. Gene regulation in Giardia lambia involves a putative microRNA derived from a small nucleolar RNA. PLoS Negl. Trop. Dis. 2011, 5, e1338. [Google Scholar] [CrossRef]
- Meningher, T.; Boleslavsky, D.; Barshack, I.; Tabibian-Keissar, H.; Kohen, R.; Gur-Wahnon, D.; Ben-Dov, I.Z.; Sidi, Y.; Avni, D.; Schwartz, E. Giardia lamblia miRNAs as a new diagnostic tool for human giardiasis. PLoS Negl. Trop. Dis. 2019, 13, e0007398. [Google Scholar] [CrossRef] [PubMed]
- Toribio, V.; Morales, S.; Lopez-Martin, S.; Cardenes, B.; Cabanas, C.; Yanez-Mo, M. Development of a quantitative method to measure EV uptake. Sci. Rep. 2019, 9, 10522. [Google Scholar] [CrossRef]
- Koudela, B.; Nohynkova, E.; Vitovec, J.; Pakandl, M.; Kulda, J. Giardia infection in pigs: Detection and in vitro isolation of trophozoites of the Giardia intestinalis group. Parasitology 1991, 102 Pt 2, 163–166. [Google Scholar] [CrossRef]
- Keister, D.B. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans. R. Soc. Trop. Med. Hyg. 1983, 77, 487–488. [Google Scholar] [CrossRef]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Willenbrock, H.; Salomon, J.; Sokilde, R.; Barken, K.B.; Hansen, T.N.; Nielsen, F.C.; Moller, S.; Litman, T. Quantitative miRNA expression analysis: Comparing microarrays with next-generation sequencing. Rna 2009, 15, 2028–2034. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, J.; Kim, K.; Chang, H.; You, K.; Kim, V.N. Bias-minimized quantification of microRNA reveals widespread alternative processing and 3’ end modification. Nucleic Acids Res. 2019, 47, 2630–2640. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]

| Isolate | sRNA | Sequence | GiardiaDB-ID |
|---|---|---|---|
| WB/1267 | rsRNA-28S | CCCGCGGAACGGCGUCGG | GL50803_r0021 |
| GS/H7 | rsRNA-28S | CCCGCGGAACGGCGUCGG | GL50803_r0021 1 |
| P15 | rsRNA-28S | CCCGCGGAACGGCGUCGG | GL50803_r0021 1 |
| WB/1267 | tsRNA-Gln | UGGGGCGUGGUGCAGCGGGAGC | GL50803_t0051 |
| GS/H7 | tsRNA-Pro | CUUUGGGUGCGAGAGGCC | GL50581_4624 |
| P15 | tsRNA-Ser | UGGGGCGUGGUGCAGCGGGAGC | GLP15_4341 |
| WB/1267 | msRNA-VSP | CGACUGCCCGAUCGAGAACUGC | GL50803_3327 |
| GS/H7 | msRNA-HP | CUCUUGUAGACCGUCGCU | GL50581_1332 |
| P15 | msRNA-IF | AUGUGAGAGUAGGUCAUC | GLP15_4980 |
| Isolate | sRNA | Sample | Ct Value ± SD |
|---|---|---|---|
| WB/1267 | rsRNA-28S | Trophozoites’ RNA ElVs’ RNA | 33.09 ± 1.89 35.72 ± 1.98 |
| tsRNA-Gln | Trophozoites’ RNA ElVs’ RNA | 26.41 ± 1.70 28.99 ± 1.61 | |
| msRNA-VSP | Trophozoites’ RNA ElVs’ RNA | 29.25 ± 1.71 30.70 ± 1.78 | |
| GS/H7 | rsRNA-28S | Trophozoites’ RNA ElVs’ RNA | 33.50 ± 0.97 35.67 ± 1.31 |
| tsRNA-Pro | Trophozoites’ RNA ElVs’ RNA | - - | |
| msRNA-HP | Trophozoites’ RNA ElVs’ RNA | 35.95 ± 0.53 19.10 ± 0.53 | |
| P15 | rsRNA-28S | Trophozoites’ RNA ElVs’ RNA | 33.50 ± 0.90 33.62 ± 1.05 |
| tsRNA-Ser | Trophozoites’ RNA ElVs’ RNA | 22.46 ± 0.70 29.05 ± 0.37 | |
| msRNA- IF | Trophozoites’ RNA ElVs’ RNA | 36.80 ± 0.37 31.34 ± 2.08 |
| Name | sRNA | Sequence |
|---|---|---|
| R1loop | rsRNA-28S | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCCGA |
| F1 | rsRNA-28S | TAATAAGGCCCGCGGAACG |
| R2loop | tsRNAGly/tsRNASer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTCCC |
| F2 | tsRNAGly/tsRNASer | ATAGTATGGGGCGTGGTGC |
| R3loop | tsRNA-Pro | GTCGTATCCAGTG GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCCTC |
| F3 | tsRNA-Pro | AAGAATTCGCTTTGGGTGCG |
| R4loop | msRNA-VSP | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAGTT |
| F4 | msRNA-VSP | AACTTCCGACTGCCCGATC |
| R5loop | msRNA-HP | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAGCG |
| F5 | msRNA-HP | AACCACTGACTCTTGTAGAC |
| R6loop | msRNA- IF | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCGATG |
| F6 | msRNA- IF | AAGCGACCAGATGTGAGAGTAG |
| Common Reverse Primer | - | GTCGTATCCAGTGCAGGGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Natali, L.; Luna Pizarro, G.; Moyano, S.; de la Cruz-Thea, B.; Musso, J.; Rópolo, A.S.; Eichner, N.; Meister, G.; Musri, M.M.; Feliziani, C.; et al. The Exosome-like Vesicles of Giardia Assemblages A, B, and E Are Involved in the Delivering of Distinct Small RNA from Parasite to Parasite. Int. J. Mol. Sci. 2023, 24, 9559. https://doi.org/10.3390/ijms24119559
Natali L, Luna Pizarro G, Moyano S, de la Cruz-Thea B, Musso J, Rópolo AS, Eichner N, Meister G, Musri MM, Feliziani C, et al. The Exosome-like Vesicles of Giardia Assemblages A, B, and E Are Involved in the Delivering of Distinct Small RNA from Parasite to Parasite. International Journal of Molecular Sciences. 2023; 24(11):9559. https://doi.org/10.3390/ijms24119559
Chicago/Turabian StyleNatali, Lautaro, Gabriel Luna Pizarro, Sofía Moyano, Benjamin de la Cruz-Thea, Juliana Musso, Andrea S. Rópolo, Norbert Eichner, Gunter Meister, Melina M. Musri, Constanza Feliziani, and et al. 2023. "The Exosome-like Vesicles of Giardia Assemblages A, B, and E Are Involved in the Delivering of Distinct Small RNA from Parasite to Parasite" International Journal of Molecular Sciences 24, no. 11: 9559. https://doi.org/10.3390/ijms24119559
APA StyleNatali, L., Luna Pizarro, G., Moyano, S., de la Cruz-Thea, B., Musso, J., Rópolo, A. S., Eichner, N., Meister, G., Musri, M. M., Feliziani, C., & Touz, M. C. (2023). The Exosome-like Vesicles of Giardia Assemblages A, B, and E Are Involved in the Delivering of Distinct Small RNA from Parasite to Parasite. International Journal of Molecular Sciences, 24(11), 9559. https://doi.org/10.3390/ijms24119559






