1. Introduction
G-quadruplexes (G4s) are among the most stable nucleic acid structures and are characterized by a large conformational diversity [
1]. Their existence in vivo and their functional roles are proved [
2] and they are formed by interactions of repetitive G-rich tracts in genomic DNA and RNA sequences [
3]. The folded G4 sequence consists of stacked G-tetrads, creating a central core of four strand segments connected by single-stranded loops. A G-tetrad is a planar arrangement of four guanine residues from different G-tracts linked by Hoogsteen hydrogen bonds. In addition to the endogenous G4s that are almost recognized as integral structures within the non-coding genome and transcriptome [
4], which play a key role in numerous biological processes mainly through their interaction with several cellular proteins [
5,
6], very promising exogenous G4s are known, due to their ability to modulate the activity of different protein target. Indeed, a significant number of aptamers selected by the combinatorial technique Systematic Evolution of Ligands by EXponential enrichment (SELEX) towards a variety of protein targets [
7] adopt G4 structures (G4 aptamers). The remarkable thermal stability of G4s provides them with a moderate resistance to nucleases, which prolongs the permanence of these aptamers in the bloodstream. Due to their advantageous properties, G4 aptamers can be considered an interesting category of molecules with several potential applications in therapeutics, diagnostics, food quality control, environmental monitoring, and drug development and delivery [
8].
Among the G4s endowed with notable biological properties, G4 aptamers can interact with some cellular proteins involved in oncogenesis and cancer progression, and can be considered particularly promising pharmacological agents. A remarkable example is AS1411, a nucleolin-targeting aptamer that reached Phase 2b clinical trials for acute myeloid leukaemia and renal cell carcinoma; however, despite a good tolerance and safety profile, its trial was terminated due to its suboptimal pharmacokinetics (rapid clearance) and low potency [
9,
10]. Further interesting aptamers belonging to this category are T40214 and T40231 (sequences (G
3C)
4 and G
2T(G
3T)
2G
3, respectively), targeting Signal Transducer and Activator of Transcription 3 (STAT3) [
11]. This protein is a key mediator of the oncogenic signaling that is frequently activated in many types of human cancer, where it contributes to tumor cell growth and resistance to apoptosis through the upregulation of genes encoding apoptosis inhibitors (Bcl-xL, Mcl-1, and survivin), cell cycle regulators (cyclin D1 and c-myc), and inducers of angiogenesis (VEGF) [
12]. Indeed, increasing evidence has shown that STAT3 is constitutively activated in 82% of prostate cancers, 70% of breast cancers, over 90% of head and neck cancers, and more than 50% of lung cancers [
13,
14,
15,
16]. In addition, further recent studies have highlighted the prognostic significance of STAT3 levels in invasive breast cancer [
17] and chordomas [
18]. Therefore, STAT3 can be considered a promising target for anticancer drug discovery.
Similarly to other STAT proteins, STAT3 is localized within the cytoplasm of resting cells and activated in response to the binding of several ligands, including cytokines and other factors, to their cognate cell surface receptors (e.g., IL-6R). STAT3 activation generates dimers, which translocate to the nucleus, where they bind to DNA-response elements in the promoters of target genes and activate specific gene-expression programs [
19]. STAT3 activation contributes to the expression of antiapoptosis proteins, such as Bcl-xL and Mcl-1, thus decreasing natural apoptotic cell death and favouring cancer cell growth [
20,
21].
Therefore, targeting the STAT3 protein through high-affinity ligands with the aim of reducing its levels or activity in cancers has noteworthy therapeutic potential. T40214 (
STAT) is a G4 aptamer [(G
3C)
4] that can influence STAT3 biological outcomes in an efficient manner in several cancer cell lines and tumor xenografts [
11,
22], forming a G4 conformation characterized by parallel strands, three G-tetrads and three propeller-shaped loops [
23]. Another G4 aptamer, namely T40231 [G
2T(G
3T)
2G
3], directly targets STAT3 protein and dramatically reduces tumor cell growth by markedly enhancing the apoptosis of prostate and breast cancer cells in which STAT3 is constitutively activated [
24]. Interestingly, a recent study [
25] showed that a further G4-forming aptamer characterized by almost the same sequence as T40231, namely T30923 [(G
3T)
4] (INT), is able to efficiently link the interleukine cell surface receptors (IL-6R) which, in turn, activate STAT3. Therefore, all these G4-forming aptamers are implicated in the same bio-molecular pathway. Furthermore, it should be noted that T30923 adopts a parallel G4 structure strictly resembling that of
STAT [
23] (
Figure 1). Concerning G4 anticancer aptamers, in a recent paper [
26], we have reported, for the first time, the antiproliferative properties of another G4-forming aptamer, namely T30175 [GTG
2(TG
3)
3T] (INTB), whose sequence and structure are closely correlated with those of T30923 [
23,
27,
28]. In fact, their sequences differ only in a T residue, and both adopt a dimeric 5′-5′ end-stacked G4 structure, in which each parallel G4 monomer of the complex contains three G-tetrads and three single-thymidine reversed-chain loops; however, T30175 is characterized by an additional bulge loop formed by the extra thymidine in the second position of the sequence (
Figure 1).
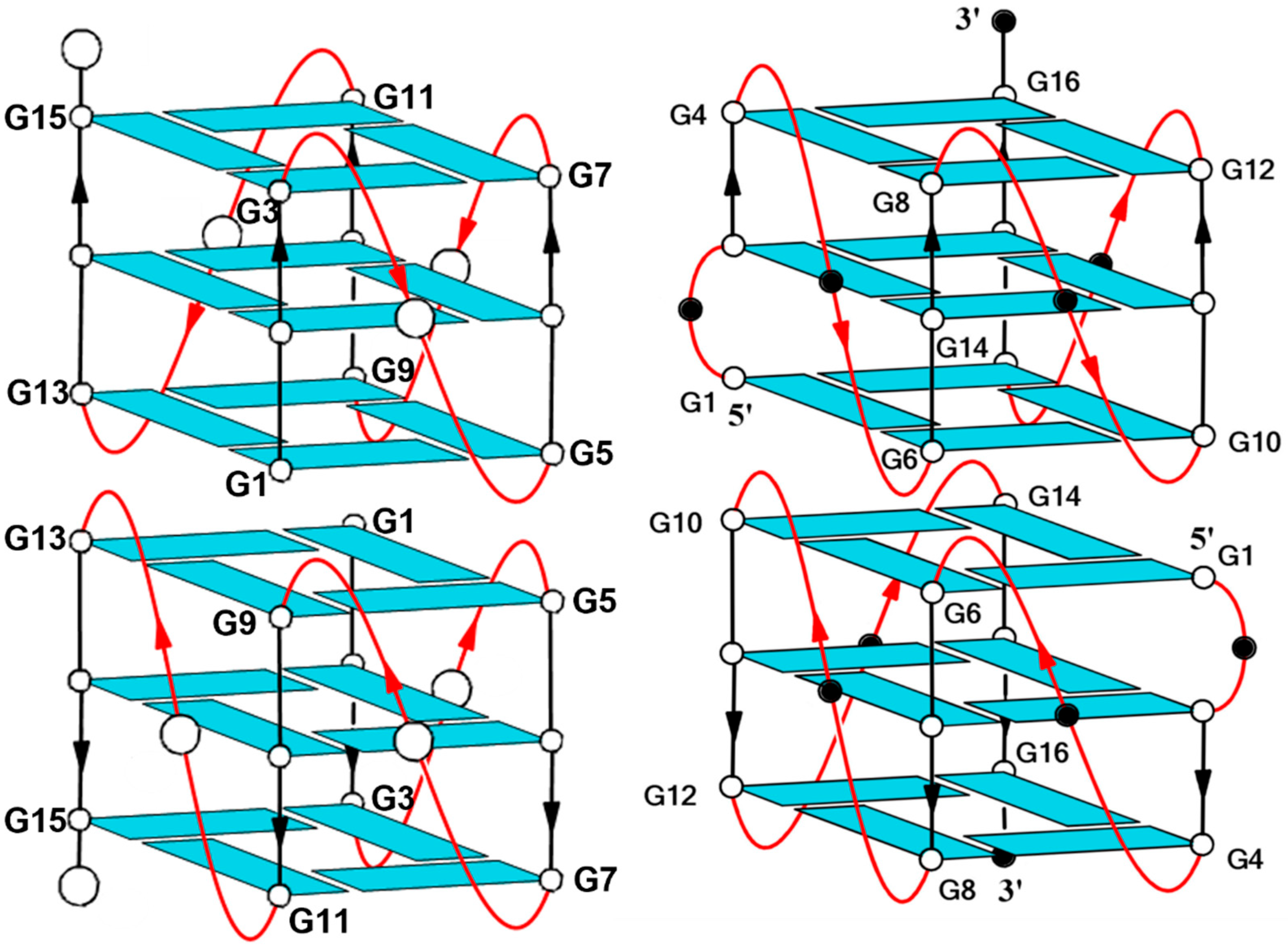
Most G4 aptamers are characterized by peculiar structural features; their stability is principally due to a compact core, formed by stacked G-tetrads, while the loop residues are mostly involved in the interaction with the target protein. Consequently, when designing modified aptamers to modulate their biological properties without affecting their stability, in most cases, modifications are focused on the loop residues directly involved in the aptamer–target interaction. Therefore, to explore the effects of single site-specific replacements of the loop residues in generating aptamers that can affect the STAT3 biochemical pathway, a series of
STAT analogues containing a thymidine (T) residue instead of the single cytidines (C) of the sequence was prepared (
Table 1). Furthermore, a second series of derivatives containing the same single-site modification was synthesized, starting from a new parent aptamer, namely
STATB [GCG
2(CG
3)
3C] (
Table 1). This oligonucleotide sequence differs from that of
STAT only by an extra cytidine in second position, similarly to T30175 versus T30923, and was designed considering the recently identified antiproliferative activity of T30175, whose structure is closely related to that of T30923, which shares the same bio-molecular pathway as
STAT. All derivatives reported in
Table 1 were investigated in terms of their structural and biological properties, with the aim of identifying new aptamers endowed with the ability to affect the biochemical pathway in which different target proteins, namely IL-6R and STAT3, are involved.
3. Discussion
The signal transducer and activator of transcription (STAT) proteins, particularly STAT3, are ideal targets for cancer therapy. STAT signalling is mainly regulated by interleukin-6 (IL-6) and its family members, although new pathways regulating STAT3 in cancer were recently identified [
32]. The continuous activation of the IL-6/STAT3 signaling pathway has been detected in cell proliferation, survival, and invasion of many human cancers, including breast cancer [
31,
33]. Recent data showed that the blockage of the IL-6/STAT3 pathway results in the activation of apoptosis [
34], thus suggesting that new regulators of STAT3 in tumours are important targets for potential therapeutic strategies in the treatment of cancer. Despite the evident potential of STAT3 as a target to obtain antitumour effects, to date, satisfying interventions aimed at inhibiting this protein have yet to be developed, due to the complexity of its biological functions.
Therefore, with the aim of designing new aptamers that can interfere in the biochemical pathway involving different target proteins, namely IL-6R and STAT3, we focused on the G4-aptamer T40214 (
STAT), which was reported to bind to STAT3 with high affinity [
11,
22], and on its similar sequence analogue
STATB, synthesizing two series of derivatives containing a thymidine (T) residue instead of the single cytidines (C) of the loop sequences.
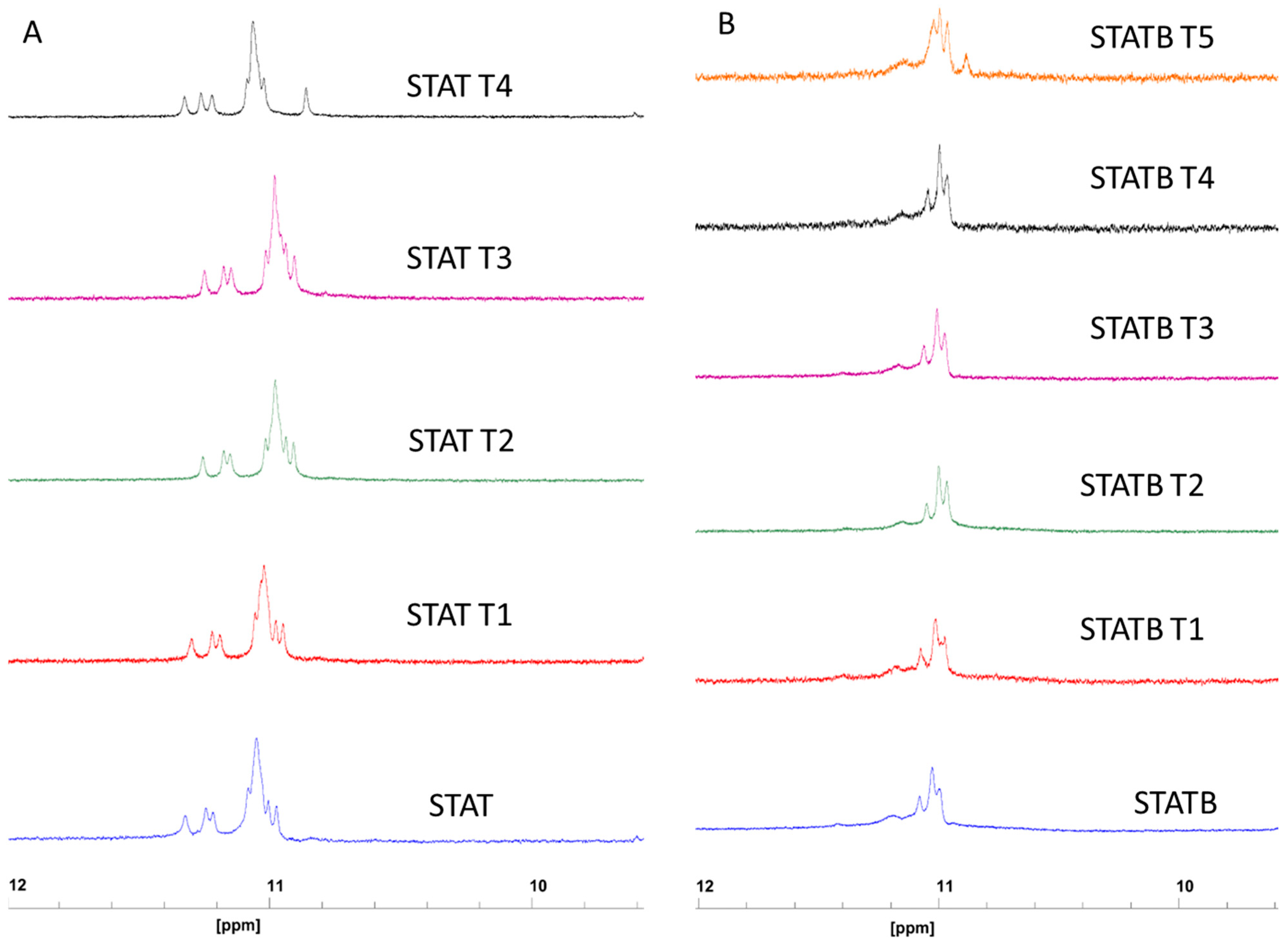
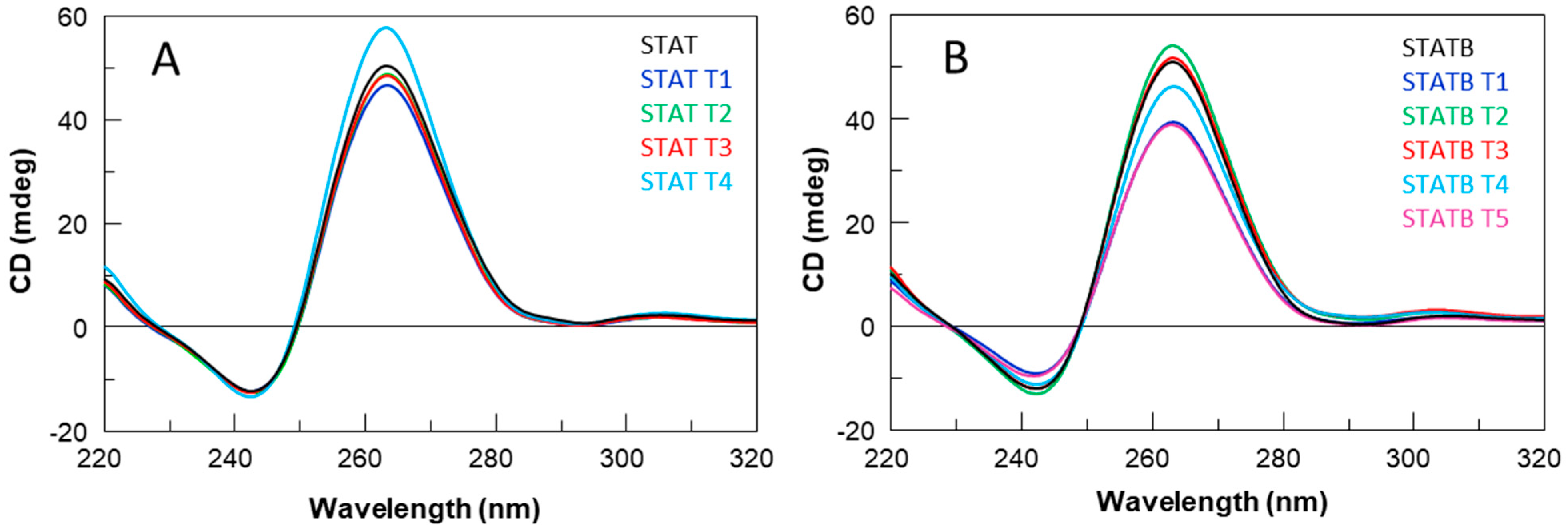
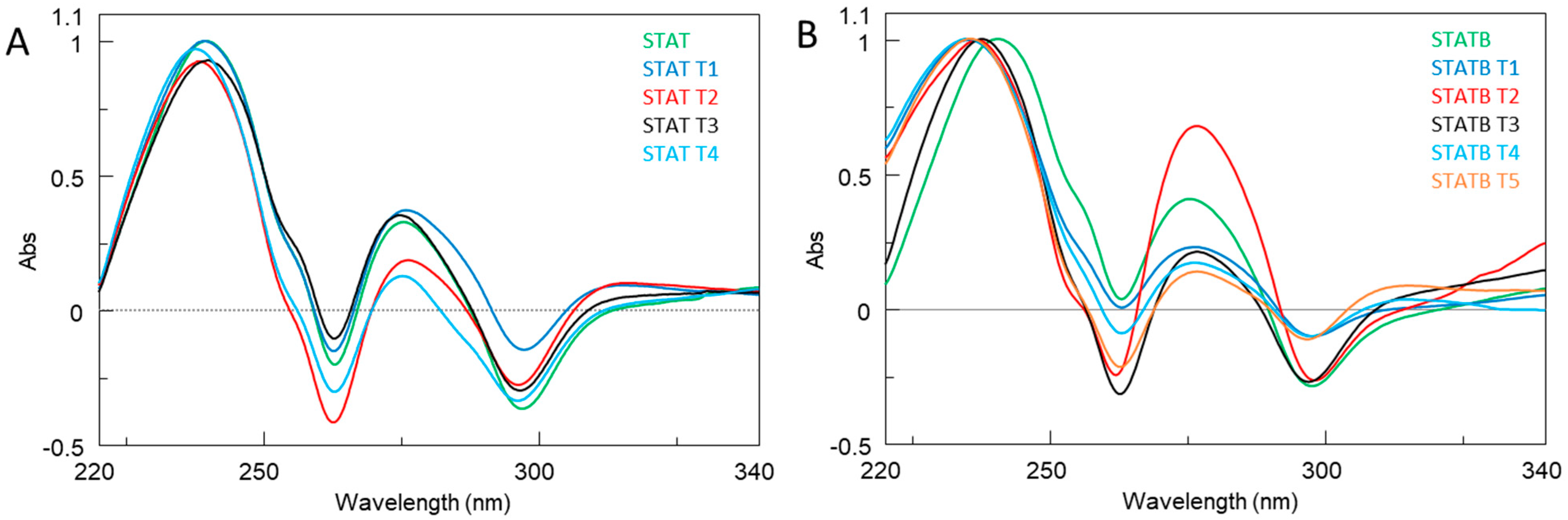
All ODNs were evaluated in terms of their structural and biological properties. The collected NMR, CD, UV, and PAGE data suggest that all derivatives adopt dimeric G4 structures strictly resembling that of the unmodified aptamer T40214, endowed with a higher thermal stability than their parent, STAT and STATB, respectively, thus indicating that the single-site replacement of a C residue with a T not only preserves the original quadruplex structure but significantly contributes to its thermal stability.
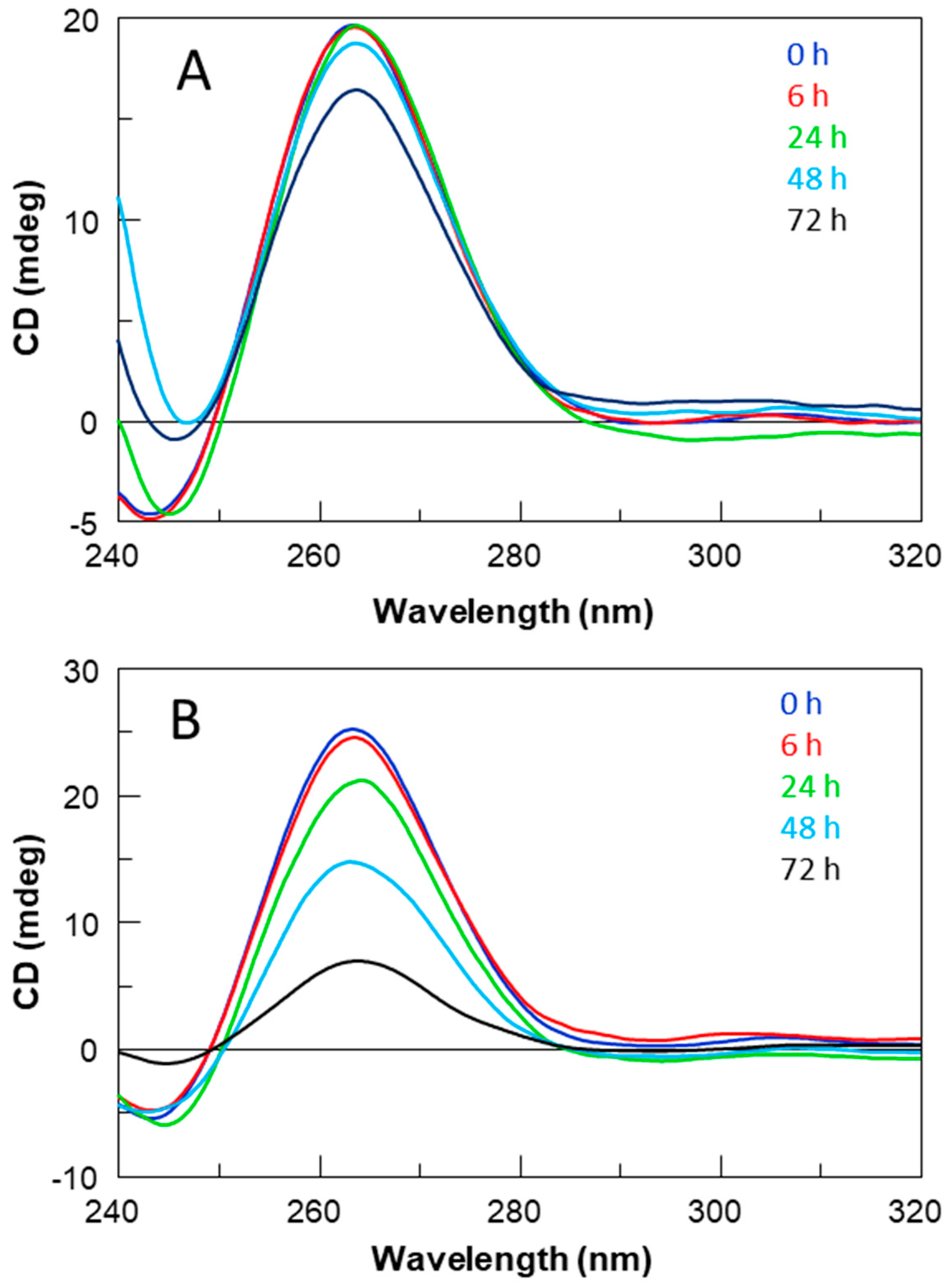
Concerning the ODN biostability, which plays a key role in their applicability in the therapeutic field, STAT series derivatives revealed a notable resistance to serum nucleases against a lower resistance of the STATB series. Single-loop modifications contributed positively, since all modified ODNs showed higher percentages of undegraded species at 72 h than the unmodified sequence, particularly in the STATB series. These data suggest a probable correlation between biostability and thermal stability: the G-quadruplexes endowed with higher thermal stability guarantee a major amount of the folded species that are biologically more stable than the thermally less stable structures in equilibrium with a larger amount of the unstructured species being more susceptible to nucleases.
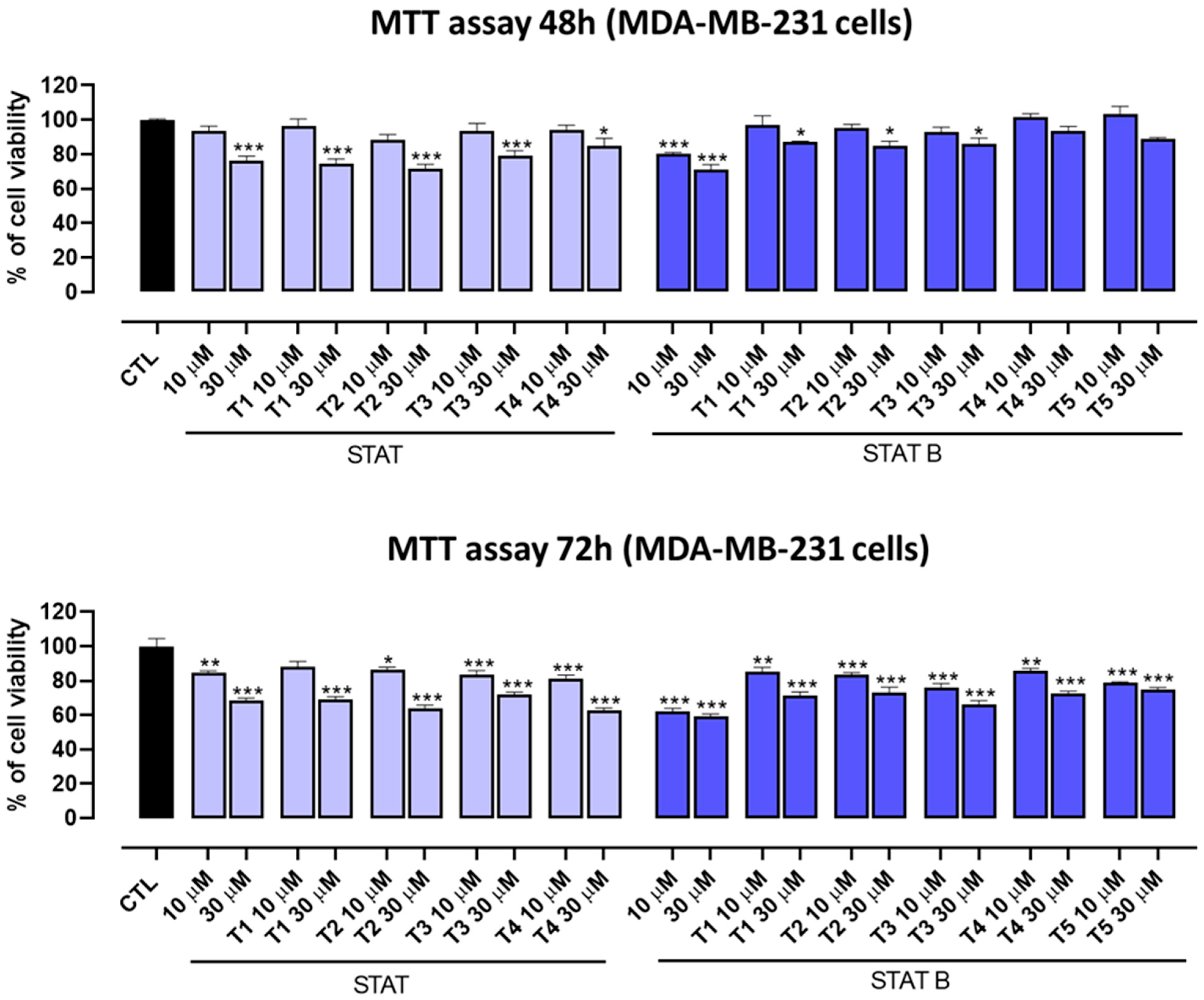
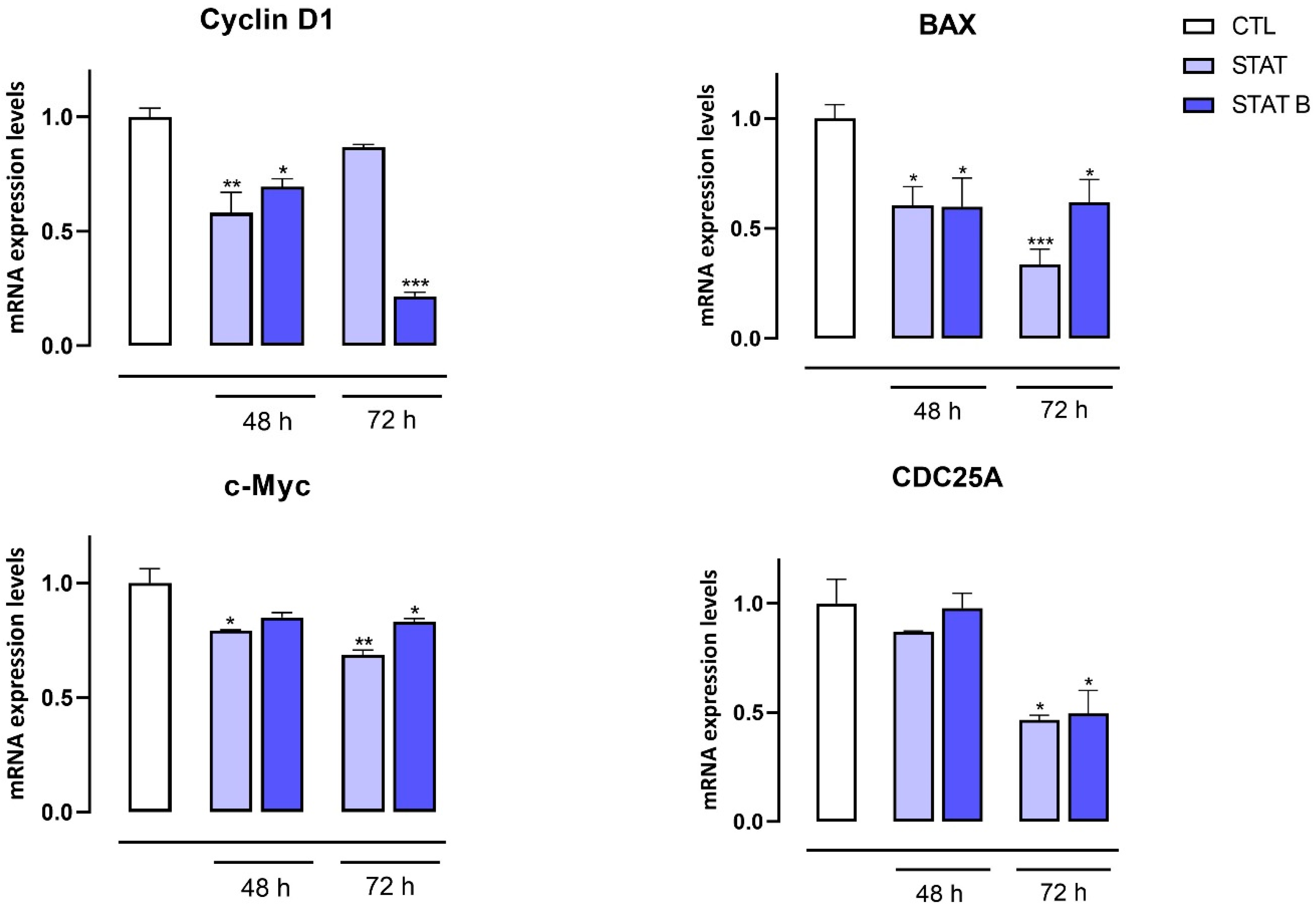
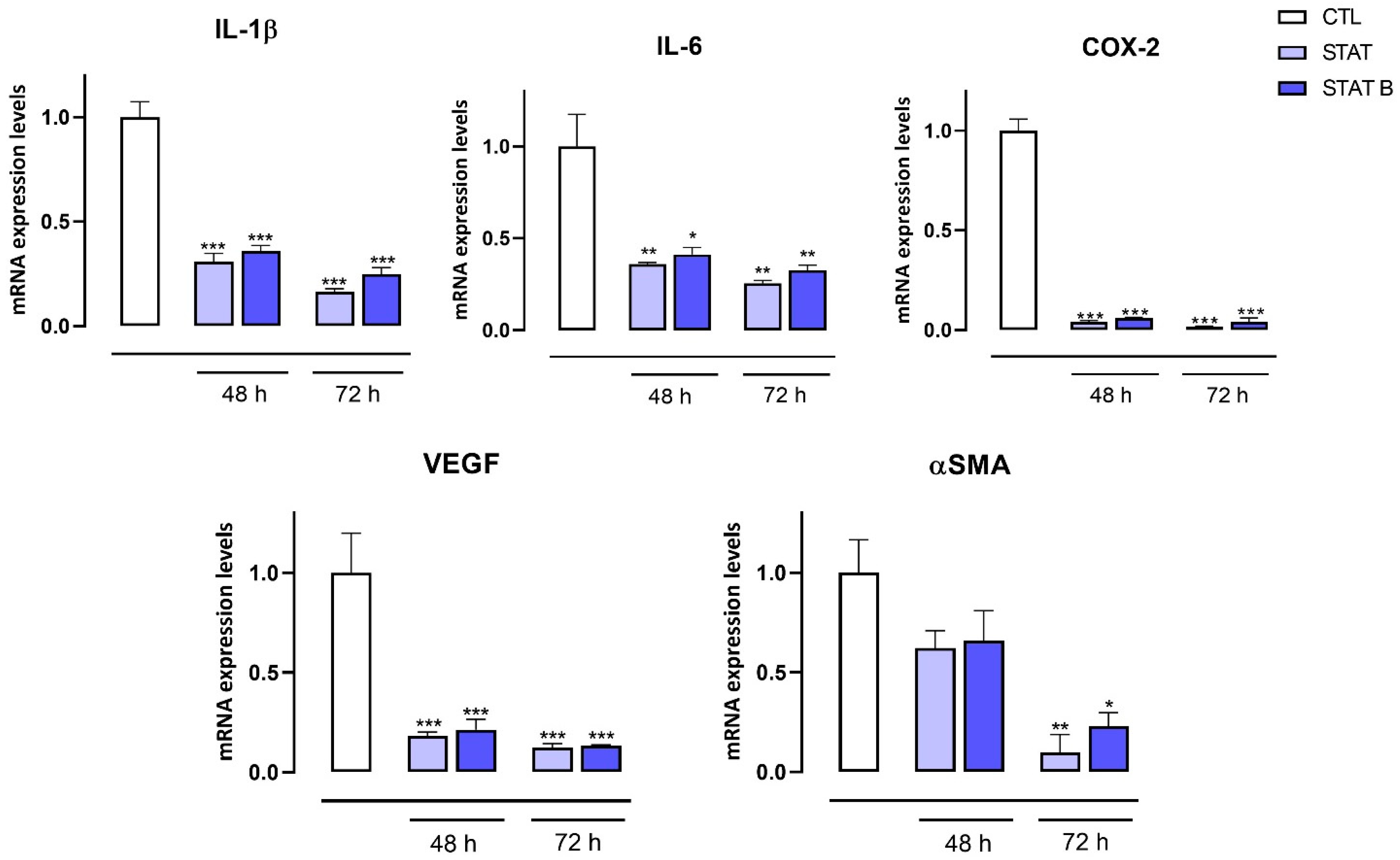
The antiproliferative activity of the investigated ODNs was tested on both human prostate (DU145) and breast (MDA-MB-231) cancer cells. Interestingly, the entire series of STAT and STATB quadruplexes reduced the proliferative capacity of the aforementioned cells, exhibiting different inhibition effects depending on time and concentration. Of note, in MDA-MB-231 cancer cells after 72 h, STAT and its derivatives showed the highest inhibition effects at 30 µM (inhibition percentages ranging from about 25 to 40%). After the same incubation time, the most pronounced antiproliferative activity (40%) was exhibited by STATB, which reached a plateau between 10 µM and 30 µM, showing the greatest efficacy at a low concentration. Next, we evaluated the effect of STAT and STATB quadruplexes on STAT3 transcriptional activity. We were able to demonstrate that both STAT and STATB compounds induce significant changes in the mRNA expression levels of genes regulating key cellular process, such as cell cycle, inflammation, and angiogenesis. In summary, this approach allowed us to demonstrate the efficacy of these novel aptamers targeting STAT3 in cancer cells.
4. Materials and Methods
4.1. Oligonucleotide Synthesis and Purification
The ODNs listed in
Table 1 were synthesized by an ABI 394 DNA synthesizer using solid-phase β-cyanoethyl phosphoramidite chemistry at the 10 µmol scale. The synthesis was carried out using normal 3′-phosphoramidites (Link Technologies, Glasgow, UK). The detachment from the support and the deprotection of the oligomers were carried out by treatment with concentrated aqueous ammonia at 55 °C overnight. The combined filtrates and washings were concentrated under reduced pressure, redissolved in H
2O, analyzed, and purified by high-performance liquid chromatography on a Nucleogel SAX column (Macherey-Nagel, Duren, Germany, 1000-8/46) using buffer A (20 mM NaH
2PO
4/Na
2HPO
4 aqueous solution (pH 7.0) containing 20% (
v/
v) CH
3CN) and buffer B (1 M NaCl, 20 mM NaH
2PO
4/Na
2HPO
4 aqueous solution (pH 7.0) containing 20% (
v/
v) CH
3CN); a linear gradient from 0% to 100% B for 45 min and a flow rate of 1 mL/min were used. The fractions of the oligomers were collected and successively desalted by Sep-pak cartridges (C-18). The isolated oligomers proved to be >98% pure by NMR.
4.2. NMR Spectroscopy
NMR samples were prepared at a concentration of approximately 1 mM in 0.6 mL (H
2O/D
2O 9:1
v/
v) of buffer solution with 10 mM KH
2PO
4/K
2HPO
4, 70 mM KCl, and 0.2 mM EDTA (pH 7.0). All the samples were heated for 5–10 min at 90 °C and slowly cooled (10–12 h) to room temperature. The solutions were equilibrated for several hours at 4 °C. The annealing process was assumed to be complete when the
1H NMR spectra were superimposable on changing time. NMR spectra were recorded at 25 °C by employing a 700 MHz Bruker spectrometer (Bruker-Biospin, Billerica, MA, USA). Proton chemical shifts were referenced to the residual water signal, resonating at 4.78 ppm (25 °C, pH 7.0). Water suppression was achieved using the excitation sculpting with the gradient routine included in the “zgesgp” pulse sequence [
35]. NMR data processing was done by using the vendor software TOPSPIN 4.1.4 (Bruker Biospin Gmbh, Rheinstetten, Germany).
4.3. CD Spectroscopy
CD samples of oligonucleotides reported in
Table 1 were prepared at an ODN concentration of 50 µM using a potassium phosphate buffer (10 mM KH
2PO
4/K
2HPO
4, 70 mM KCl, pH 7.0) and submitted to the annealing procedure (heating at 90 °C and slowly cooling at room temperature). CD spectra of all quadruplexes and CD melting curves were registered on a Jasco 715 CD spectrophotometer (Jasco, Tokyo, Japan). For the CD spectra, the wavelength varied from 220 to 320 nm at 100 nm min
−1 scan rate, and the spectra recorded with a response of 4 s, at 1.0 nm bandwidth and normalized by subtraction of the background scan with buffer. The temperature was kept constant at 20 °C with a thermoelectrically controlled cell holder (Jasco PTC-348). CD melting curves were registered as a function of temperature (range: 20 °C–95 °C) for all G-quadruplexes, annealed as previously reported, at their maximum Cotton effect wavelengths. To test the G-quadruplex thermal stabilities at low potassium concentration, samples of all ODNs were prepared at an ODN concentration of 25 µM, using a potassium phosphate buffer 1 mM KH
2PO
4/K
2HPO
4, 5 mM KCl, pH 7.0) and submitted to the annealing procedure as previously described. CD melting curves were registered as a function of temperature (range: 20 °C–95 °C) for all G-quadruplexes at their maximum Cotton effect wavelengths. The CD data were recorded in a 0.1 cm pathlength cuvette with a scan rate of 30 °C/h.
4.4. UV Thermal Difference Spectra (TDS)
UV samples of investigated oligonucleotides were prepared using a buffer solution: KH2PO4/K2HPO4 (1 mM, pH 7.0), KCl (5 mM). For each oligonucleotide sample, a UV spectrum was recorded above and below its melting temperature (Tm). All experiments were performed on a Jasco V 750 UV/Vis spectrophotometer (Jasco, Tokyo, Japan) using quartz cuvettes with an optical path of 1 cm and at 25 μM strand concentration. Absorbance spectra were recorded in the 220–340 nm range, with a scan speed of 200 nm min−1 and a data interval of 1 nm. The difference between the UV spectra at high (95 °C) and low (20 °C) temperatures was defined as the TDS; this represents the spectral difference between the unfolded and folded forms. The temperature (20 or 95 °C) was kept constant with a thermostatic circulating water bath for cell holders (Jasco CTU-100). The thermal difference spectra were normalized (+1 for the highest positive peak).
4.5. Gel Electrophoresis
All oligonucleotides were analyzed by non-denaturing PAGE. All oligonucleotide samples were prepared at an ODN concentration of 1 mM using a potassium phosphate buffer (10 mM KH2PO4/K2HPO4, 70 mM KCl, pH 7.0) and submitted to the annealing procedure (heating at 90 °C and slowly cooling at room temperature). Each oligonucleotide was loaded on a 20% polyacrylamide gel containing Tris–Borate-EDTA (TBE) 2.5× and KCl 20 mM. The run buffer was TBE 1× containing 50 mM KCl. For all samples, a solution of glycerol/TBE 10× was added just before loading. Electrophoresis was performed at 8 V/cm at a temperature close to 10 °C. Bands were visualized by UV shadowing.
4.6. Nuclease Stability Assay
Nuclease stability assay of all ODNs was conducted in 10% Fetal Bovine Serum (FBS) diluted with Dulbecco’s Modified Eagle’s Medium (DMEM) at 37 °C and studied by CD analysis. Approximately 7 nmol of stock solution of each ODN (~1 O.D.U.) was evaporated to dryness under reduced pressure and then incubated with 250 μL 10% FBS at 37 °C. The degradation patterns were analyzed by monitoring the CD signal decrease in each sample at 37 °C, as a function of time. CD spectra at 0, 6, 24, 48 and 72 h for all ODNs were recorded at 37 °C using a Jasco 715 spectrophotometer equipped with a Peltier temperature control system (Jasco, Tokyo, Japan). Data were collected from 240 to 320 nm with a 1 s response time and a 1 nm bandwidth using a 0.1 cm quartz cuvette. Each spectrum shown is corrected for the spectrum of the reaction medium (10% FBS in DMEM).
4.7. Cell Cultures
Human breast cancer cell line MDA-MB-231 (cat. no. HTB-26) and human prostate cancer cell line DU145 (cat. no. HTB-81) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM (Sigma-Aldrich, Milan, Italy; cat. no. D6546) supplemented with 10% fetal bovine serum (FBS) (Gibco, Milan, Italy; cat. no. A4736301), penicillin (100 U/mL) and streptomycin (100 μg/mL) (cat. no. 30-002-CI), 2 mmol/L L-glutamine (cat. no. 25-005-CI) and 0.01 M HEPES buffer (cat. no. 25-060-CI) (all from Corning, Manassas, VA, USA) and placed at 37 °C in a humidified incubator containing 5% CO2.
4.8. MTT Assay
Cell proliferation was measured by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. MDA-MB-231 and DU145 cells were seeded on 96-well plates (6 × 103 cells/well). After 24 h, cells were treated with STAT, STAT T1, STAT T2, STAT T3, STAT T4, STATB, STATB T1, STATB T2, STATB T3, STATB T4 and STATB T5 (10 and 30 µM) for 48 and 72 h before adding MTT (cat. M5655, Merk, Italy) (final concentration 5 mg/mL in PBS). Cells were incubated for an additional 3 h at 37 °C. After this time, cells were lysed, and formazan salts resulting from MTT reduction were solubilized with a solution containing 50% (v/v) N,N-dimethyl formamide, 20% (w/v) sodium dodecylsulfate with an adjusted pH of 4.5. The absorbance was measured using a microplate spectrophotometer (Thermo Scientific Multiskan GO, Thermo Fisher Scientific, Waltham, MA, USA) equipped with a 570-nm filter.
4.9. RNA Purification and Quantitative Real-Time PCR
MDA-MB-231 and DU145 cells were seeded (106 cells/well) in 100 mm culture dishes and, after 24 h, treated or not with STAT and STATB (30 µM) for 48 and 72 h. Total RNA was isolated from cells using the QIAzol Lysis Reagent according to the manufacturer’s instructions (Cat. 79306, Qiagen, Hilden, Germany). The purity and quantity of each purified RNA were evaluated considering the ratio between readings at 260/280 nm using an Eppendorf BioPhotometer and Nanodrop apparatus (Thermo Fisher Scientific, MA, USA). Purified mRNA was reverse-transcribed using iScript Reverse Transcription Supermix for RT-qPCR (cat. 1708841, Bio-Rad, Segrate (MI), Italy). qPCR was carried out in a CFX96 real-time PCR detection system (Bio-Rad) with the use of specific primers (Cyclin D1 5′-GCTGCGAAGTGGAAACCATC-3′, 5′-CCTTCTGCACACATTTGAA-3′; BAX 5′-CCCGAGAGGTCTTTTTCCGAG-3′, 5′-CCAGCCCATGATGGTTCTGAT-3′; c-Myc 5′-GGCTCCTGGCAAAAGGTCA-3′, 5′-CTGCGTAGTTGTGCTGATGT-3′; CDC25A 5′-CTCCTCCGAGTCAACAGATTCA-3′, 5′-CAACAGCTTCTGAGGTAGGGA-3′; IL-1β 5′-ATGATGGCTTATTACAGTGGCAA-3′, 5′-GTCGGAGATTCGTAGCTGGA-3′; IL-6 5′-AGACAGCCACTCACCTCTTCAG-3′, 5′-TTCTGCCAGTGCCTCTTTGCTG-3′; COX-2 5′-TAAGTGCGATTGTACCCGGAC-3′, 5′-TTTGTAGCCATAGTCAGCATTGT-3′; VEGF 5′-AGGGCAGAATCATCACGAAGT-3′, 5′-AGGGTCTCGATTGGATGGCA-3′; αSMA 5′-CTATGCCTCTGGACGCACAACT-3′, 5′-CAGATCCAGACGCATGATGGCA-3′) and SYBR Green master mix kit (cat. 1725271, Bio-Rad). Real-Time PCR cycling protocol was: polymerase activation and DNA denaturation 95 °C for 30 s, amplification 40 cycles, denaturation 95 °C for 15 s, annealing and extension 60 °C for 30 s, plate read at 60 °C, melt-curve analysis 65–95 °C 0.5 °C increment, 5 s/step. The housekeeping gene (ribosomal protein S16) was used as an internal control to normalize the CT values using the 2−ΔΔCt formula.
4.10. Statistical Analysis
Data were expressed as mean ± SEM of n = 3 experiments. Data were analyzed and presented using GraphPad Prism 8.2.0 software (San Diego, CA, USA). Significance was determined using Student’s two-tailed t-test. Results were considered significant at p values less than 0.05 and are labelled with a single asterisk. In addition, p-values lower than 0.01 and 0.001 are indicated with double and triple asterisks, respectively.
5. Conclusions
In summary, we investigated the structural and biological features of two series of G4 forming oligonucleotides, STAT and STATB sequences, and their respective analogues, which contain a single site-specific replacement of the C loop residues with a T one. This study aims to identify new aptamers modulating their biological properties, particularly their ability to affect the IL6/STAT3 biochemical pathway, without affecting their stability. The evaluation of the structural features indicated that all derivatives preserve the ability to fold in very stable G4 structures that are strictly similar to that of the unmodified aptamer T40214. Furthermore, the antiproliferative activities against MDA-MB-231 and DU145 cancer cell lines, and biological degradability of STAT and STATB series were analyzed. The most interesting antiproliferative effects were registered at 72 h at 30 µM; under these conditions, all investigated ODNs showed a similar biological activity on both cell lines, revealing a marked antiproliferative power. The results concerning the nuclease stability indicated that, after 72 h in fetal bovine serum, STAT series derivatives mostly retain their structures, while the STATB series analogues show a lower percentage of undegraded species. Anyway, their nuclease resistance, to some extent, suggests that the structured species for all ODNs, which are probably most responsible for the antiproliferative activity, persist over time. Finally, a transcriptomic analysis aimed to evaluate STAT and STATB’s influence on the expression of a large plethora of genes in MDA-MB-231 cells, suggested the potential involvement of both aptamers in the modulation of IL-6/STAT3 pathway, leading to their interfering in different biological processes. These data provide new tools that could affect an interesting biochemical pathway and help to develop novel anticancer and anti-inflammatory drugs.