Abstract
The efflux pumps, beside the class D carbapenem-hydrolysing enzymes (CHLDs), are being increasingly investigated as a mechanism of carbapenem resistance in Acinetobacter baumannii. This study investigates the contribution of efflux mechanism to carbapenem resistance in 61 acquired blaCHDL-genes-carrying A. baumannii clinical strains isolated in Warsaw, Poland. Studies were conducted using phenotypic (susceptibility testing to carbapenems ± efflux pump inhibitors (EPIs)) and molecular (determining expression levels of efflux operon with regulatory-gene and whole genome sequencing (WGS)) methods. EPIs reduced carbapenem resistance of 14/61 isolates. Upregulation (5–67-fold) of adeB was observed together with mutations in the sequences of AdeRS local and of BaeS global regulators in all 15 selected isolates. Long-read WGS of isolate no. AB96 revealed the presence of AbaR25 resistance island and its two disrupted elements: the first contained a duplicate ISAba1-blaOXA-23, and the second was located between adeR and adeA in the efflux operon. This insert was flanked by two copies of ISAba1, and one of them provides a strong promoter for adeABC, elevating the adeB expression levels. Our study for the first time reports the involvement of the insertion of the ΔAbaR25-type resistance island fragment with ISAba1 element upstream the efflux operon in the carbapenem resistance of A. baumannii.
1. Introduction
Acinetobacter baumannii is a non-fermentative, Gram-negative coccobacillus, responsible for numerous opportunistic and nosocomial infections. It contributes to a variety of disease states, including ventilator-associated pneumonia (VAP), meningitidis, bloodstream, skin and soft tissues infections. It has been implicated in wound and surgical site contamination, as well as contributing to catheter-borne infections amongst others []. During of the SARS-CoV-2 (COVID-19) pandemic, carbapenem-resistant A. baumannii (CRAB) was a leading cause of VAP in SARS-CoV-2 infected patients []. A. baumannii is resistant to treatment by many antibiotics due to its intrinsic and acquired resistance mechanisms. Current treatment strategies include certain aminoglycosides, tigecycline, colistin, β-lactams with sulbactam and the “last-resort” antibiotics—carbapenems []. Of major concern is the increased incidence of carbapenem resistance, which has risen dramatically over the last few decades. According to the European Centre for Disease Prevention and Control report, in 2020, laboratories from 29 EU/EEA (European Union/European Economy Area) countries reported the detection of 7622 invasive isolates belonging to the genus Acinetobacter, of which 7542 (99%), showed resistance to carbapenems []. The multidrug resistance (MDR) of the phenotype of hospital strains to a wide spectrum of antibiotics and chemotherapeutic agents, including carbapenems, is now a very significant public health problem. In the last six years, only two new drugs (eravacycline and cefiderocol—siderophore cephalosporin) which showed activity against A. baumannii have been approved by the European Medicines Agency (EMA) [].
Intrinsic carbapenem resistance mechanisms in A. baumannii include production of chromosomally encoded carbapenemases [], decreased outer membrane permeability [] and the presence of various efflux systems [] combined with genomic plasticity and ability to easily adapt to more demanding conditions by modulating gene expression can give rise to an extensively drug-resistant isolates (XDR) with no therapeutic options []. By far, the most common carbapenemases in A. baumannii are the carbapenem-hydrolysing class D enzymes (CHDLs). The CHDL enzyme families identified in A. baumannii are intrinsic OXA-51-like and acquired OXA-23-like, OXA-24-like, OXA-58-like, OXA-143-like and OXA-235-like [,]. Apart from OXA-type enzymes, A. baumannii strains have been found to contain plasmids carrying other carbapenemases, such as metallo-β-lactamases (MBLs), e.g., NDM-1 or class A serine carbapenemases including Klebsiella pneumoniae carbapenemases (KPCs) []. However, similar to other Gram-negative rods, carbapenem resistance is often a result of multiple resistance mechanisms including efflux pumps [].
The following superfamilies of multidrug resistant (MDR) efflux pumps have been identified in A. baumannii so far: RND (resistance-nodulation-cell division), MATE (multidrug and toxic compound extrusion), MFS (major facilitator superfamily) and SMR (small multidrug resistance) []. Pumps belonging to RND family are ubiquitous in A. baumannii and demonstrate the broadest substrate range, including antibiotics, chemotherapeutics, non-antibiotic drugs, dyes, biocides, detergents and antiseptics [,]. The overproduction of the MDR efflux pumps is associated with drug resistance. AdeABC, AdeIJK and AdeFGH are the three major RND efflux systems which have been identified in A. baumannii []. It has been shown that overproduction of AdeABC is responsible for reduction of susceptibility to aminoglycosides, fluoroquinolones and tigecycline amongst clinical strains [,]. Its substrate range also encompasses chloramphenicol and β-lactams, including carbapenems [,]. This efflux system is composed of a tripartite proteins assembly spanning the inner and outer membranes: the AdeA membrane fusion component, the AdeB multidrug transporter and the AdeC outer membrane component []. Amongst the three mentioned RND efflux systems, only AdeABC has been implicated in carbapenem resistance [,,]. AdeABC as well as other efflux systems are encoded by genes organized in operons that are located in bacterial chromosomes [].
Two-component regulatory systems (TCSs) function as an important mechanism which enables bacteria to recognize, response and adapt to various environmental stimuli [,,,,]. AdeRS is a local TCS which regulates the expression of the adeABC operon [,,]. The gene encoding AdeRSs are located immediately upstream and are divergently transcribed to adeABC []. AdeS is the sensor kinase which, in response to environmental stimulus autophosphorylates, transfers the phosphate group to the response regulator AdeR []. Between adeRS and adeABC operons, there is a 133 bp intercistronic spacer with a direct-repeat motif to which AdeR binds, regulating expression of adeABC []. Changes in adeRS, including substitutions, deletions and insertions may lead to overexpression of adeABC operon []. However, AdeABC overproduction is not determined only by the mutations within AdeRS sequences. Lin et al. [] suggested that regulation of adeABC expression is controlled by another TCS—BaeSR. It is a global TCS and responses to environmental stress stimuli. It was shown that BaeSR contributed to regulation of adeABC expression by influencing susceptibility to tigecycline, an antibiotic whose susceptibility is also governed by adeABC overexpression []. However, the relationship between BaeSR and AdeABC, is not yet clear and needs further investigation.
AbeM (MATE family) was suggested as another efflux pump which may influence carbapenem susceptibility []. Apart from carbapenems, it also has fluoroquinolones, gentamicin, doxorubicin and triclosan within its substrate range []. AbeM is a prevalent pump in A. baumannii and can be found in up to 98% of isolates [].
In our previous paper, we performed phenotypic and molecular characterisation of the collection of 61 imipenem-non-susceptible A. baumannii clinical isolates, focusing on carbapenemases and insertion sequences (ISs). That study revealed the presence of blaOXA-24-like in 39/61 isolates, ISAba1-blaOXA-23-like in 14/61 isolates and ISAba3-blaOXA-58-like in 6/61 isolates []. The location of an IS upstream of the blaCHDL gene should lead to the overproduction of CHDL enzyme []. However, when performed on strains where the IS preceded blaCHDL, the CarbAcineto NP Test revealed non-obvious and uninterpretable levels of carbapenemase activity. High carbapenem MIC values together with the uninterpretable CarbAcineto NP test results for the isolates with acquired blaCHDL genes suggested additional resistance mechanisms affecting carbapenem activity and prompted us to investigate the contribution of efflux pumps [].
The objective of this study was to determine the impact of the most important efflux system, AdeABC, and additionally the AbeM pump on carbapenem resistance in clinical isolates of acquired blaCHDL-gene-carrying A. baumannii. The goal was to be achieved by relying on phenotypic (susceptibility testing to carbapenems with efflux pump inhibitors (EPIs)) and molecular methods (determining expression levels of adeB pump gene with adeSR regulatory genes, whole genome sequencing and analysing changes of the following genes: abeM, baeSR and adeSRABC, with special attention to genetic environment of the latter).
2. Results
2.1. Susceptibility Profiles and EPI Effects on Carbapenem Resistance
The susceptibility results of the tested 61 isolates to eight antibiotics (meropenem, imipenem, cefepime, ceftazidime, ciprofloxacin, levofloxacin, gentamicin and tobramicin) that are substrates of the AdeABC and AdeM efflux pumps are presented in Table S1 in the Supplementary Materials. The results showed that 100% of isolates were resistant to meropenem, and 98.4% (60/61) to imipenem. One isolate was intermediately resistant to imipenem. At the same time, 93.4% isolates showed resistance to ceftazidime, and the remaining 6.6% showed intermediate susceptibility to it. More than 50% were susceptible and/or intermediately susceptible to cefepime. All the isolates were ciprofloxacin resistant, and about 50% showed resistance to levofloxacin. About 75% of the isolates were resistant to aminoglycosides gentamicin and tobramycin.
The MIC values of imipenem and meropenem were evaluated for all 61 isolates in the presence and the absence of two efflux pump inhibitors, Phe-Arg-β-naphthylamide (PAβN) and Carbonyl cyanide 3-chlorophenylhydrazone (CCCP), to determine the potential role of efflux pumps in carbapenem resistance (Table 1, Table S2 in the Supplementary Materials). At least a 4-fold reduction in meropenem MIC values in the presence of PAβN was observed for 21% (13/61) of studied isolates, whereas the addition of CCCP did not produce any effect on meropenem MIC in any of the isolates tested. Over 50% (7/12) of isolates which exhibited a meropenem MIC reduction in the presence of PAβN carried the ISAba1-blaOXA-23-like gene. The remaining PAβN-affected strains consisted of one ISAba3-blaOXA-58-like-carrying isolate and four blaOXA-24-like-carrying isolates. Imipenem MIC values were significantly reduced in the presence of PAβN in only three isolates (two isolates with a ISAba1-blaOXA-23-like gene and one with a blaOXA-24-like gene). CCCP reduced imipenem MICs in only two isolates, one isolate belonged to the blaOXA-24-like group and one isolate to the ISAba3-blaOXA-58-like-possessing group. It should be emphasized that only 6.6% (4/61) of isolates demonstrated simultaneous imipenem and meropenem MIC reduction in the presence of at least one inhibitor. A summary of the MIC reduction of both carbapenems after the addition of EPIs and the presence of genes encoding the acquired CHDL enzymes and the activity of carbapenemases in the CarbAcineto NP assay [] for all 61 tested isolates are presented in Table S2 in the Supplementary Materials.

Table 1.
Effect of CCCP and PAβN on the carbapenem MIC values among the blaCHDL-carrying A. baumannii isolates (n = 14).
2.2. Presence and Expression of Efflux Pump Genes
PCR was used to show that all 61 A. baumannii clinical isolates contained genes coding the AdeB and AbeM efflux pumps.
In order to determine the contribution of carbapenemase and efflux activity upon carbapenem resistance, three aspects of the A. baumannii isolates were taken into account when selecting isolates for further analysis: OXA-type production, CarbAcineto NP test result [] and EPIs impact on the meropenem MIC value. This resulted in a group of 15 isolates selected for further testing. Isolates were divided into three groups depending on the type of blaOXA gene carriage: (I) ISAba3-blaOXA-58-like (the number of selected isolates is two (n = 2), (II) ISAba1-blaOXA-23-like (n = 7), (III) blaOXA-24-like (n = 6). Each group consisted of isolates with and isolates without at least a 4-fold reduction in meropenem MIC value observed in the presence of PAβN.
To define the role of AdeABC and its regulators AdeRS in carbapenem resistance in the selected 15 isolates, expression of adeB, adeR and adeS was analysed by quantitative PCR (qPCR). For each isolate, a significant (at least a 3-fold) upregulation of adeB was observed (from 5- to 67-fold) compared with A. baumannii ATCC 17978 which was used as a baseline (Table 2). Isolates with the highest expression level of adeB (49-, 58- and 67-fold) all belonged to the ISAba1-blaOXA-23-like group (isolate nos. AB129, AB185 and AB96, respectively), whilst no upregulation (>2-fold) of adeSR regulatory genes was observed. However, for two isolates (no. AB87 and AB165), a significant (between three- and four-fold) upregulation of adeR was observed (Table 2). Moreover, in the case of seven other isolates (nos. AB43, AB86, AB118, AB76, AB81, AB159 and AB176), a 2-fold increase in adeR expression was obtained. Only one isolate (A. baumannii 165) showed upregulation of the adeS gene expression.

Table 2.
Characteristics of A. baumannii isolates (n = 15) selected for further analysis focusing on expression levels of adeB, adeR and adeS genes, as well as on changes in efflux pump proteins and their regulators in relation to the effect of PAβN on carbapenem MIC values.
2.3. Amino Acid Sequence analysis of Efflux Pumps and their TCS Regulators
All 15 isolates were short-read whole genome sequenced (WGS) and screened for mutations of adeRS, adeA, adeB, baeS, baeR and abeM. Carbapenem-sensitive A. baumannii ATCC 17978 was used as a reference for sequence comparison and identification of single-nucleotide-polymorphisms (SNPs). Sequence analysis of adeSR revealed nucleotide changes in adeS in all isolates compared to the reference isolate, which resulted in changes in 7 amino acids located in positions 172, 186, 268, 303, 7348, 356 and 357 (Table 2, Figure 1 and Figure S1 in the Supplementary Materials). Furthermore, all isolates were missing four terminal amino acids: E358, E359, I360 and G361. In addition, in three isolates (nos. AB96, AB129 and AB185), there was a deletion of two amino acids (H102-G103) in AdeS.

Figure 1.
Sequence comparison of the adeS gene of clinical isolate A. baumannii 96 (in red font) and reference strain A. baumannii ATCC 17978 (in black font). The nucleotide positions where the change occurred are underlined. Nucleotide mutations that led to changes in amino acids (substitution and deletion) are shown in the green boxes. The positions of the amino acid changes are given according to the AdeS protein of the reference strain. The STOP codon is shown in the yellow box.
The same three isolates nos. AB96, AB129 and AB185 have an ISAba1 insertion within adeR gene sequence, which results in a truncated AdeR protein. All 15 isolates had SNPs mutations leading to the amino acid changes in positions 120 and 136 in AdeR sequence when compared to A. baumannii ATCC 17978. Isolate no. AB165 had an additional amino acid substitution in position 115 (Table 2). The nucleotide sequence of adeR of isolate no. AB165 in comparison to the reference strain A. baumannii ATCC 17978 is presented in Figure 2.

Figure 2.
Sequence comparison of the adeR gene of clinical isolate A. baumannii 165 (in red font) and reference strain A. baumannii ATCC 17978 (in black font). The nucleotide positions where the change occurred are underlined. Nucleotide mutations that led to changes in amino acids are shown in the green boxes. The positions of the amino acid changes are given according to the AdeS protein of the reference strain.
Nucleotide sequence analysis of adeAB revealed changes in the amino acid sequence of AdeA in three positions K25E, K352R and T391A in each studied isolate. As for the global TCS, a mutation was found that caused one change in amino acid position S437T in BaeS in all isolates. There were no changes in amino acid sequence of AdeB and BaeR in any of the clinical isolates. In addition, only A. baumannii 185 had truncated AbeM protein due to mutation L434STOP. The whole genome datasets of the 15 strains generated and analysed during the current study were previously deposited in the NCBI GeneBank (Submission ID: SUB9082120, BioProject ID:PRJNA701882).
2.4. Core-Genome SNP Phylogenetic Analysis
To explore more how closely are related all the selected 15 A. baumannii isolates, they were genotyped by the core-genome SNP-based phylogenetic analysis. This study revealed the selected isolates to form three clusters (Figure 3). Due to the obtained results of the short-read WGS analysis and thus the similarities of changes in the adeSR-adeABC region, the most interesting were isolates nos. AB96, AB129 and AB185. The use of A. baumannii 96 as a reference allowed the most visible graphical presentation of the genetic diversity of the tested isolates and showed that it clusters together with isolates nos. AB129 and AB185, all of which showed the same nucleotide sequence changes in adeRS.
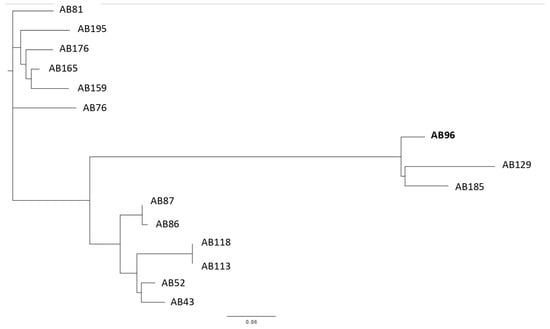
Figure 3.
Core genome SNP-based phylogenetic tree analysis of A. baumannii clinical isolates (n = 15), where isolate A. baumannii 96 (AB96) was used as a reference.
2.5. Analysis of Genetic Structure of adeRS and adeABC Region in Isolate A. baumannii 96
Short-read WGS and bioinformatic analysis of adeSR-adeABC operon in all 15 isolates revealed disruption of this region, resulting in truncated adeR and an ISAba1 insertion upstream of adeA in the three isolates from the same cluster (nos. AB96, AB129 and AB185). A. baumannii 96 was selected for further genetic analysis using nanopore sequencing. This isolate showed the highest upregulation of the adeB gene (67-fold) and resistance to both carbapenems (MIC = 32 mg/L for meropenem and imipenem) and a reduction (4-fold) in the MIC value of imipenem in the presence of PAβN. The comparison of the obtained final genomic sequence of isolate no. AB96 with the database of sequences deposited in NCBI GenBank revealed the presence of a complete sequence of a resistance island AbaR25 (46.4 kb) and located elsewhere in the genome, two disrupted elements of mentioned island: 13.5 kb long first element which will now be referred to as Δ1AbaR25-fragment and 16.5 kb long second element referred to as Δ2AbaR25-fragment. The first element of this island, Δ1AbaR25-fragment, was found in the region between the adeR and adeA genes (Figure 4). This insertion resulted in both a truncated AdeR protein and loss of the greater part of intercistronic spacer with the AdeR binding site. The ISAba1 located upstream of adeA was oriented in the opposite direction providing a strong promoter for the adeABC operon. Comparison of Δ1AbaR25-fragment with AbaR25 revealed several nucleotide changes” a substitution within tniC which resulted in new protein TniC1 (E174K), double nucleotide deletion ΔTC within tniA which resulted in the presence of two open reading frames tniAΔ1 and tniAΔ2, single nucleotide deletion ΔC within tniE which resulted in a truncated TniE protein (ΔTniE), double nucleotide deletion ΔAG within sup which resulted in two open reading frames supΔ1 and supΔ2. The second element of this island, Δ2AbaR25-fragment, was present in another place in the genome of isolates no. AB96. This fragment had Tn2006 carrying blaOXA-23-like gene, which led to the duplication of the ISAba1-blaOXA-23-like region in isolate no. AB96’s genome. This genome sequence is available at the NCBI BioProject repository (Submission ID: SUB12923155, BioProject ID: PRJNA939738).
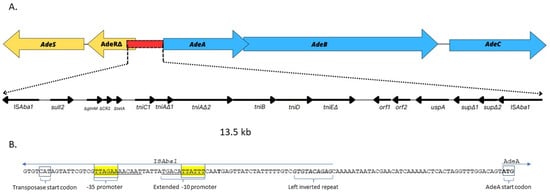
Figure 4.
Genetic structure of adeRS and adeABC region in isolate A. baumannii 96. (A) Scheme of structure of adeABC-adeRS disrupted by Δ1AbaR25-fragment, i.e., 13.5 kb-long fragment of the AbaR25 resistance island, inserted between adeRΔ and adeA. (B) Nucleotide sequence of region upstream of adeA containing the promotor of ISAba1 which flanks Δ1AbaR25-fragment. The genes and open reading frames are shown by labelled arrows, and the arrowheads indicate the direction of transcription.
3. Discussion
Previous studies have shown that carbapenems are substrates for two pumps AdeABC and AbeM in A. baumannii [,]. Roy et al. confirmed the ability of several carbapenems (imipenem, meropenem, doripenem and ertapenem) to bind to the active site in AdeB using a sequence-structure based in-silico approach []. The ability of EPI PAβN to bind to AdeB and its activity as a competitive inhibitor of this efflux pump were also confirmed []. Our study sheds new light on the involvement of efflux pumps in carbapenem resistance in A. baumannii, regardless of the presence and activity of various acquired CHDL enzymes. All 61 isolates used in this study were previously phenotypically and molecularly characterized for carriage of various acquired blaCHDL genes []. The contribution of efflux to carbapenem resistance was investigated in a subset of these strains in this study. Phenotypic tests detecting the contribution of efflux pumps in drug resistance in several Gram-negative bacteria use EPIs such as PAβN and CCCP [,,,,]. At least a 4-fold reduction in antibiotic MIC values in the presence of EPI indicates efflux-related resistance [,,]. PAβN is a specific competitive inhibitor for efflux pumps belonging to the RND family [,]. The second inhibitor CCCP is widely and successfully used in Acinetobacter efflux studies with different pumps substrates such as tigecycline, ciprofloxacin, but also meropenem [,,] and imipenem [,]. In our study the reduction of carbapenem MIC in the presence of EPIs was observed for only a few isolates, regardless of the acquired CHDL enzymes they produced. The production of CHDL carbapenemases was previously confirmed by the CarbAcineto NP test for all of these isolates, except for the OXA-58-positive isolate no. AB43 []. It may suggest that increased efflux is the leading carbapenem resistance mechanism in this isolate.
AdeABC, belonging to RND family, is the most important MDR efflux pump in A. baumannii. Its substrate range is extensive and encompasses many substances including several β-lactams [,,]. Its role in carbapenem resistance is still unclear since resistance to carbapenems is mainly associated with carbapenemase production []. Previous studies indicate that efflux activity may contribute to carbapenem resistance in A. baumannii [,,]. Zhang et al. reported that a imipenem-selected stress in A. baumannii strain led to overproduction of AdeABC and consequently to the reduction of susceptibility to a variety of antibiotics including carbapenems []. Moreover, the adeB expression levels were upregulated in A. baumannii isolates with a reduction in the MIC of meropenem in the presence of PAβN []. However, Salehi et al. did not observe imipenem MIC reduction in the presence of PAβN in A. baumannii, even though expression of adeB was significantly upregulated []. In our study, all 15 isolates, chosen for adeB expression level analysis, had significant (≥3-fold) upregulation of this gene, regardless of PAβN effect on meropenem MIC and the CarbAcineto NP result. However, three isolates nos. AB96, AB129 and AB185 (each possessing an ISAba1-blaOXA-23-like) presented noticeably the highest adeB expression, up to 67-fold upregulation. A significant reduction of the meropenem MIC value in the presence of PAβN was observed only for A. baumannii 96, while for the other two isolates, there was no difference. The isolate no. AB96 was the only one of the three where a positive CarbAcineto NP test result was observed, suggesting higher carbapenemase activity for this isolate. On the other hand, it was presented by Salehi et al. that despite the lack of influence of PAβN on carbapenem MIC values, upregulation of adeABC in A. baumannii was observed [].
The most common cause of adeABC overexpression are changes in nucleotide sequences of regulatory genes. To date, both the local and global regulators of the AdeABC efflux system have been identified. BaeSR is a global regulatory system which was showed to regulate expression of adeA and adeB genes []. One mutation found within BaeS amino acid sequence, S437T, was associated with upregulation of adeB gene by Salehi et al. []. It is worth noting that all of 15 isolates analysed in our study were positive for this mutation, and simultaneously all of them presented overexpression of adeB, while Salehi et al. reported only one isolate with this mutation. However, as with the efflux pumps of other Gram-negative bacilli, the influence of local regulators is better documented. Various studies reported that specific mutations in the adeRS TCS local regulatory system of AdeABC resulted in overproduction of AdeB leading to decreased antimicrobial susceptibility [,]. Within AdeS, the following substitutions of amino acid sequence were observed: T153M in histidine box, D30G in periplasmic loop, G186V in the αhelix of the dimerization and DHp domain, G103D in HAMP linker domain [,,,]. An ISAba1 insertion within adeS resulting in a truncated AdeS may also be responsible for enhancing expression of efflux pump operons []. In all 15 sequenced isolates, we have identified in AdeS a G186V point mutation that can alter the conformation of AdeS DHp domain and then restore expression [,]. However, the other observed six amino acid substitutions (L172P, N268H, Y303F, V348I, G356N, H357N) and four terminal amino acid deletions in AdeS considered as may-be silent, not associated with the overexpression of adeABC operon. Furthermore, three isolates (A. baumannii numbers 96, 129 and 185) were missing H102 and G103 in the HAMP linker domain of AdeS. This, to our knowledge, is the first deletion observed at this location in AdeS, although the substitution G103D has been reported before []. Compared to AdeS, changes in the amino acid sequence of AdeR were less frequently observed. Mutations in AdeR which can boost adeB expression are D20N, D26N, A136V in the phosphorylation site, A91V of signal-receiving domain and P116L at the first residue of helix α5 [,,,]. Sequence comparison of our tested strains revealed two mutations, V120I and A136V, in AdeR in all of them. Haeili et al. reported both of these substitutions in isolates with elevated adeABC pump expression [].
Most importantly, our short-read WGS results revealed that the highest levels of upregulation of adeB in the three isolates (nos. AB96, AB129 and AB185) were due to the presence of additional ISAba1 sequences both upstream of adeA and into adeR. An ISAba1 insertion identified upstream of adeA provides a strong promoter for adeABC operon. On the other hand, an ISAba1 insertion starting downstream of adeR resulted in disruption of the gene and forming a truncated protein. To date, only one A. baumannii clinical strain (no. Ab209) has been reported to have an insertion of ISAba1 upstream of the abaABC operon []. The authors concluded that ISAba1 governs the expression of the adeABC operon despite disruption of adeR by the second ISAba1 in the same isolate. This is because ISAba1 provides a strong promoter upstream of the gene adeA. However, unlike the Polish isolates with upregulation from 49- to 67-fold, only 2.56-fold upregulation of the adeB gene of strain Ab209 compared to A. baumannii ATCC 19606 was obtained []. Moreover, Zang et al. showed that ISAba1 insertion upstream membrane fusion protein AdeI encoding gene increased transcription of a different RND efflux pump operon adeIJK []. However, in our three isolates, the rest of the adeR-adeA region remained unknown; therefore, a long-read WGS was performed. Finally, the obtained complete sequence of isolate no. AB96’s genome revealed the presence of the AbaR25 resistance island described previously [] and located elsewhere in the genome and two disrupted fragments of this island. It should be emphasized that AbaR-type islands (AbaRs) are important genetic elements responsible for antimicrobial resistance in A. baumannii strains [,]. The AbaR25 belongs to the AbaR4-like group of resistance islands that carry the most important ISAba1-blaOXA-23 gene and also other resistance genes such as sul2, tetA, strA and strB. The island of AbaR25 was located in chromosome of isolate no. AB96 as most of the previously documented cases [].
Most importantly, the unknown region adeR-adeA of the tested isolate no. AB96 turned out to be a 13.5 kb-long element of the AbaR25 island (the Δ1AbaR25-fragment), which was flanked by two copies of ISAba1. We suppose that this fragment of the AbaR25 island was most likely duplicated in the genome of isolate no. AB96 by homologous recombination. This insertion resulted not only in truncated AdeR protein but also in the loss of the intercistronic spacer; an AdeR binding site located between adeR and adeA. Chang et al. demonstrated that a direct repeat motif (“AAGTGTGGAG” separated with an “A” nucleotide) is the minimal element to enable binding of AdeR []. As this region was deleted in A. baumannii 96, AdeR is unable to regulate adeABC operon expression. Taking the above into account, the location and orientation of the second ISAba1 sequence flanking the island fragment became crucial for abaABC expression in tested isolate no. AB96. It is known that ISAba1 provides a strong promoter with both -10 and -35 sites located within IS sequence. However, its orientation is crucial to actually elevate expression []. The nucleotide sequence analysis of A. baumannii 96 genome revealed the presence of a strong promoter with an extended –10 motif “TGACATTATTT”, indicating that it can play a significant role in controlling the expression of adeABC. It is the first time the insertion of a fragment of the resistance island AbaR25 in the region between adeR and adeA is detected and analysed regarding the adeB overexpression.
Interestingly, apart from the AbaR25 resistance island itself and the Δ1AbaR25-fragment, an additional second element of AbaR25 (Δ2AbaR25-fragment) was found in the genome of isolate no. AB96. It contained a Tn2006 carrying ISAba1-blaOXA-23-like gene. ISAba1 provides a strong promoter, elevating the blaOXA-23-like expression levels [].
It is generally believed that the resistance of acquired blaCHDL-genes-carrying strains results from the production of CHDL enzymes, especially when dealing with the presence of a strong promoter as is the case in ISAba1-blaOXA-23-like gene. Moreover, the wide prevalence in A. baumannii clinical strains of AbaR4-like resistance islands contributes to the presence of the ISAba1-blaOXA-23-like gene. Our study for the first time reports that despite the duplication of the ISAba1-blaOXA-23 gene in the genome, the insertion of an AbaR25-island fragment with the ISAba1 sequence as a strong promoter of the adeABC operon is important for the contribution of the efflux system to carbapenem resistance.
4. Materials and Methods
4.1. Bacterial Strains
A collection of 61 non-repetitive imipenem-insensitive A. baumannii strains were isolated in the period 2009–2014 from patients hospitalised in one tertiary hospital in Warsaw, Poland. The isolates were recovered from the following clinical specimens: respiratory tract samples (n = 15), wound swabs (n = 15), urine samples (n = 15), rectal swabs (n = 4), blood samples (n = 3), fistula swabs (n = 3), stoma swabs (n = 3) and one isolate from each, peritoneal fluid, surgical drain swab and catheter tip. All studied strains were characterized (clinical material type, antimicrobial susceptibility determination, phenotypic carbepenemase detection, presence of genes encoding carbapenemases, genetic relatedness between strains by PFGE method, MLST sequence type) in our previous paper []. All strains carried blaCHDL genes relevant for carbapenem resistance, including 59 isolates with acquired blaCHDL genes and the other 2 isolates with ISAba1-blaOXA-51-like genes []. Epidemiological data of isolates and the blaCHDL-gene groups they possess are presented in Table S1 in the Supplementary Materials. All strains were stored at −80 °C in LB broth supplemented with glycerol until analysis. Prior to testing, each strain was sub-cultured on tryptic soy agar (TSA) (bioMérieux, Mercy l’Etoile, France) medium for 24 h at 35 °C to ensure viability.
4.2. Determination of the MICs of AdeABC and AbeM Efflux Pumps Substrates
Minimal inhibitory concentrations (MICs) of carbapenems (imipenem and meropenem), cephalosporins (cefepime and ceftazidime), fluoroquinolones (ciprofloxacin and levofloxacin), and aminoglycosides (gentamicin and tobramicin), were determined using the VITEK 2 system (bioMérieux, Mercy l’Etoile, France). Interpretation of the MIC results was performed according to CLSI breakpoints [].
4.3. Determination of the MICs of Carbapenems with and without Efflux Pump Inhibitors
In order to determine strains with active efflux pumps extruding carbapenems, the MIC values of imipenem and meropenem (both from Sigma, St. Louis, MO, USA) in the presence or absence of efflux pump inhibitors (EPIs) were estimated in Mueller Hinton II (MH II) broth medium (Becton, Dickinson and Company, Franklin Lakes, NJ, USA), using double agent dilutions method, according to the CLSI guideline []. The following two RND efflux pump inhibitors (EPIs) Phe-Arg-β-naphthylamide—PAβN and Carbonyl cyanide 3-chlorophenylhydrazone—CCCP (both from Sigma, St. Louis, MO, USA) were used. The final concentrations of PAβN in broth MH II medium were 50 mg/L. Our previous research showed that the MIC values of PAβN were ≥250 mg/L for all tested strains. The second inhibitor CCCP was used at the concentration of one-fourth of MIC of each isolate (i.e., in the range from 0.75 to 5 mg/L). Solutions of PAβN were prepared in deionized water whilst CCCP in dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO, USA). In order to provide bacterial cell’s outer membrane stabilization, the broth MH II medium was supplemented with 1 mM MgSO4 (Sigma, St. Louis, MO, USA) []. The plates were incubated for 18 h at 35 °C. At least a 4-fold change in MIC values after addition of EPI was considered significant []. Such significant reduction in the MIC values of meropenem or imipenem in the presence of at least one of the EPIs was interpreted as the likely contribution of efflux pumps to carbapenem resistance of the studied isolate.
The assay was validated by the MIC determination of selected antimicrobial agents against reference strain E. coli ATCC 25922 and comparison of the experimental MIC values with the CLSI guidelines []. The MIC breakpoints for imipenem and meropenem were ≤2, 4 and ≥8 mg/L to designate susceptible, intermediate and resistant strains, respectively [].
4.4. Detection of Genes Encoding AdeABC and AbeM Efflux Pumps
Detection of adeB and abeM genes was carried out with a singleplex PCR. The total DNA of the clinical isolates was extracted using a Genomic Mini Kit (A&A Biotechnology, Gdynia, Poland). PCR was performed by using the Hypernova polymerase (Blirt, DNA, Gdańsk, Poland) with the following amplification parameters: 95 °C for 4 min, following 25 cycles of 30 s at 95 °C, annealing 30 s at 55 °C (adeB gene) and at 54 °C (abeM gene), 45 s at 72 °C and a final extension of 3 min at 72 °C. Sequences of the primers are presented in Table 3.

Table 3.
Sequences of primers used for the efflux pump genes’ amplification by classic PCR and for the analysis of efflux pump gene and its regulatory genes’ expression by qPCR.
4.5. Whole Genome Sequencing
Genomic DNA of bacterial strains was isolated using a Genomic Mini AX Bacteria Spin Kit (A&A Biotechnology, Gdynia, Poland). Whole genome sequencing was carried out with the following two methods: (I) using Public Health England—Genomic Services and Development Unit (PHEGSDU) on the HiSeq 2500 System (Illumina, Cambridge, UK) with paired end read lengths of 150 bp, for 15 selected isolates, (II) using GridION sequencer (Oxford Nanopore Technologies, Oxford, UK) in the Laboratory of DNA Sequencing and Oligonucleotide Synthesis IBB PAS in Warsaw for one isolate, A. baumannii 96. The procedure of short-read WGS by the HiSeq 2500 System was performed as described in our previous paper []. To carry out the long-read WGS, the Illumina data of the strain no. AB96 was downloaded from the BioProject no. PRJNA701882. Sequence quality metrics were assessed using FASTQC tool [] and quality trimmed using fastp []. Prior to long-read library preparation genomic DNA was sheared into ~30kb fragments using 26G needle followed by size selection using Short Read Eliminator kit (Circulomics, Baltimore, MD, USA). A total of 3 µg of recovered DNA was taken for 1D library construction using SQK-LSK109 kit, and 0.8 µg of final library was loaded into R9.4.1 flow cell and sequenced on GridION sequencer (Oxford Nanopore Technologies, Oxford, UK). The methodology of the genome assembly of the A. baumannii 96 strain is presented in the Supplementary Materials.
Obtained sequences of studied isolates were compared with A. baumannii ATCC 17978 isolate with the use of SnapGene, version 6.0.2. Finally using the WGS data, the amino acid changes within adeABC, adeRS, baeRS, abeM, were determined. The obtained genomic sequence of isolate no AB96 was compared with those deposited in GenBank by BLASTn.
4.6. Core-Genome SNP Phylogenetic Analysis
The Illumina data of the A. baumanii strains downloaded from the BioProject no. PRJNA701882 were assembled into contigs using Unicycler. Phylogenetic analysis and SNP tree construction was conducted using kSNP3.0 []. Phylogenetic tree was visualized using Figtree []. Phylogenetic analysis was performed in cooperation with the Laboratory of DNA Sequencing and Oligonucleotide Synthesis IBB PAS in Warsaw, Poland.
5. Conclusions
Our study for the first time reports the involvement of the insertion of the ΔAbaR25-type resistance island fragment with ISAba1 element upstream of the efflux operon in the carbapenem resistance of A. baumannii clinical isolates. This insert was flanked by two copies of ISAba1, and one of them provides a strong promoter for adeABC, elevating the adeB expression levels. Interestingly, our study also revealed that AbaR25 resistance island is the reservoir of resistance genes, and they may be transferred to another part of the bacterial genome. Apart from the island of resistance, two of its fragments were found in the chromosome of A. baumannii strain: the first contained a duplicate ISAba1-blaOXA-23 gene and the second contained an IS element located upstream of the efflux operon. It is worth noting that the analysis of the AdeABC-regulatory genes of the Polish isolate with adeB-overexpression showed many point mutations leading to amino acid changes in the AdeRS local regulator as well as in the BaeS global regulator. However, the presence of ISAba1 as a strong promoter plays a major role in the high upregulation of adeABC. Therefore, our data indicate that in some isolates both efflux systems and CHDL enzymes may participate independently and equally in meropenem resistance.
Supplementary Materials
The supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms24119525/s1. References [,,,,,,] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, A.S. and A.E.L.; methodology, A.S., M.E.W., L.J.B. and A.E.L.; software, A.S., M.E.W., L.J.B. and A.E.L.; validation, A.S., M.E.W. and L.J.B.; formal analysis, A.S., M.E.W., L.J.B., S.T. and A.E.L.; investigation, A.S. and A.E.L.; resources, A.E.L. and S.T.; data curation, A.S. and M.E.W.; writing—original draft preparation, A.S., M.E.W., L.J.B., S.T. and A.E.L.; writing—review and editing, A.S., M.E.W., L.J.B., S.T. and A.E.L.; visualization, A.S.; supervision, S.T. and A.E.L.; project administration, A.E.L.; funding acquisition, S.T. and A.E.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially supported by grant from the Medical University of Warsaw (grant no. FW15/PM1/18) and by the Foundation for the Development of Diagnostics and Therapy, Warsaw, Poland (REGON: 006220910, NIP: 5262173856 and KRS: 0000195643). Moreover, an article processing charge was partially funded by the Medical University of Warsaw and the National Medicines Institute in Warsaw.
Institutional Review Board Statement
The clinical samples were part of the routine diagnostic procedure in microbiology laboratories, and therefore the Institutional Review Board Statement was not applicable.
Informed Consent Statement
The clinical samples were part of the routine diagnostic procedure in microbiology laboratories, and therefore the informed consent statement was not applicable.
Data Availability Statement
The complete whole genome sequence of clinical isolate A. baumannii 96 obtained from additional long-read WGS analysis is available at the NCBI BioProject repository (Submission ID: SUB12923155, BioProject ID: PRJNA939738). The whole genome datasets of the 15 strains analysed during the current study were previously deposited in the NCBI GeneBank, and are available at the NCBI BioProject repository (Submission ID: SUB9082120, BioProject ID: PRJNA701882).
Acknowledgments
We thank Mark Sutton for the possibility of Alicja Słoczyńska’s work as part of the ERAZMUS + project at Public Health England, National Infection Service, Porton Down, Salisbury, Wiltshire, UK. We also thank Jan Gawor from Institute of Biochemistry and Biophysics, Polish Academy of Sciences in Warszawa, Poland, for assistance with genome assembly and phylogenetic analysis. This research was carried out partially with the use of the CePT infrastructure financed by the European Union through the European Regional Development Fund as part of the Operational Program “Innovative Economy” for 2007–2013.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McConnell, M.J.; Actis, L.; Pachon, J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef] [PubMed]
- De Benedetto, I.; Lupia, T.; Shbaklo, N.; Bianchi, A.; Concialdi, E.; Penna, M.; Corcione, S.; De Rosa, F.G. Prognostic evaluation of Acinetobacter baumannii ventilator-associated pneumonia in COVID-19. Infez. Med. 2022, 30, 570–576. [Google Scholar] [PubMed]
- Nasr, P. Genetics, epidemiology, and clinical manifestations of multidrug-resistant Acinetobacter baumannii. J. Hosp. Infect. 2020, 104, 4–11. [Google Scholar] [CrossRef] [PubMed]
- WHO Regional Office for Europe/European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2022—2020 Data. Copenhagen: WHO Regional Office for Europe 2022. Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2022-2020-data (accessed on 10 May 2023).
- Krajewska, J.; Laudy, A.E. The European Medicines Agency approved the new antibacterial drugs—Response to the 2017 WHO report on the global problem of multi-drug resistance. Adv. Microbiol. 2021, 60, 249–264. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- Rumbo, C.; Gato, E.; López, M.; Ruiz de Alegría, C.; Fernández-Cuenca, F.; Martínez-Martínez, L.; Vila, J.; Pachón, J.; Cisneros, J.M.; Rodríguez-Baño, J. Contribution of efflux pumps, porins, and beta-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013, 57, 5247–5257. [Google Scholar] [CrossRef]
- Freire, M.P.; de Oliveira Garcia, D.; Garcia, C.P.; Campagnari Bueno, M.F.; Camargo, C.H.; Kono Magri, A.S.G.; Francisco, G.R.; Reghini, R.; Vieira, M.F.; Ibrahim, K.Y.; et al. Bloodstream infection caused by extensively drug-resistant Acinetobacter baumannii in cancer patients: High mortality associated with delayed treatment rather than with the degree of neutropenia. Clin. Microbiol. Infect. 2016, 22, 352–358. [Google Scholar] [CrossRef]
- Słoczyńska, A.; Wand, M.E.; Tyski, S.; Laudy, A.E. Analysis of blaCHDL genes and insertion sequences related to carbapenem resistance in Acinetobacter baumannii clinical strains isolated in Warsaw, Poland. Int. J. Mol. Sci. 2021, 22, 2486. [Google Scholar] [CrossRef]
- Roy, S.; Junghare, V.; Dutta, S.; Hazra, S.; Basu, S. Differential binding of carbapenems with the AdeABC efflux pump and modulation of the expression of AdeB linked to novel mutations within Two-Component System AdeRS in Carbapenem-Resistant Acinetobacter baumannii. mSystems 2022, 7, e0021722. [Google Scholar] [CrossRef] [PubMed]
- Abdi, S.N.; Ghotaslou, R.; Ganbarov, K.; Mobed, A.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Kafil, H.S. Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect. Drug Resist. 2020, 13, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Laudy, A.E. Non-antibiotics, efflux pumps and drug resistance of Gram-negative rods. Pol. J. Microbiol. 2018, 67, 129–135. [Google Scholar] [CrossRef]
- Verma, P.; Tiwari, M.; Tiwari, V. Efflux pumps in multidrug-resistant Acinetobacter baumannii: Current status and challenges in the discovery of efflux pumps inhibitors. Microb. Pathog. 2021, 152, 104766. [Google Scholar] [CrossRef] [PubMed]
- Coyne, S.; Courvalin, P.; Perichon, B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011, 55, 947–953. [Google Scholar] [CrossRef]
- Gerson, S.; Nowak, J.; Zander, E.; Ertel, J.; Wen, Y.; Krut, O.; Seifert, H.; Higgins, P.G. Diversity of mutations in regulatory genes of resistance-nodulation-cell division efflux pumps in association with tigecycline resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2018, 73, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Bilya, S.R.; Xu, W. adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 2019, 30, 100549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, B.; Luo, Y.; Tao, Z.; Nie, Y.; Wang, Y.; Ding, F.; Li, Y.; Gu, D. Comparative analysis of carbapenemases, RND family efflux pumps and biofilm formation potential among Acinetobacter baumannii strains with different carbapenem susceptibility. BMC Infect. Dis. 2021, 21, 841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; He, X.; Ding, F.; Wu, W.; Luo, Y.; Fan, B.; Cao, H. Overproduction of efflux pumps caused reduced susceptibility to carbapenem under consecutive imipenem-selected stress in Acinetobacter baumannii. Infect. Drug Resist. 2017, 11, 457–467. [Google Scholar] [CrossRef]
- Lin, M.F.; Lin, Y.Y.; Yeh, H.W.; Lan, C.Y. Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol. 2014, 14, 119. [Google Scholar] [CrossRef]
- Chang, T.Y.; Huang, B.J.; Sun, J.R.; Perng, C.L.; Chan, M.C.; Yu, C.P.; Chiueh, T.S. AdeR protein regulates adeABC expression by binding to a direct-repeat motif in the intercistronic spacer. Microbiol. Res. 2016, 183, 60–67. [Google Scholar] [CrossRef]
- Sun, J.R.; Perng, C.L.; Chan, M.C.; Morita, Y.; Lin, J.C.; Su, C.M.; Wang, W.Y.; Chang, T.Y.; Chiueh, T.S. A truncated AdeS kinase protein generated by ISAba1 insertion correlates with tigecycline resistance in Acinetobacter baumannii. PLoS ONE 2012, 7, e49534. [Google Scholar] [CrossRef]
- Hammerstrom, T.G.; Beabout, K.; Clements, T.P.; Saxer, G.; Shamoo, Y. Acinetobacter baumannii repeatedly evolves a hypermutator phenotype in response to tigecycline that effectively surveys evolutionary trajectories to resistance. PLoS ONE 2015, 10, e0140489. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.F.; Chen, X.Y.; Yan, G.F.; Wang, Y.P.; Ying, C.M. Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy 2012, 58, 152–158. [Google Scholar] [CrossRef]
- Su, X.Z.; Chen, J.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 2005, 49, 4362–4364. [Google Scholar] [CrossRef] [PubMed]
- Rafiei, E.; Shahini Shams Abadi, M.; Zamanzad, B.; Gholipour, A. The frequency of efflux pump genes expression in Acinetobacter baumannii isolates from pulmonary secretions. AMB Express 2022, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Nordmann, P. Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin. Microbiol. Infect. 2006, 12, 826–836. [Google Scholar] [CrossRef]
- Zając, O.M.; Tyski, S.; Laudy, A.E. The contribution of efflux systems to levofloxacin resistance in Stenotrophomonas maltophilia clinical strains isolated in Warsaw, Poland. Biology 2022, 11, 1044. [Google Scholar] [CrossRef]
- Pannek, S.; Higgins, P.G.; Steinke, P.; Jonas, D.; Akova, M.; Bohnert, J.A.; Seifert, H.; Kern, W.V. Multidrug efflux inhibition in Acinetobacter baumannii: Comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide. J. Antimicrob. Chemother. 2006, 57, 970–974. [Google Scholar] [CrossRef]
- Lee, Y.; Yum, J.H.; Kim, C.K.; Yong, D.; Jeon, E.H.; Jeong, S.H.; Ahn, J.Y.; Lee, K. Role of OXA-23 and AdeABC efflux pump for acquiring carbapenem resistance in an Acinetobacter baumannii strain carrying the blaOXA-66 gene. Ann. Clin. Lab. Sci. 2010, 40, 43–48. [Google Scholar]
- Laudy, A.E.; Kulińska, E.; Tyski, S. The impact of efflux pump inhibitors on the activity of selected non-antibiotic medicinal products against Gram-negative bacteria. Molecules 2017, 22, 114. [Google Scholar] [CrossRef]
- Laudy, A.E.; Róg, P.; Smolińska-Król, K.; Ćmiel, M.; Słoczyńska, A.; Patzer, J.; Dzierżanowska, D.; Wolinowska, R.; Starościak, B.; Tyski, S. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PLoS ONE 2017, 12, e0180121. [Google Scholar] [CrossRef]
- Huang, L.; Sun, L.; Xu, G.; Xia, T. Differential susceptibility to carbapenems due to the AdeABC efflux pump among nosocomial outbreak isolates of Acinetobacter baumannii in a Chinese hospital. Diagn. Microbiol. Infect. Dis. 2008, 62, 326–332. [Google Scholar] [CrossRef]
- Ardebili, A.; Lari, A.R.; Talebi, M. Correlation of ciprofloxacin resistance with the AdeABC efflux system in Acinetobacter baumannii clinical isolates. Ann. Lab. Med. 2014, 34, 433–438. [Google Scholar] [CrossRef]
- Ardehali, S.H.; Azimi, T.; Fallah, F.; Owrang, M.; Aghamohammadi, N.; Azimi, L. Role of efflux pumps in reduced susceptibility to tigecycline in Acinetobacter baumannii. New Microbes New Infect. 2019, 30, 100547. [Google Scholar] [CrossRef]
- Amiri, G.; Abbasi Shaye, M.; Bahreini, M.; Mafinezhad, A.; Ghazvini, K.; Sharifmoghadam, M.R. Determination of imipenem efflux-mediated resistance in Acinetobacter spp., using an efflux pump inhibitor. Iran J. Microbiol. 2019, 11, 368–372. [Google Scholar] [CrossRef]
- Sanchez-Carbonel, A.; Mondragon, B.; Lopez-Chegne, N.; Pena-Tuesta, I.; Huayan-Davila, G.; Blitchtein, D.; Carrillo-Ng, H.; Silva-Caso, W.; Aguilar-Luis, M.A.; Del Valle-Mendoza, J. The effect of the efflux pump inhibitor Carbonyl Cyanide m-Chlorophenylhydrazone (CCCP) on the susceptibility to imipenem and cefepime in clinical strains of Acinetobacter baumannii. PLoS ONE 2021, 16, e0259915. [Google Scholar] [CrossRef] [PubMed]
- Marchand, I.; Damier-Piolle, L.; Courvalin, P.; Lambert, T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 2004, 48, 3298–3304. [Google Scholar] [CrossRef]
- Nemec, A.; Maixnerova, M.; van der Reijden, T.J.; van den Broek, P.J.; Dijkshoorn, L. Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J. Antimicrob. Chemother. 2007, 60, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Higgins, P.G.; Wisplinghoff, H.; Stefanik, D.; Seifert, H. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J. Antimicrob. Chemother. 2004, 54, 821–823. [Google Scholar] [CrossRef]
- Salehi, B.; Ghalavand, Z.; Yadegar, A.; Eslami, G. Characteristics and diversity of mutations in regulatory genes of resistance-nodulation-cell division efflux pumps in association with drug-resistant clinical isolates of Acinetobacter baumannii. Antimicrob. Resist. Infect. Control 2021, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.R.; Perng, C.L.; Lin, J.C.; Yang, Y.S.; Chan, M.C.; Chang, T.Y.; Lin, F.M.; Chiueh, T.S. AdeRS combination codes differentiate the response to efflux pump inhibitors in tigecycline-resistant isolates of extensively drug-resistant Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2141–2147. [Google Scholar] [CrossRef]
- Hornsey, M.; Ellington, M.J.; Doumith, M.; Thomas, C.P.; Gordon, N.C.; Wareham, D.W.; Quinn, J.; Lolans, K.; Livermore, D.M.; Woodford, N. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J. Antimicrob. Chemother. 2010, 65, 1589–1593. [Google Scholar] [CrossRef]
- Sun, J.R.; Jeng, W.Y.; Perng, C.L.; Yang, Y.S.; Soo, P.C.; Chiang, Y.S.; Chiueh, T.S. Single amino acid substitution Gly186Val in AdeS restores tigecycline susceptibility of Acinetobacter baumannii. J. Antimicrob. Chemother. 2016, 71, 1488–1492. [Google Scholar] [CrossRef]
- Haeili, M.; Abdollahi, A.; Ahmadi, A.; Khoshbayan, A. Molecular characterization of tigecycline non-susceptibility among extensively drug-resistant Acinetobacter baumannii isolates of clinical origin. Chemotherapy 2021, 66, 99–106. [Google Scholar] [CrossRef]
- Lari, A.R.; Ardebili, A.; Hashemi, A. AdeR-AdeS mutations & overexpression of the AdeABC efflux system in ciprofloxacin-resistant Acinetobacter baumannii clinical isolates. Indian J. Med. Res. 2018, 147, 413–421. [Google Scholar] [PubMed]
- Higgins, P.G.; Schneiders, T.; Hamprecht, A.; Seifert, H. In vivo selection of a missense mutation in adeR and conversion of the novel bla(OXA-164) gene into bla(OXA-58) in carbapenem-resistant Acinetobacter baumannii isolates from a hospitalized patient. Antimicrob. Agents Chemother. 2010, 54, 5021–5027. [Google Scholar] [CrossRef] [PubMed]
- Lopes, B.S.; Amyes, S.G. Insertion sequence disruption of adeR and ciprofloxacin resistance caused by efflux pumps and gyrA and parC mutations in Acinetobacter baumannii. Int. J. Antimicrob. Agents 2013, 41, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Zang, M.; Adams, F.G.; Hassan, K.A.; Eijkelkamp, B.A. The Impact of omega-3 fatty acids on the evolution of Acinetobacter baumannii drug resistance. Microbiol. Spectr. 2021, 9, e0145521. [Google Scholar] [CrossRef] [PubMed]
- Saule, M.; Samuelsen, O.; Dumpis, U.; Sundsfjord, A.; Karlsone, A.; Balode, A.; Miklasevics, E.; Karah, N. Dissemination of a carbapenem-resistant Acinetobacter baumannii strain belonging to international clone II/sequence type 2 and harboring a novel AbaR4-like resistance island in Latvia. Antimicrob. Agents Chemother. 2013, 57, 1069–1072. [Google Scholar] [CrossRef]
- Bi, D.; Xie, R.; Zheng, J.; Yang, H.; Zhu, X.; Ou, H.Y.; Wei, Q. Large-scale identification of AbaR-Type genomic islands in Acinetobacter baumannii reveals diverse insertion sites and clonal lineage-specific antimicrobial resistance gene profiles. Antimicrob. Agents Chemother. 2019, 63, e02526-18. [Google Scholar] [CrossRef]
- Kim, D.H.; Choi, J.Y.; Kim, H.W.; Kim, S.H.; Chung, D.R.; Peck, K.R.; Thamlikitkul, V.; So, T.M.; Yasin, R.M.; Hsueh, P.R.; et al. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob. Agents Chemother. 2013, 57, 5239–5246. [Google Scholar] [CrossRef]
- Segal, H.; Jacobson, R.K.; Garny, S.; Bamford, C.M.; Elisha, B.G. Extended -10 promoter in ISAba-1 upstream of blaOXA-23 from Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 3040–3041. [Google Scholar] [CrossRef] [PubMed]
- Nigro, S.J.; Hall, R.M. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 2016, 71, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. M100: Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI: Wayne, PA, USA, 2022. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard, Document M07-A9, 9th ed.; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Lamers, R.P.; Cavallari, J.F.; Burrows, L.L. The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAbetaN) permeabilizes the outer membrane of gram-negative bacteria. PLoS ONE 2013, 8, e60666. [Google Scholar] [CrossRef] [PubMed]
- Fernando, D.; Kumar, A. Growth phase-dependent expression of RND efflux pump- and outer membrane porin-encoding genes in Acinetobacter baumannii ATCC 19606. J. Antimicrob. Chemother. 2012, 67, 569–572. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 May 2023).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Gardner, S.N.; Slezak, T.; Hall, B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. 2010. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 10 May 2023).
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Meric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus long-read assemblies for bacterial genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef]
- Available online: https://github.com/rrwick/Polypolish (accessed on 10 May 2023).
- Wick, R.R.; Holt, K.E. Polypolish: Short-read polishing of long-read bacterial genome assemblies. PLoS Comput. Biol. 2022, 18, e1009802. [Google Scholar] [CrossRef]
- Zimin, A.V.; Puiu, D.; Luo, M.C.; Zhu, T.; Koren, S.; Marcais, G.; Yorke, J.A.; Dvorak, J.; Salzberg, S.L. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 2017, 27, 787–792. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).