α-Tocotrienol and Redox-Silent Analogs of Vitamin E Enhances Bortezomib Sensitivity in Solid Cancer Cells through Modulation of NFE2L1
Abstract
1. Introduction
2. Results
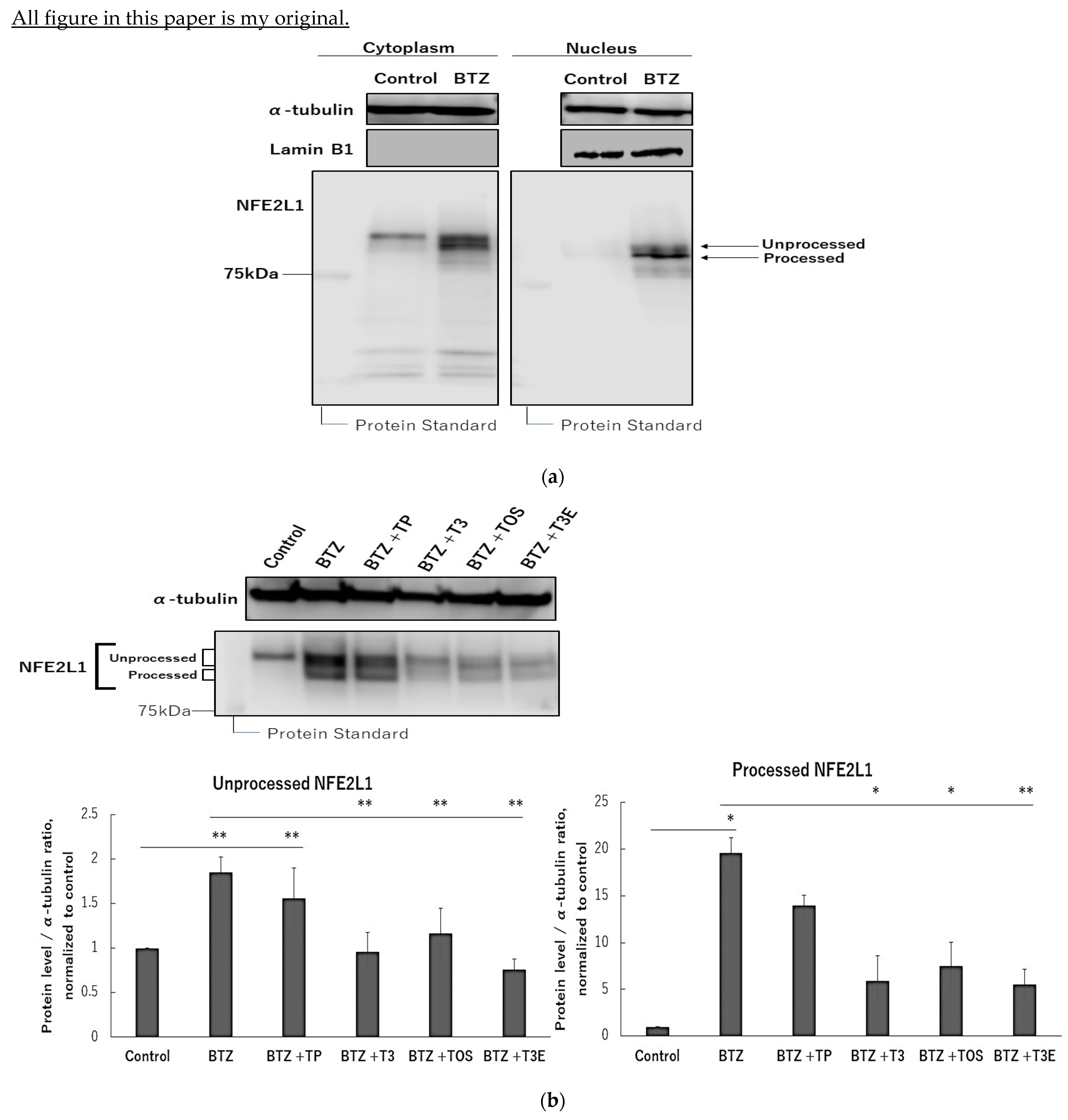
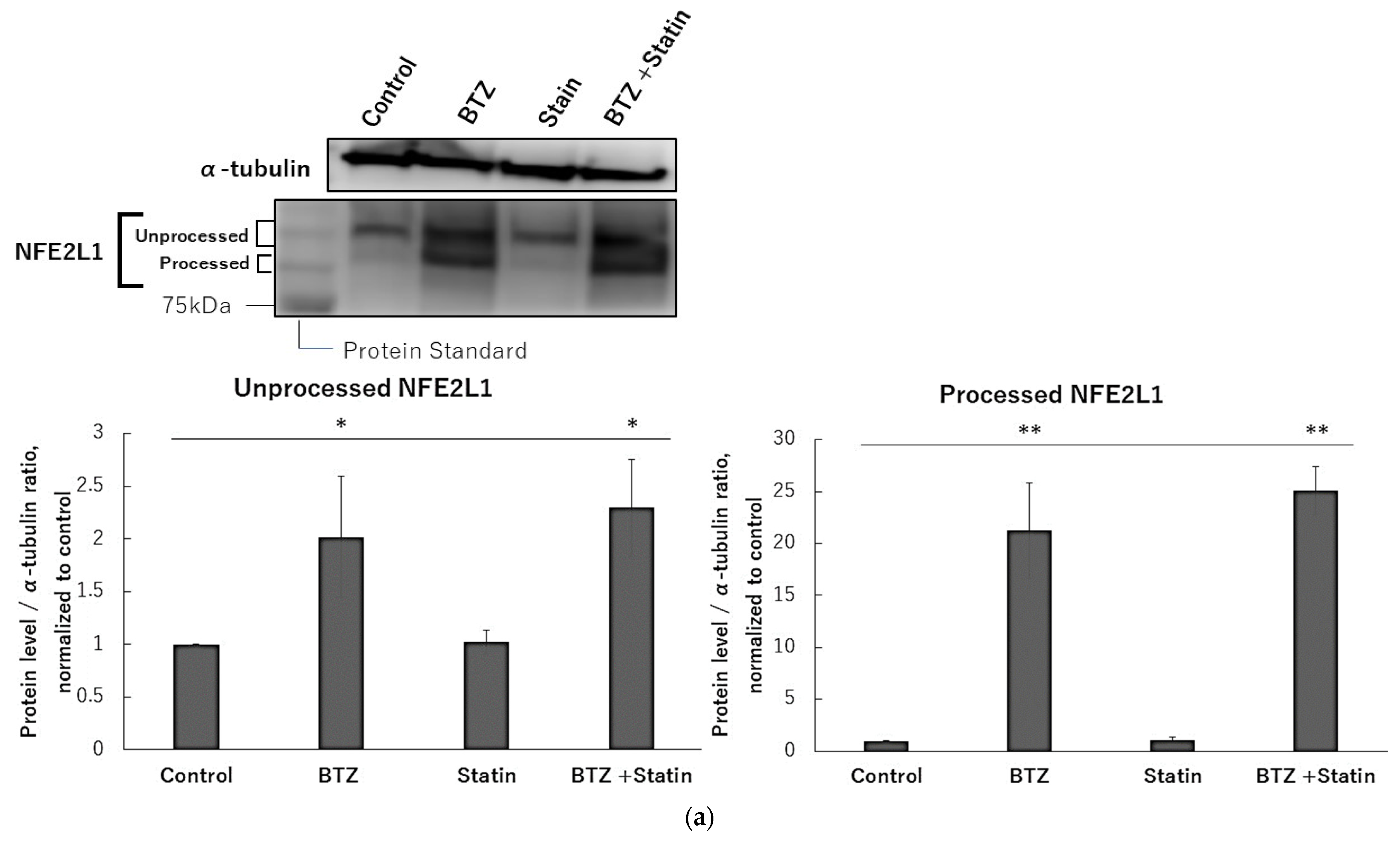
2.1. T3, TOS, and T3E Suppressed the Increase in Protein Levels of NFE2L1 Induced by Bortezomib Treatment
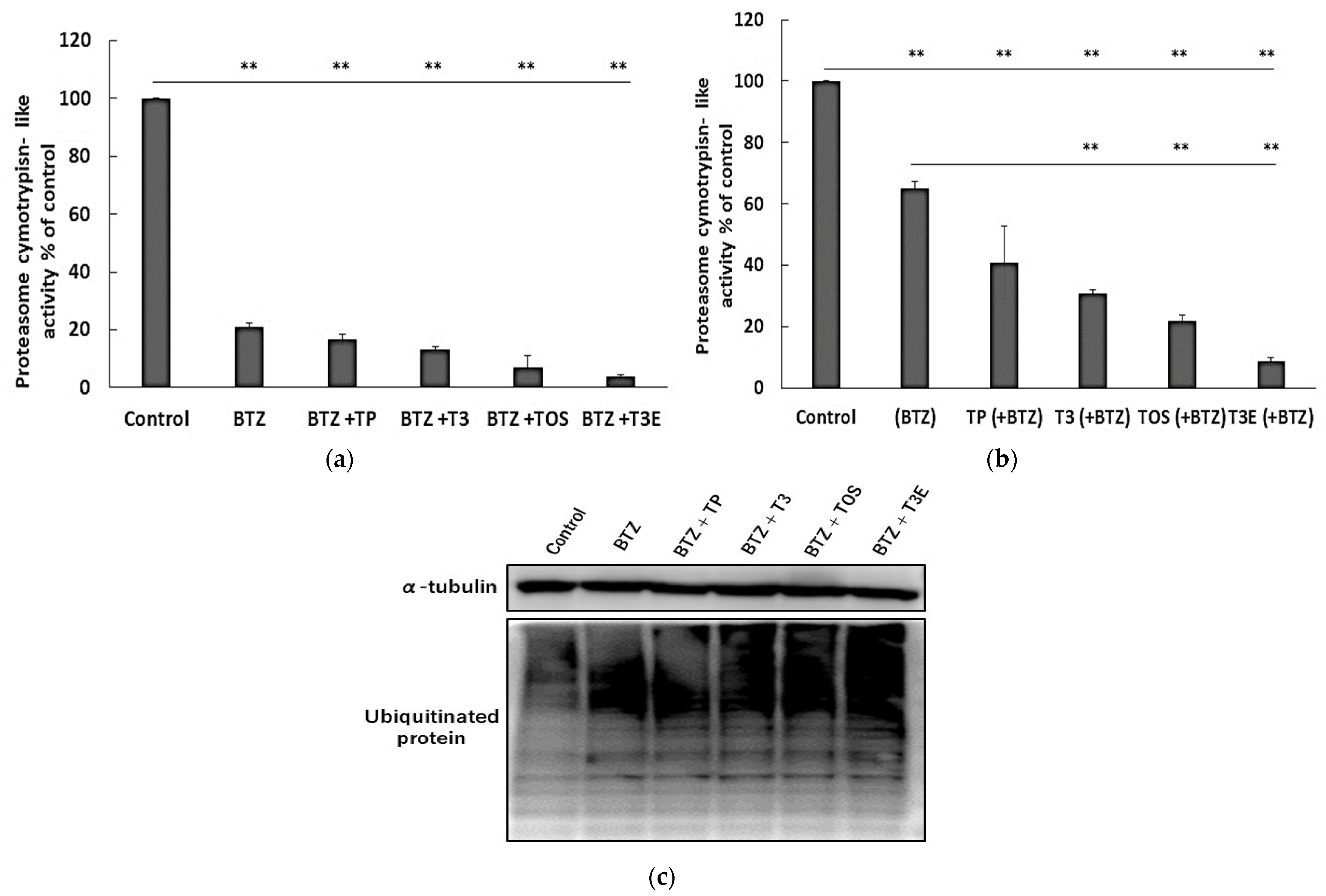
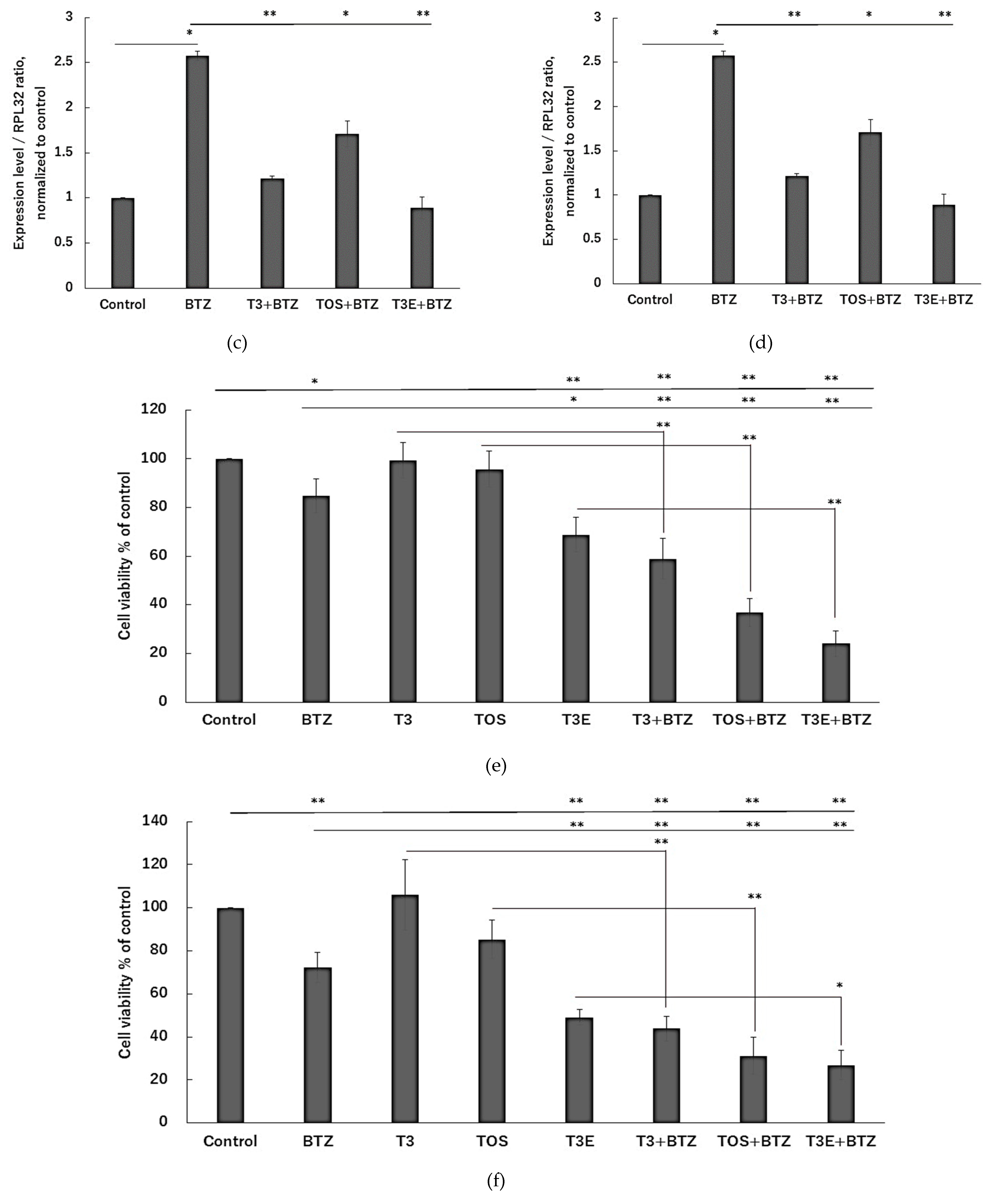
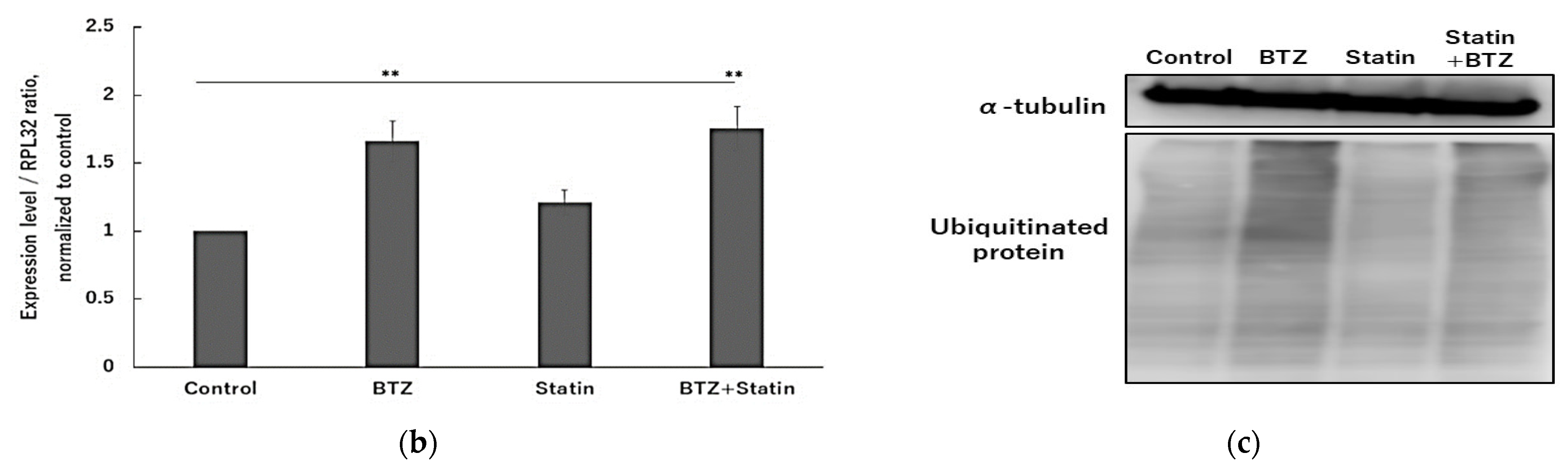
2.2. T3, TOS, and T3E Suppressed the Increase in Expression Levels of Proteasome-Related Proteins Induced by Bortezomib Treatment
2.3. T3, TOS, and T3E Suppressed the Recovery of Proteasome Activity under and after Bortezomib Treatment
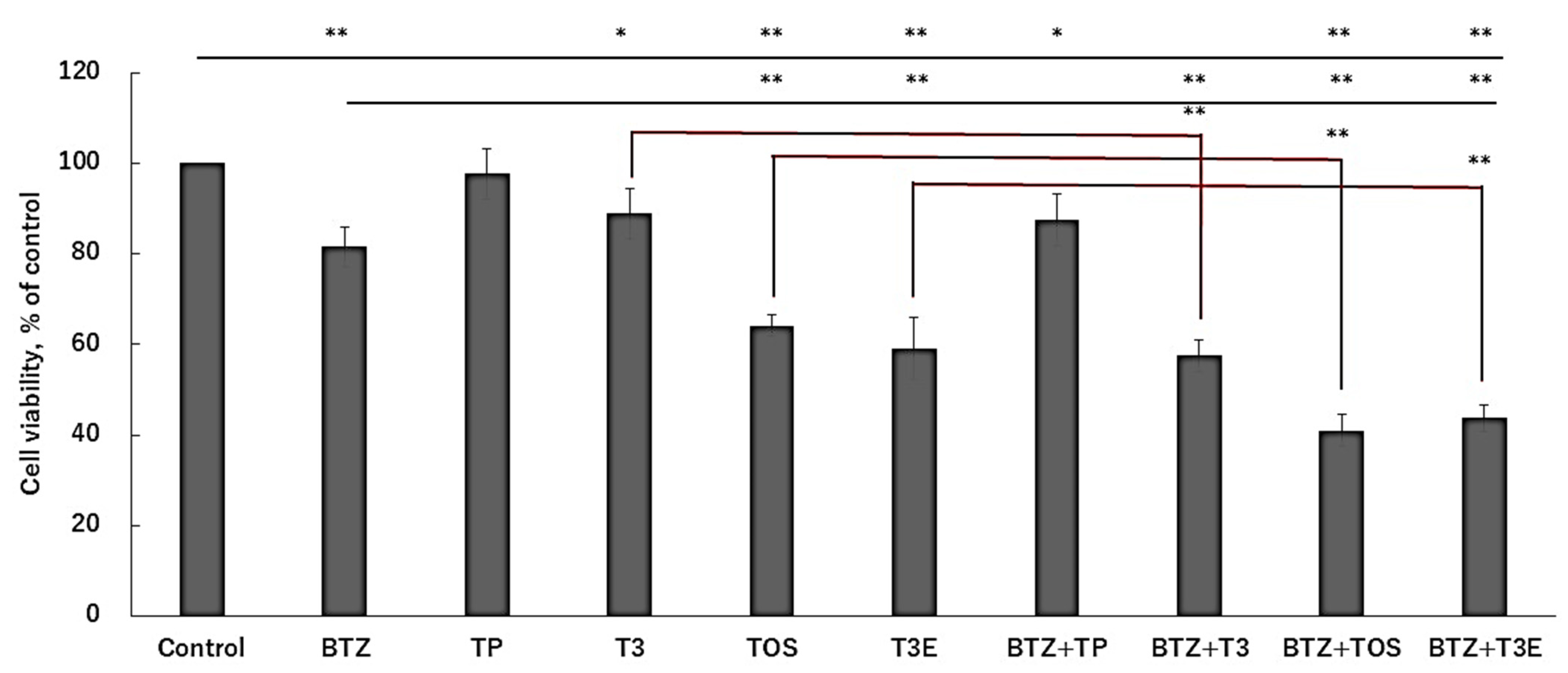
2.4. T3, TOS, and T3E Enhanced Sensitivity to Bortezomib in H2452
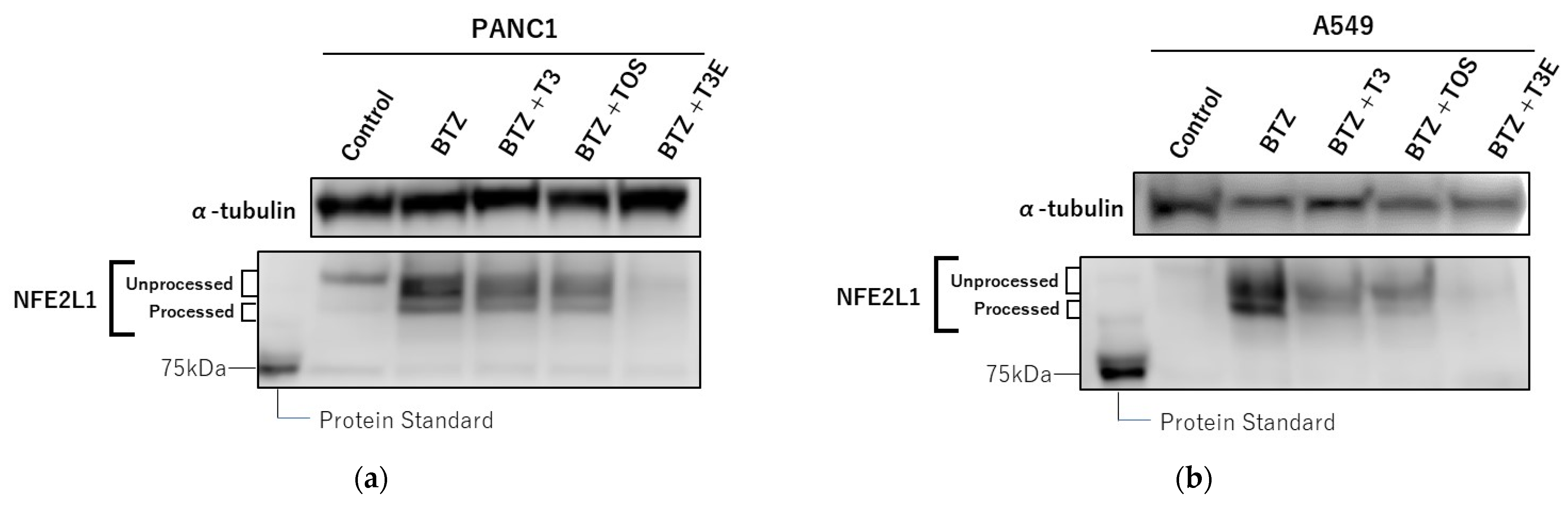
2.5. T3, TOS, and T3E Also Enhanced the Sensitivity to Bortezomib through Protein Levels of NFE2L1 and NRF3 in Other Solid Cancer Cell Lines
2.6. Atorvastatin Did Not Affect NFE2L1 and NRF3 under Bortezomib Treatment
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. α-T3E Synthesis
4.3. Cell Culture
4.4. Reagent Dissolution
4.5. Cellular Fractionation
4.6. Cell Viability
4.7. Proteasome Activity
4.8. Isolation of mRNA and Real-Time Quantitative PCR
4.9. Immunoblotting
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shen, M.; Schmitt, S.; Buac, N.; Dou, P. Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opin. Ther. Targets 2013, 17, 1091–1108. [Google Scholar] [CrossRef]
- Arlt, A.; Bauer, I.; Schafmayer, C.; Tepel, J.; Sebens, M.S.; Brosch, M.; Röder, C.; Kalthoff, H.; Hampe, J.; Moyer, M.P.; et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (NFE2L2). Oncogene 2009, 28, 3983–3996. [Google Scholar] [CrossRef]
- Chen, L.; Madura, K. Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res. 2005, 65, 5599–5606. [Google Scholar] [CrossRef]
- Stoebner, P.-E.; Lavabre-Bertrand, T.; Henry, L.; Guiraud, I.; Carillo, S.; Dandurand, M.; Joujoux, J.-M.; Bureau, J.-P.; Meunier, L. High plasma proteasome levels are detected in patients with metastatic malignant melanoma. Br. J. Dermatol. 2005, 152, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Walter, R.F.H.; Sydow, S.R.; Berg, E.; Kollmeier, J.; Christoph, D.C.; Christoph, S.; Eberhardt, W.E.E.; Mairinger, T.; Wohlschlaeger, J.; Schmid, K.W.; et al. Bortezomib sensitivity is tissue dependent and high expression of the 20S proteasome precludes good response in malignant pleural mesothelioma. Cancer Manag. Res. 2019, 11, 8711–8720. [Google Scholar] [CrossRef]
- Dutaud, D.; Aubry, L.; Henry, L.; Levieux, D.; Hendil, K.B.; Kuehn, L.; Bureau, J.P.; Ouali, A. Development and evaluation of a sandwich ELISA for quantification of the 20S proteasome in human plasma. J. Immunol. Methods 2002, 260, 183–193. [Google Scholar] [CrossRef]
- Fricker, D.L. Proteasome Inhibitor Drugs. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 457–476. [Google Scholar] [CrossRef]
- Daniel, J.E. The ubiquitin-proteasome system: Opportunities for therapeutic intervention in solid tumors. Endocr. Relat. Cancer 2015, 22, T1. [Google Scholar]
- Roeten, M.S.F.; Cloos, J.; Jansen, G. Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemother. Pharmacol. 2018, 81, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Hainsworth, J.D.; Meluch, A.A.; Spigel, D.R.; Barton, J.; Simons, L.; Meng, C.; Gould, B.; Greco, F.A. Weekly docetaxel and bortezomib as first-line treatment for patients with hormone-refractory prostate cancer: A Minnie Pearl Cancer Research Network phase II trial. Clin. Genitourin. Cancer 2007, 5, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cao, Q.; Dudek, A.Z. Phase II study of panobinostat and bortezomib in patients with pancreatic cancer progressing on gemcitabine-based therapy. Anticancer. Res. 2012, 32, 1027–1031. [Google Scholar] [PubMed]
- Fanucchi, M.P.; Fossella, F.V.; Belt, R.; Natale, R.; Fidias, P.; Carbone, D.P.; Govindan, R.; Raez, L.E.; Robert, F.; Ribeiro, M.; et al. Randomized phase II study of bortezomib alone and bortezomib in combination with docetaxel in previously treated advanced non-small-cell lung cancer. J. Clin. Oncol. 2006, 24, 5025–5033. [Google Scholar] [CrossRef]
- Fennell, D.A.; McDowell, C.; Busacca, S.; Webb, G.; Moulton, B.; Cakana, A.; O’Byrne, K.J.; Meerbeeck, J.V.; Donnellan, P.; McCaffrey, J.; et al. Phase II clinical trial of first or second-line treatment with bortezomib in patients with malignant pleural mesothelioma. J. Thorac. Oncol. 2012, 7, 1466–1470. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, S.K.; Lee, C.S.; Young, P.; Beskow, A.; Chan, J.Y.; Deshaies, R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 2010, 38, 17–28. [Google Scholar] [CrossRef]
- Op, M.; Ribeiro, S.T.; Chavarria, C.; Gassart, A.D.; Zaffalon, L.; Martinon, F. The aspartyl protease DDI2 drives adaptation to proteasome inhibition in multiple myeloma. Cell Death Dis. 2022, 13, 475. [Google Scholar] [CrossRef]
- Chen, T.; Ho, M.; Briere, J.; Moscvin, M.; Czarnecki, P.G.; Anderson, K.C.; Blackwell, T.K.; Bianchi, G. Multiple myeloma cells depend on the DDI2/NRF1-mediated proteasome stress response for survival. Blood Adv. 2022, 6, 429–440. [Google Scholar] [CrossRef]
- Sekine, H.; Okazaki, K.; Kato, K.; Alam, M.M.; Shima, H.; Katsuoka, F.; Tsujita, T.; Suzuki, N.; Kobayashi, A.; Igarashi, K.; et al. O-GlcNAcylation Signal Mediates Proteasome Inhibitor Resistance in Cancer Cells by Stabilizing NRF1. Mol. Cell. Biol. 2018, 38, e00252-18. [Google Scholar] [CrossRef]
- Tomlin, F.M.; Gerling-Driessen, U.I.M.; Liu, Y.; Flynn, R.A.; Vangala, J.R.; Lentz, C.S.; Clauder-Muenster, S.; Jakob, P.; Mueller, W.F.; Ordoñez-Rueda, D.; et al. Inhibition of NGLY1 Inactivates the Transcription Factor Nrf1 and Potentiates Proteasome Inhibitor Cytotoxicity. ACS Cent. Sci. 2017, 3, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Steffen, J.; Seeger, M.; Koch, A.; Krüger, E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 2010, 40, 147–158. [Google Scholar] [CrossRef]
- Radhakrishnan, S.K.; Besten, W.D.; Deshaies, R.J. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. elife 2014, 3, e01856. [Google Scholar] [CrossRef]
- Koizumi, S.; Hamazaki, J.; Murata, S. Transcriptional regulation of the 26S proteasome by Nrf1. Phys. Biol. Sci. 2018, 94, 325–336. [Google Scholar] [CrossRef]
- Abraham, A.; Kattoor, A.J.; Saldeen, T.; Mehta, J.L. Vitamin E and its anticancer effects. Crit. Rev. Food Sci. Nutr. 2019, 59, 2831–2838. [Google Scholar] [CrossRef]
- Kaneko, S.; Sato, C.; Shiozawa, N.; Sato, A.; Sato, H.; Virgona, N.; Yano, T. Suppressive Effect of Delta-Tocotrienol on Hypoxia Adaptation of Prostate Cancer Stem-like Cells. Anticancer. Res. 2018, 38, 1391–1399. [Google Scholar]
- Rajendran, P.; Li, F.; Manu, A.K.; Shanmugam, K.M.; Loo, Y.S.; Kumar, P.A.S. Gγ-Tocotrienol is a novel inhibitor of constitutive and inducible STAT3 signalling pathway in human hepatocellular carcinoma: Potential role as an antiproliferative, pro-apoptotic and chemosensitizing agent. Br. J. Pharmacol. 2011, 163, 283–298. [Google Scholar] [CrossRef]
- Savitskaya, M.A.; Onischenko, G.E. α-Tocopheryl Succinate Affects Malignant Cell Viability, Proliferation, and Differentiation. Biochemistry 2016, 81, 806–818. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, K.; Virgona, N.; Harada, K.; Kido, W.; Yano, Y.; Ando, A.; Hagiwara, K.; Yano, T. A redox-silent analogue of tocotrienol acts as a potential cytotoxic agent against human mesothelioma cells. Life Sci. 2009, 84, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, N.; Sugahara, R.; Namiki, K.; Sato, C.; Ando, A.; Sato, A.; Virgona, N.; Yano, T. Inhibitory effect of a redox-silent analogue of tocotrienol on hypoxia adaptation in prostate cancer cells. Anticancer. Drugs 2017, 28, 289–297. [Google Scholar] [CrossRef]
- Ramdas, P.; Radhakrishnan, A.K.; Sani, A.A.; Kumari, M.; Rao, J.S.; Rahman, P.S. Advancing the Role of Gamma-Tocotrienol as Proteasomes Inhibitor: A Quantitative Proteomic Analysis of MDA-MB-231 Human Breast Cancer Cells. Biomolecules 2019, 10, 19. [Google Scholar] [CrossRef]
- Ishii, K.; Fusegi, M.; Mori, T.; Teshima, K.; Ninomiya, N.; Kohno, K.; Sato, A.; Ishida, T.; Miyakoshi, Y.; Yano, T. A Redox-Silent Analogue of Tocotrienol May Break the Homeostasis of Proteasomes in Human Malignant Mesothelioma Cells by Inhibiting STAT3 and NRF1. Int. J. Mol. Sci. 2022, 23, 2655. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.; Taubert, R.M.; Haberecht, S.; Venz, S.; Krüger, E. Inhibition of calpain-1 stabilizes TCF11/Nrf1 but does not affect its activation in response to proteasome inhibition. Biosci. Rep. 2018, 38, BSR20180393. [Google Scholar] [CrossRef]
- Arlt, A.; Sebens, S.; Krebs, S.; Geismann, C.; Grossmann, M.; Kruse, M.L.; Schreiber, S.; Schäfer, H. Inhibition of the NRF2 transcription factor by the alkaloid trigonelline renders pancreatic cancer cells more susceptible to apoptosis through decreased proteasomal gene expression and proteasome activity. Oncogene 2013, 32, 4825–4835. [Google Scholar] [CrossRef]
- Waku, T.; Katayama, H.; Hiraoka, M.; Hatanaka, A.; Nakamura, N.; Tanaka, Y.; Tamura, N.; Watanabe, A.; Kobayashi, A. NFE2L1 and NFE2L3 Complementarily Maintain Basal Proteasome Activity in Cancer Cells through CPEB3-Mediated Translational Repression. Mol. Cell Biol. 2020, 40, e00010-20. [Google Scholar] [CrossRef]
- Widenmaier, B.S.; Snyder, A.N.; Nguyen, B.T.; Arduini, A.; Lee, Y.G.; Arruda, P.A.; Saksi, J.; Bartelt, A.; Hotamisligil, S.G. NRF1 Is an ER Membrane Sensor that Is Central to Cholesterol Homeostasis. Cell. 2017, 171, 1094–1109. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, A.M.; Katoh, H.; Hatanaka, A.; Iwanari, H.; Nakamura, N.; Hamakubo, T.; Natsume, T.; Waku, T.; Kobayashi, A. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci. Rep. 2017, 7, 12494. [Google Scholar] [CrossRef] [PubMed]
- Waku, T.; Nakamura, N.; Koji, M.; Watanabe, H.; Katoh, H.; Tatsumi, C.; Tamura, N.; Hatanaka, A.; Hirose, S.; Katayama, H.; et al. NRF3-POMP-20S Proteasome Assembly Axis Promotes Cancer Development via Ubiquitin-Independent Proteolysis of p53 and Retinoblastoma Protein. J. Mol. Cell Biol. 2020, 40, e00597-19. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Yoshida, Y.; Nishio, K.; Hayakawa, M.; Niki, E. Characterization of cellular uptake and distribution of vitamin E. Ann. N. Y. Acad. Sci. 2004, 1031, 368–375. [Google Scholar] [CrossRef]
- Irías-mata, A.; Sus, N.; Flory, S.; Stock, D.; Woerner, D.; Podszun, M.; Frank, J. α-tocopherol transfer protein does not regulate the cellular uptake and intracellular distribution of α- and γ-tocopherols and -tocotrienols in cultured liver cells. Redox Biol. 2018, 19, 28–36. [Google Scholar] [CrossRef]
- Fassmannová, D.; Sedlák, F.; Sedláček, J.; Špička, I.; Šašková, G.K. Nelfinavir Inhibits the TCF11/Nrf1-Mediated Proteasome Recovery Pathway in Multiple Myeloma. Cancers 2020, 12, 1065. [Google Scholar] [CrossRef]
- Yano, Y.; Satoh, H.; Fukumoto, K.; Kumadaki, I.; Ichikawa, T.; Yamada, K.; Hagiwara, K.; Yano, T. Induction of cytotoxicity in human lung adenocarcinoma cells by 6-O-carboxypropyl-alpha-tocotrienol, a redox-silent derivative of alpha-tocotrienol. Int. J. Cancer 2005, 115, 839–846. [Google Scholar] [CrossRef]

| Gene Name | Primer | Sequence |
|---|---|---|
| 60S ribosomal protein L32 (RPL32) | Forward | AACCCTGTTGTCAATGCCTC |
| Reverse | CATCTCCTTCTCGGCATCA | |
| Proteasome 20S Subunit Alpha 7 (PSMA7) | Forward | CTGTGCTTTGGATGACAACG |
| Reverse | CGATGTAGCGGGTGATGTACT | |
| Proteasome 20S Subunit Beta 4 (PSMB4) | Forward | TCAGTCCTCGGCGTTAAGTT |
| Reverse | GCTTAGCACTGGCTGCTTCT | |
| Proteasome 20S Subunit Beta 7 (PSMB7) | Forward | CGGCTGTGTCGGTGTATG |
| Reverse | GCCAGTTTTCCGGACCTT | |
| Proteasome 26S Subunit ATPase4 (PSMC4) | Forward | GGAAGACCATGTTGGCAAAG |
| Reverse | AAGATGATGGCAGGTGCATT | |
| Proteasome maturation protein (POMP) | Forward | AGGCAGTGCAGCAGGTTC |
| Reverse | GGCTCTCCCATGACTTCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishii, K.; Hido, M.; Sakamura, M.; Virgona, N.; Yano, T. α-Tocotrienol and Redox-Silent Analogs of Vitamin E Enhances Bortezomib Sensitivity in Solid Cancer Cells through Modulation of NFE2L1. Int. J. Mol. Sci. 2023, 24, 9382. https://doi.org/10.3390/ijms24119382
Ishii K, Hido M, Sakamura M, Virgona N, Yano T. α-Tocotrienol and Redox-Silent Analogs of Vitamin E Enhances Bortezomib Sensitivity in Solid Cancer Cells through Modulation of NFE2L1. International Journal of Molecular Sciences. 2023; 24(11):9382. https://doi.org/10.3390/ijms24119382
Chicago/Turabian StyleIshii, Kyota, Mayuko Hido, Misaki Sakamura, Nantiga Virgona, and Tomohiro Yano. 2023. "α-Tocotrienol and Redox-Silent Analogs of Vitamin E Enhances Bortezomib Sensitivity in Solid Cancer Cells through Modulation of NFE2L1" International Journal of Molecular Sciences 24, no. 11: 9382. https://doi.org/10.3390/ijms24119382
APA StyleIshii, K., Hido, M., Sakamura, M., Virgona, N., & Yano, T. (2023). α-Tocotrienol and Redox-Silent Analogs of Vitamin E Enhances Bortezomib Sensitivity in Solid Cancer Cells through Modulation of NFE2L1. International Journal of Molecular Sciences, 24(11), 9382. https://doi.org/10.3390/ijms24119382








