Integrative Analysis Reveals the Diverse Effects of 3D Stiffness upon Stem Cell Fate
Abstract
1. Introduction
2. Results and Discussion
2.1. Characterization of GelMA Stiffness
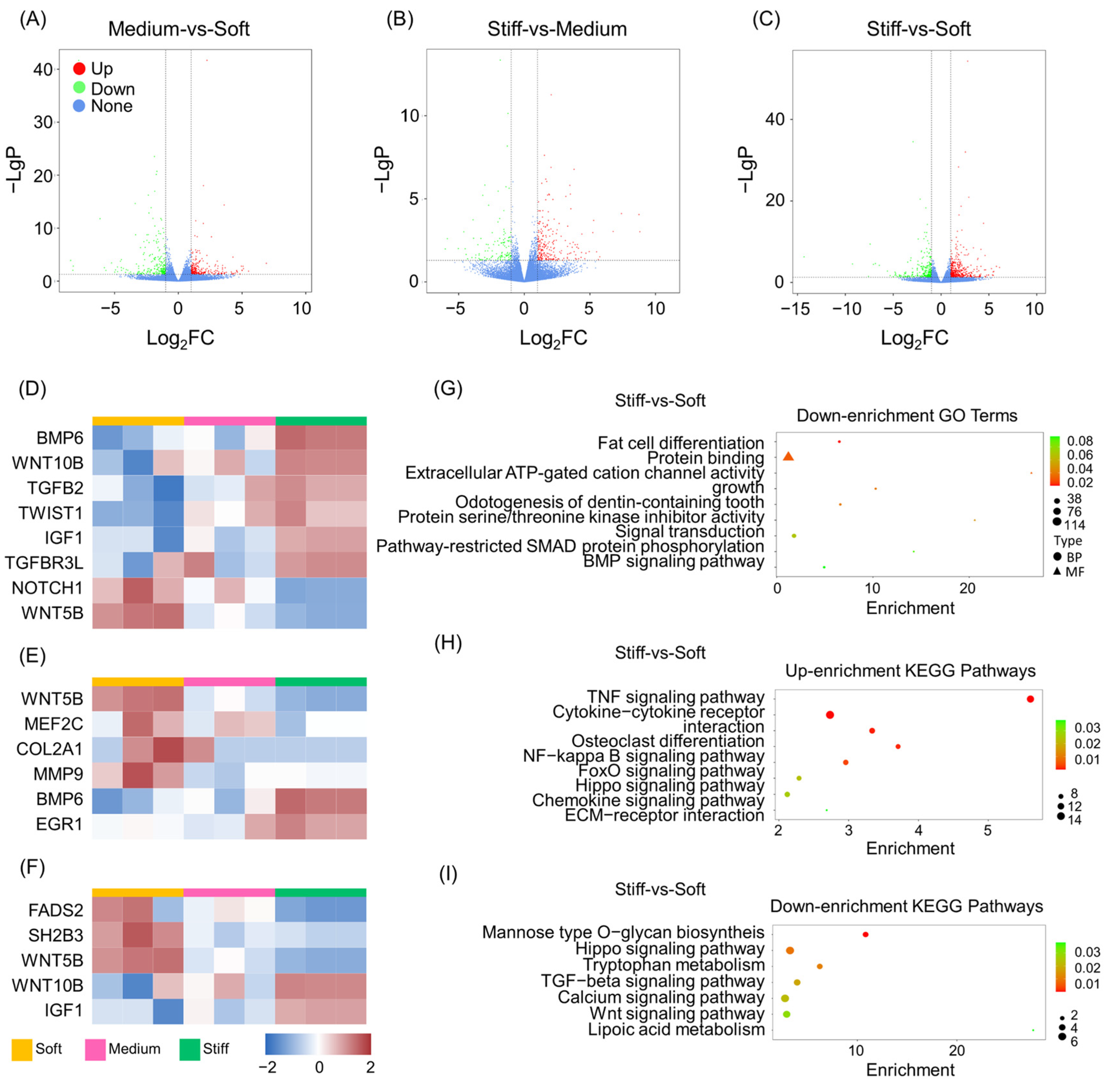
2.2. Overview of Transcriptome Analysis
2.3. GO and KEGG Pathway Analysis of DEGs
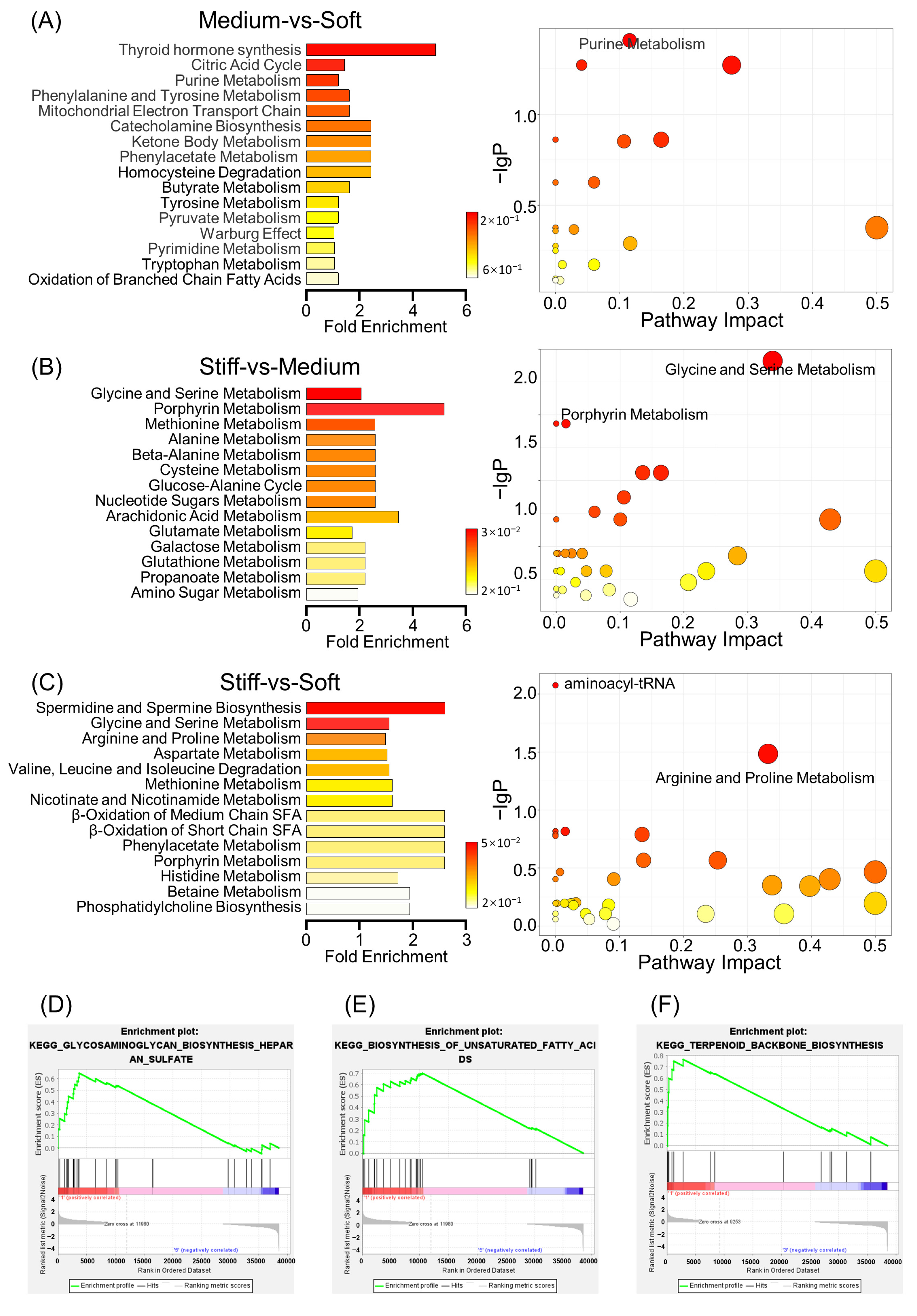
2.4. Overview of Metabolomics Analysis
2.5. Metabolite Set Enrichment Analysis and Metabolic Pathway Analysis
2.6. Comprehensive Analysis of Metabolomics and Transcriptomics
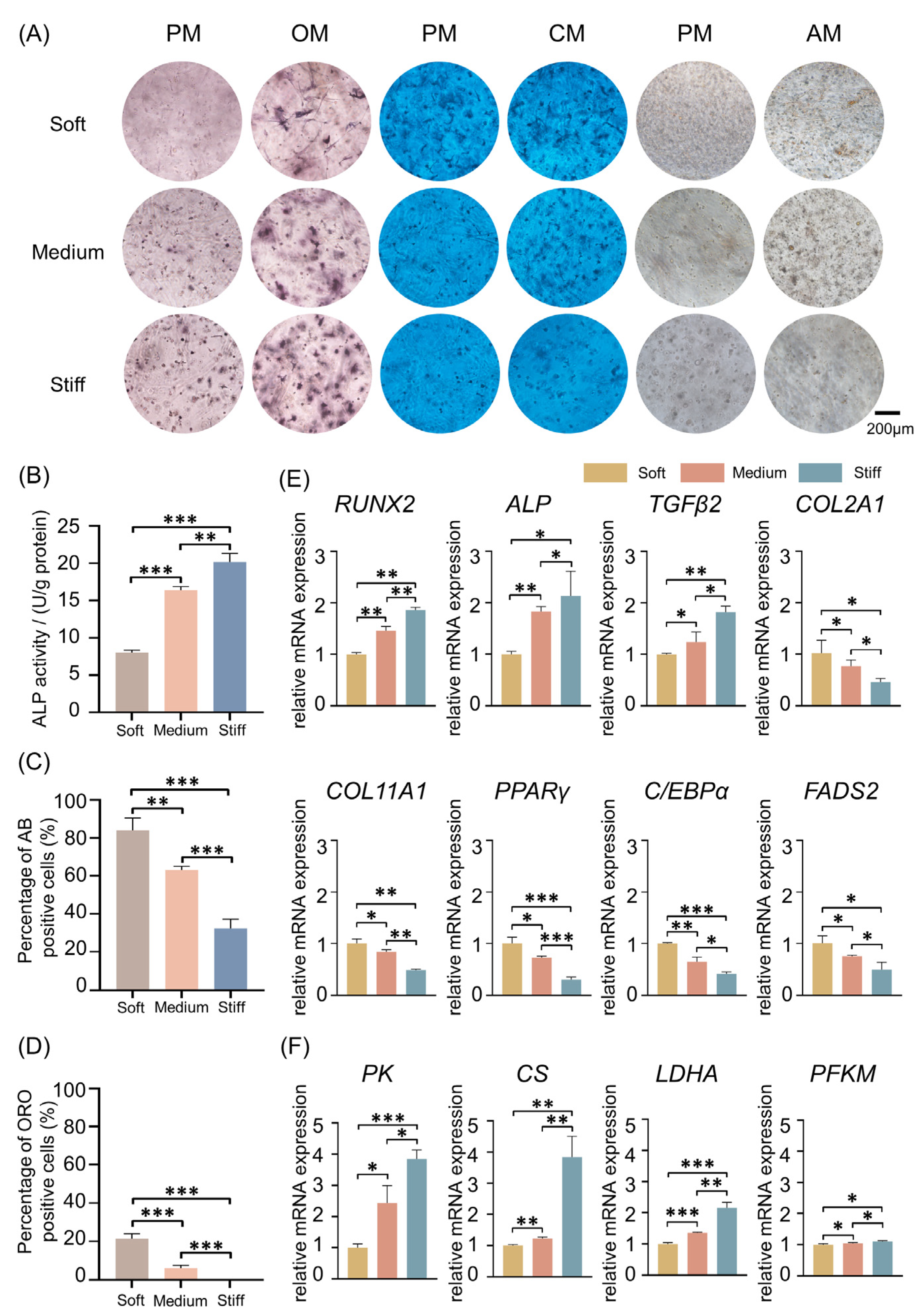
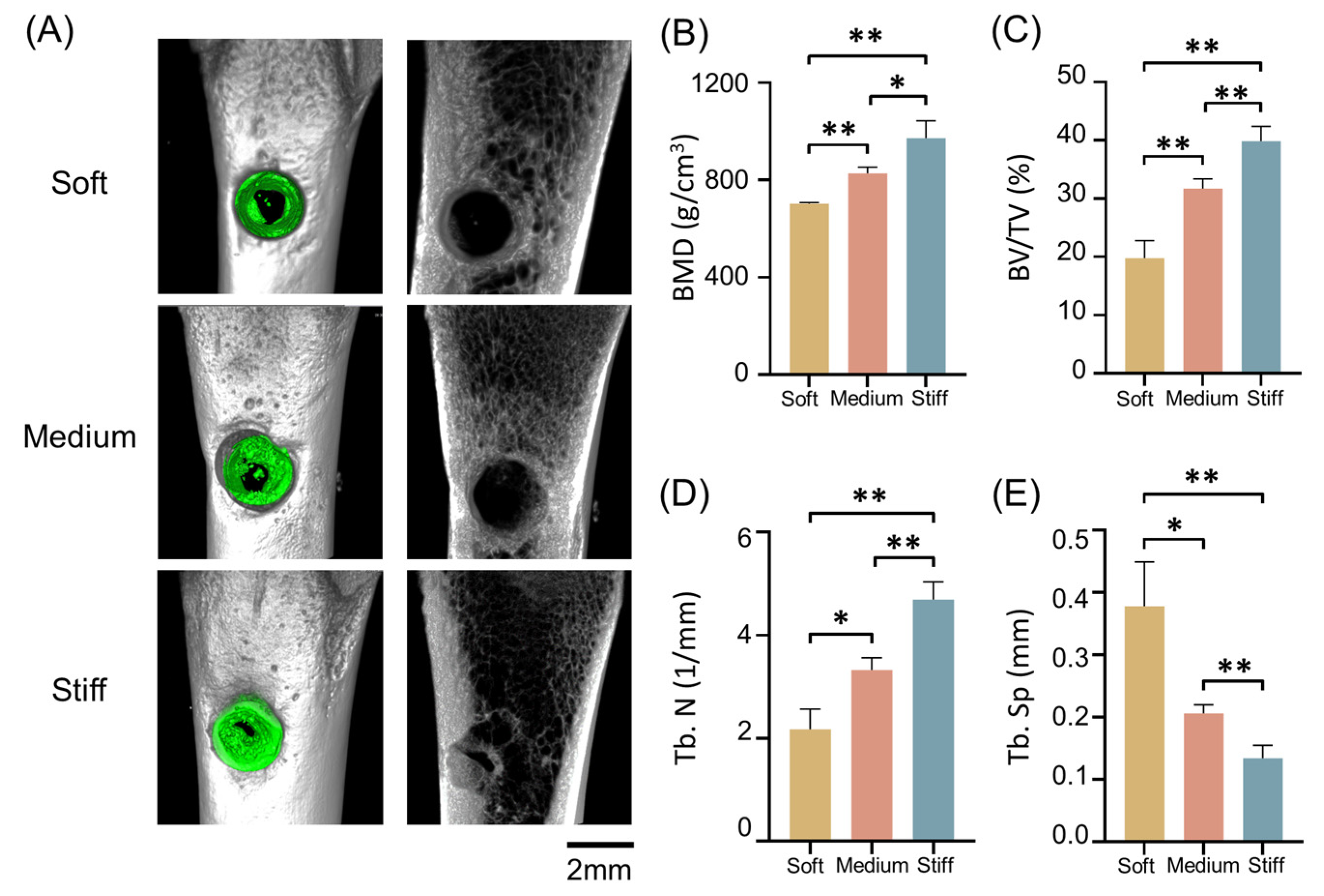
2.7. Validation In Vitro and In Vivo
3. Materials and Methods
3.1. Physicochemical Properties of GelMA
3.2. Cell Culture
3.3. Sequencing and Data Analysis
3.4. ALP Staining and Quantification
3.5. Oil Red O Staining and Quantification
3.6. Alcian Blue Staining and Quantification
3.7. Live/Dead Cell Staining
3.8. Immunostaining of Cell Culture
3.9. qRT-PCR Analysis
3.10. Animal Model
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crowder, S.W.; Leonardo, V.; Whittaker, T.; Papathanasiou, P.; Stevens, M.M. Material Cues as Potent Regulators of Epigenetics and Stem Cell Function. Cell Stem Cell 2016, 18, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.L.; McDevitt, T.C.; Engler, A.J. Materials as stem cell regulators. Nat. Mater. 2014, 13, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, S.; Tahmasebi Birgani, Z.; Habibovic, P. Biomaterial-induced pathway modulation for bone regeneration. Biomaterials 2022, 283, 121431. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.R.; Cho, S.; Discher, D.E. Stem Cell Differentiation is Regulated by Extracellular Matrix Mechanics. Physiology 2018, 33, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Tian, M.; Pei, Q.; Tan, F.; Pei, H. Extracellular Matrix Stiffness: New Areas Affecting Cell Metabolism. Front. Oncol. 2021, 11, 631991. [Google Scholar] [CrossRef] [PubMed]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742. [Google Scholar] [CrossRef]
- Darnell, M.; Gu, L.; Mooney, D. RNA-seq reveals diverse effects of substrate stiffness on mesenchymal stem cells. Biomaterials 2018, 181, 182–188. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Meng, Z.; Qiu, Y.; Lin, K.C.; Kumar, A.; Placone, J.K.; Fang, C.; Wang, K.C.; Lu, S.; Pan, M.; Hong, A.W.; et al. RAP2 mediates mechanoresponses of the Hippo pathway. Nature 2018, 560, 655–660. [Google Scholar] [CrossRef]
- Huebsch, N.; Arany, P.R.; Mao, A.S.; Shvartsman, D.; Ali, O.A.; Bencherif, S.A.; Rivera-Feliciano, J.; Mooney, D.J. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat. Mater. 2010, 9, 518–526. [Google Scholar] [CrossRef]
- Darnell, M.; Young, S.; Gu, L.; Shah, N.; Lippens, E.; Weaver, J.; Duda, G.; Mooney, D. Substrate Stress-Relaxation Regulates Scaffold Remodeling and Bone Formation In Vivo. Adv. Healthc. Mater. 2017, 6, 1601185. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Xie, J.; Huck, W.T.S. Recent Advances in Engineering the Stem Cell Microniche in 3D. Adv. Sci. 2018, 5, 1800448. [Google Scholar] [CrossRef] [PubMed]
- Pek, Y.S.; Wan, A.C.A.; Ying, J.Y. The effect of matrix stiffness on mesenchymal stem cell differentiation in a 3D thixotropic gel. Biomaterials 2010, 31, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Kamperman, T.; Willemen, N.G.A.; Kelder, C.; Koerselman, M.; Becker, M.; Lins, L.; Johnbosco, C.; Karperien, M.; Leijten, J. Steering Stem Cell Fate within 3D Living Composite Tissues Using Stimuli-Responsive Cell-Adhesive Micromaterials. Adv. Sci. 2023, 10, e2205487. [Google Scholar] [CrossRef] [PubMed]
- Kamperman, T.; Henke, S.; Crispim, J.F.; Willemen, N.G.A.; Dijkstra, P.J.; Lee, W.; Offerhaus, H.L.; Neubauer, M.; Smink, A.M.; de Vos, P. Tethering Cells via Enzymatic Oxidative Crosslinking Enables Mechanotransduction in Non-Cell-Adhesive Materials. Adv. Mater. 2021, 33, e2102660. [Google Scholar] [CrossRef]
- Lin, C.; He, Y.; Feng, Q.; Xu, K.; Chen, Z.; Tao, B.; Li, X.; Xia, Z.; Jiang, H.; Cai, K. Self-renewal or quiescence? Orchestrating the fate of mesenchymal stem cells by matrix viscoelasticity via PI3K/Akt-CDK1 pathway. Biomaterials 2021, 279, 121235. [Google Scholar] [CrossRef]
- Sinha, S.; Ayushman, M.; Tong, X.; Yang, F. Dynamically Crosslinked Poly(ethylene-glycol) Hydrogels Reveal a Critical Role of Viscoelasticity in Modulating Glioblastoma Fates and Drug Responses in 3D. Adv. Healthc. Mater. 2023, 12, e2202147. [Google Scholar] [CrossRef]
- Baek, J.; Lopez, P.A.; Lee, S.; Kim, T.S.; Kumar, S.; Schaffer, D.V. Egr1 is a 3D matrix-specific mediator of mechanosensitive stem cell lineage commitment. Sci. Adv. 2022, 8, eabm4646. [Google Scholar] [CrossRef]
- Cossu, G.; Birchall, M.; Brown, T.; De Coppi, P.; Culme-Seymour, E.; Gibbon, S.; Hitchcock, J.; Mason, C.; Montgomery, J.; Morris, S.; et al. Lancet Commission: Stem cells and regenerative medicine. Lancet 2018, 391, 883–910. [Google Scholar] [CrossRef]
- Meleshina, A.V.; Dudenkova, V.V.; Bystrova, A.S.; Kuznetsova, D.S.; Shirmanova, M.V.; Zagaynova, E.V. Two-photon FLIM of NAD(P)H and FAD in mesenchymal stem cells undergoing either osteogenic or chondrogenic differentiation. Stem Cell Res. Ther. 2017, 8, 15. [Google Scholar] [CrossRef]
- Yang, H.; Cheam, N.M.J.; Cao, H.; Lee, M.K.H.; Sze, S.K.; Tan, N.S.; Tay, C.Y. Materials Stiffness-Dependent Redox Metabolic Reprogramming of Mesenchymal Stem Cells for Secretome-Based Therapeutic Angiogenesis. Adv. Healthc. Mater. 2019, 8, e1900929. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Liu, S.; Yang, B.; Wang, R.; Man, G.; Wang, D.E.; Xu, H. Update on the effects of energy metabolism in bone marrow mesenchymal stem cells differentiation. Mol. Metab. 2022, 58, 101450. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Cooper-White, J.; Janmey, P.A.; Mooney, D.J.; Shenoy, V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 2020, 584, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Chu, K.; Shrestha, A.; Revelo, X.S.; Zhang, X.; Gold, M.J.; Khan, S.; Lee, M.; Huang, C.; Akbari, M.; et al. Mechanical Stiffness Controls Dendritic Cell Metabolism and Function. Cell Rep. 2021, 34, 108609. [Google Scholar] [CrossRef]
- Park, J.S.; Burckhardt, C.J.; Lazcano, R.; Solis, L.M.; Isogai, T.; Li, L.; Chen, C.S.; Gao, B.; Minna, J.D.; Bachoo, R.; et al. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature 2020, 578, 621–626. [Google Scholar] [CrossRef]
- Zhou, Q.; Ren, X.; Oberoi, M.K.; Bedar, M.; Caprini, R.M.; Dewey, M.J.; Kolliopoulos, V.; Yamaguchi, D.T.; Harley, B.A.C.; Lee, J.C. β-Catenin Limits Osteogenesis on Regenerative Materials in a Stiffness-Dependent Manner. Adv. Healthc. Mater. 2021, 10, e2101467. [Google Scholar] [CrossRef]
- Niehrs, C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 767–779. [Google Scholar] [CrossRef]
- Brommage, R.; Liu, J.; Hansen, G.M.; Kirkpatrick, L.L.; Potter, D.G.; Sands, A.T.; Zambrowicz, B.; Powell, D.R.; Vogel, P. High-throughput screening of mouse gene knockouts identifies established and novel skeletal phenotypes. Bone Res. 2014, 2, 14034. [Google Scholar] [CrossRef]
- Sales, A.; Khodr, V.; Machillot, P.; Chaar, L.; Fourel, L.; Guevara-Garcia, A.; Migliorini, E.; Albigès-Rizo, C.; Picart, C. Differential bioactivity of four BMP-family members as function of biomaterial stiffness. Biomaterials 2022, 281, 121363. [Google Scholar] [CrossRef]
- De Ugarte, D.A.; Morizono, K.; Elbarbary, A.; Alfonso, Z.; Zuk, P.A.; Zhu, M.; Dragoo, J.L.; Ashjian, P.; Thomas, B.; Benhaim, P.; et al. Comparison of multi-lineage cells from human adipose tissue and bone m arrow. Cells Tissues Organs 2003, 174, 101–109. [Google Scholar] [CrossRef]
- Vukicevic, S.; Grgurevic, L. BMP-6 and mesenchymal stem cell differentiation. Cytokine Growth Factor Rev. 2009, 20, 441–448. [Google Scholar] [CrossRef] [PubMed]
- McDermott, A.M.; Herberg, S.; Mason, D.E.; Collins, J.M.; Pearson, H.B.; Dawahare, J.H.; Tang, R.; Patwa, A.N.; Grinstaff, M.W.; Kelly, D.J.; et al. Recapitulating bone development through engineered mesenchymal condensations and mechanical cues for tissue regeneration. Sci. Transl. Med. 2019, 11, eaav7756. [Google Scholar] [CrossRef] [PubMed]
- Palomares, K.T.; Gleason, R.E.; Mason, Z.D.; Cullinane, D.M.; Einhorn, T.A.; Gerstenfeld, L.C.; Morgan, E.F. Mechanical stimulation alters tissue differentiation and molecular expression during bone healing. J. Orthop. Res. 2009, 27, 1123–1132. [Google Scholar] [CrossRef]
- Xuan, Y.; Wang, H.; Yung, M.M.; Chen, F.; Chan, W.S.; Chan, Y.S.; Tsui, S.K.; Ngan, H.Y.; Chan, K.K.; Chan, D.W. SCD1/FADS2 fatty acid desaturases equipoise lipid metabolic activity and redox-driven ferroptosis in ascites-derived ovarian cancer cells. Theranostics 2022, 12, 3534–3552. [Google Scholar] [CrossRef] [PubMed]
- Mu, M.; Gao, P.; Yang, Q.; He, J.; Wu, F.; Han, X.; Guo, S.; Qian, Z.; Song, C. Alveolar Epithelial Cells Promote IGF-1 Production by Alveolar Macrophages through TGF-β to Suppress Endogenous Inflammatory Signals. Front. Immunol. 2020, 11, 1585. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.; Zhang, Y.; Ellyard, J.I.; Vinuesa, C.G.; Murphy, J.M.; Laktyushin, A.; Kershaw, N.J.; Babon, J.J. Structural and functional analysis of target recognition by the lymphocyte adaptor protein LNK. Nat. Commun. 2021, 12, 6110. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Lane, M.D. Adipogenesis: From stem cell to adipocyte. Annu. Rev. Biochem. 2012, 81, 715–736. [Google Scholar] [CrossRef]
- Hu, L.; Yang, G.; Hägg, D.; Sun, G.; Ahn, J.M.; Jiang, N.; Ricupero, C.L.; Wu, J.; Rodhe, C.H.; Ascherman, J.A.; et al. IGF1 Promotes Adipogenesis by a Lineage Bias of Endogenous Adipose Stem/Progenitor Cells. Stem Cells 2015, 33, 2483–2495. [Google Scholar] [CrossRef]
- Suthon, S.; Perkins, R.S.; Bryja, V.; Miranda-Carboni, G.A.; Krum, S.A. WNT5B in Physiology and Disease. Front. Cell Dev. Biol. 2021, 9, 667581. [Google Scholar] [CrossRef]
- Bagchi, D.P.; MacDougald, O.A. Wnt Signaling: From Mesenchymal Cell Fate to Lipogenesis and Other Mature Adipocyte Functions. Diabetes 2021, 70, 1419–1430. [Google Scholar] [CrossRef]
- Kanazawa, A.; Tsukada, S.; Sekine, A.; Tsunoda, T.; Takahashi, A.; Kashiwagi, A.; Tanaka, Y.; Babazono, T.; Matsuda, M.; Kaku, K.; et al. Association of the gene encoding wingless-type mammary tumor virus integration-site family member 5B (WNT5B) with type 2 diabetes. Am. J. Hum. Genet. 2004, 75, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Han, W.; Qin, A.; Wang, Z.; Xu, J.; Qian, Y. The emerging role of Hippo signaling pathway in regulating osteoclast formation. J. Cell. Physiol. 2018, 233, 4606–4617. [Google Scholar] [CrossRef] [PubMed]
- Saidova, A.A.; Vorobjev, I.A. Lineage Commitment, Signaling Pathways, and the Cytoskeleton Systems in Mesenchymal Stem Cells. Tissue Eng. Part B Rev. 2020, 26, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.X.; Xiong, L.; Zhao, K.; Zeng, P.; Wang, B.; Tang, F.L.; Sun, D.; Guo, H.H.; Yang, X.; Cui, S.; et al. YAP promotes osteogenesis and suppresses adipogenic differentiation by regulating β-catenin signaling. Bone Res. 2018, 6, 18. [Google Scholar] [CrossRef]
- Zhang, N.; Kandalai, S.; Zhou, X.; Hossain, F.; Zheng, Q. Applying multi-omics toward tumor microbiome research. iMeta 2023, 2, e73. [Google Scholar] [CrossRef]
- Tharp, K.M.; Higuchi-Sanabria, R.; Timblin, G.A.; Ford, B.; Garzon-Coral, C.; Schneider, C.; Muncie, J.M.; Stashko, C.; Daniele, J.R.; Moore, A.S.; et al. Adhesion-mediated mechanosignaling forces mitohormesis. Cell Metab. 2021, 33, 1322–1341.e13. [Google Scholar] [CrossRef]
- Dupont, S.; Wickström, S.A. Mechanical regulation of chromatin and transcription. Nat. Rev. Genet. 2022, 23, 624–643. [Google Scholar] [CrossRef]
- Xie, J.; Bao, M.; Hu, X.; Koopman, W.J.H.; Huck, W.T.S. Energy expenditure during cell spreading influences the cellular response to matrix stiffness. Biomaterials 2021, 267, 120494. [Google Scholar] [CrossRef]
- Wan, M.C.; Tang, X.Y.; Li, J.; Gao, P.; Wang, F.; Shen, M.J.; Gu, J.T.; Tay, F.; Chen, J.H.; Niu, L.N.; et al. Upregulation of mitochondrial dynamics is responsible for osteogenic differentiation of mesenchymal stem cells cultured on self-mineralized collagen membranes. Acta Biomater. 2021, 136, 137–146. [Google Scholar] [CrossRef]
- Bertero, T.; Oldham, W.M.; Cottrill, K.A.; Pisano, S.; Vanderpool, R.R.; Yu, Q.; Zhao, J.; Tai, Y.; Tang, Y.; Zhang, Y.Y.; et al. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J. Clin. Investig. 2016, 126, 3313–3335. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Oldham, W.M.; Grasset, E.M.; Bourget, I.; Boulter, E.; Pisano, S.; Hofman, P.; Bellvert, F.; Meneguzzi, G.; Bulavin, D.V.; et al. Tumor-Stroma Mechanics Coordinate Amino Acid Availability to Sustain Tumor Growth and Malignancy. Cell Metab. 2019, 29, 124–140.e10. [Google Scholar] [CrossRef] [PubMed]
- Torrino, S.; Grasset, E.M.; Audebert, S.; Belhadj, I.; Lacoux, C.; Haynes, M.; Pisano, S.; Abélanet, S.; Brau, F.; Chan, S.Y.; et al. Mechano-induced cell metabolism promotes microtubule glutamylation to force metastasis. Cell Metab. 2021, 33, 1342–1357.e10. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Newman, H.; Shen, L.; Sharma, D.; Hu, G.; Mirando, A.J.; Zhang, H.; Knudsen, E.; Zhang, G.F.; Hilton, M.J.; et al. Glutamine Metabolism Regulates Proliferation and Lineage Allocation in Skeletal Stem Cells. Cell Metab. 2019, 29, 966–978.e964. [Google Scholar] [CrossRef] [PubMed]
- Donat, A.; Knapstein, P.R.; Jiang, S.; Baranowsky, A.; Ballhause, T.M.; Frosch, K.H.; Keller, J. Glucose Metabolism in Osteoblasts in Healthy and Pathophysiological Conditions. Int. J. Mol. Sci. 2021, 22, 4120. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Iwata, J. Amino acid metabolism and autophagy in skeletal development and homeostasis. Bone 2021, 146, 115881. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef]
- Ramsay, A.L.; Alonso-Garcia, V.; Chaboya, C.; Radut, B.; Le, B.; Florez, J.; Schumacher, C.; Fierro, F.A. Modeling Snyder-Robinson Syndrome in multipotent stromal cells reveals impaired mitochondrial function as a potential cause for deficient osteogenesis. Sci. Rep. 2019, 9, 15395. [Google Scholar] [CrossRef]
- Ohtaka, Y.; Spiegelman, S. Translational Control of Protein Synthesis in a Cell-Free System Directed by a Polycistronic Viral RNA. Science 1963, 142, 493–497. [Google Scholar] [CrossRef]
- Nováková, S.; Danchenko, M.; Okajčeková, T.; Baranovičová, E.; Kováč, A.; Grendár, M.; Beke, G.; Pálešová, J.; Strnádel, J.; Janíčková, M.; et al. Comparative Proteomic and Metabolomic Analysis of Human Osteoblasts, Differentiated from Dental Pulp Stem Cells, Hinted Crucial Signaling Pathways Promoting Osteogenesis. Int. J. Mol. Sci. 2021, 22, 7908. [Google Scholar] [CrossRef]
- Bartolák-Suki, E.; Imsirovic, J.; Nishibori, Y.; Krishnan, R.; Suki, B. Regulation of Mitochondrial Structure and Dynamics by the Cytoskeleton and Mechanical Factors. Int. J. Mol. Sci. 2017, 18, 1812. [Google Scholar] [CrossRef] [PubMed]
- Vercellino, I.; Sazanov, L.A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell Biol. 2022, 23, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Dobson, P.F.; Dennis, E.P.; Hipps, D.; Reeve, A.; Laude, A.; Bradshaw, C.; Stamp, C.; Smith, A.; Deehan, D.J.; Turnbull, D.M.; et al. Mitochondrial dysfunction impairs osteogenesis, increases osteoclast activity, and accelerates age related bone loss. Sci. Rep. 2020, 10, 11643. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X.; Zhang, C.; Wang, J.; Wang, S.; Hu, L. Dysfunction of metabolic activity of bone marrow mesenchymal stem cells in aged mice. Cell Prolif. 2022, 55, e13191. [Google Scholar] [CrossRef]
- Wei, J.; Hu, M.; Huang, K.; Lin, S.; Du, H. Roles of Proteoglycans and Glycosaminoglycans in Cancer Development and Progression. Int. J. Mol. Sci. 2020, 21, 5983. [Google Scholar] [CrossRef]
- Belting, M. Glycosaminoglycans in cancer treatment. Thromb. Res. 2014, 133 (Suppl. 2), S95–S101. [Google Scholar] [CrossRef]
- Tcw, J.; Qian, L.; Pipalia, N.H.; Chao, M.J.; Liang, S.A.; Shi, Y.; Jain, B.R.; Bertelsen, S.E.; Kapoor, M.; Marcora, E.; et al. Cholesterol and matrisome pathways dysregulated in astrocytes and microglia. Cell 2022, 185, 2213–2233.e25. [Google Scholar] [CrossRef]
- Bellavia, D.; Caradonna, F.; Dimarco, E.; Costa, V.; Carina, V.; De Luca, A.; Raimondi, L.; Gentile, C.; Alessandro, R.; Fini, M.; et al. Terpenoid treatment in osteoporosis: This is where we have come in research. Trends Endocrinol. Metab. 2021, 32, 846–861. [Google Scholar] [CrossRef]
- Wu, L.; Magaz, A.; Wang, T.; Liu, C.; Darbyshire, A.; Loizidou, M.; Emberton, M.; Birchall, M.; Song, W. Stiffness memory of indirectly 3D-printed elastomer nanohybrid regulates chondrogenesis and osteogenesis of human mesenchymal stem cells. Biomaterials 2018, 186, 64–79. [Google Scholar] [CrossRef]
- Serpooshan, V.; Julien, M.; Nguyen, O.; Wang, H.; Li, A.; Muja, N.; Henderson, J.E.; Nazhat, S.N. Reduced hydraulic permeability of three-dimensional collagen scaffolds attenuates gel contraction and promotes the growth and differentiation of mesenchymal stem cells. Acta Biomater. 2010, 6, 3978–3987. [Google Scholar] [CrossRef]
- Zhan, X. Effect of matrix stiffness and adhesion ligand density on chondrogenic differentiation of mesenchymal stem cells. J. Biomed. Mater. Res. A 2020, 108, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Cheng, X.; Jansen, J.A.; Leeuwenburgh, S.G.C.; Mao, J.; Yang, F.; Chen, L. The molecular conformation of silk fibroin regulates osteogenic cell behavior by modulating the stability of the adsorbed protein-material interface. Bone Res. 2021, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cheng, C.; Cuellar-Camacho, J.L.; Li, M.; Xia, Y.; Li, W. Thermally Responsive Microfibers Mediated Stem Cell Fate via Reversibly Dynamic Mechanical Stimulation. Adv. Funct. Mater. 2018, 28, 1804773. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 2019, 120, 2782–2790. [Google Scholar] [CrossRef] [PubMed]
- Paauwe, M.; Schoonderwoerd, M.J.A.; Helderman, R.F.C.P.; Harryvan, T.J.; Groenewoud, A.; van Pelt, G.W.; Bor, R.; Hemmer, D.M.; Versteeg, H.H.; Snaar-Jagalska, B.E.; et al. Endoglin Expression on Cancer-Associated Fibroblasts Regulates Invasion and Stimulates Colorectal Cancer Metastasis. Clin. Cancer Res. 2018, 24, 6331–6344. [Google Scholar] [CrossRef]
- Dauer, P.; Zhao, X.; Gupta, V.K.; Sharma, N.; Kesh, K.; Gnamlin, P.; Dudeja, V.; Vickers, S.M.; Banerjee, S.; Saluja, A. Inactivation of Cancer-Associated-Fibroblasts Disrupts Oncogenic Signa ling in Pancreatic Cancer Cells and Promotes Its Regression. Cancer Res. 2018, 78, 1321–1333. [Google Scholar] [CrossRef]
- Derynck, R.; Akhurst, R.J. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat. Cell Biol. 2007, 9, 1000–1004. [Google Scholar] [CrossRef]
- Geiser, A.G.; Hummel, C.W.; Draper, M.W.; Henck, J.W.; Cohen, I.R.; Rudmann, D.G.; Donnelly, K.B.; Adrian, M.D.; Shepherd, T.A.; Wallace, O.B.; et al. A new selective estrogen receptor modulator with potent uterine antagonist activity, agonist activity in bone, and minimal ovarian stimulation. Endocrinology 2005, 146, 4524–4535. [Google Scholar] [CrossRef]
- Dünker, N.; Krieglstein, K. Tgfbeta2 -/- Tgfbeta3 -/- double knockout mice display severe midline fusion defects and early embryonic lethality. Anat. Embryol. 2002, 206, 73–83. [Google Scholar] [CrossRef]
- Li, S.-N.; Wu, J.-F. TGF-β/SMAD signaling regulation of mesenchymal stem cells in adipocyte commitment. Stem Cell Res. Ther. 2020, 11, 41. [Google Scholar] [CrossRef]
- Finnson, K.W.; Parker, W.L.; ten Dijke, P.; Thorikay, M.; Philip, A. ALK1 opposes ALK5/Smad3 signaling and expression of extracellular matrix components in human chondrocytes. J. Bone Miner Res. 2008, 23, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Blaney Davidson, E.N.; Remst, D.F.G.; Vitters, E.L.; van Beuningen, H.M.; Blom, A.B.; Goumans, M.J.; van den Berg, W.B.; van der Kraan, P.M. Increase in ALK1/ALK5 ratio as a cause for elevated MMP-13 expression in osteoarthritis in humans and mice. J. Immunol. 2009, 182, 7937–7945. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-F.; Darowish, M.; Zuscik, M.J.; Chen, D.; Schwarz, E.M.; Rosier, R.N.; Drissi, H.; O’Keefe, R.J. Smad3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. J. Bone Miner. Res. 2006, 21, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, L.; Xu, X.; Li, C.; Huang, C.; Deng, C.X. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J. Cell. Biol. 2001, 153, 35–46. [Google Scholar] [CrossRef]
- Ferguson, C.M.; Schwarz, E.M.; Reynolds, P.R.; Puzas, J.E.; Rosier, R.N.; O’Keefe, R.J. Smad2 and 3 mediate transforming growth factor-beta1-induced inhibition of chondrocyte maturation. Endocrinology 2000, 141, 4728–4735. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-O.; Sampson, E.R.; Maynard, R.D.; O’Keefe, R.J.; Chen, D.; Drissi, H.; Rosier, R.N.; Hilton, M.J.; Zuscik, M.J. Ski inhibits TGF-β/hosphor-Smad3 signaling and accelerates hypertrophic differentiation in chondrocytes. J. Cell. Biochem. 2012, 113, 2156–2166. [Google Scholar] [CrossRef]
- Bielajew, B.J.; Hu, J.C.; Athanasiou, K.A. Collagen: Quantification, biomechanics, and role of minor subtypes in cartilage. Nat. Rev. Mater. 2020, 5, 730–747. [Google Scholar] [CrossRef]
- Deng, H.; Huang, X.; Yuan, L. Molecular genetics of the COL2A1-related disorders. Mutat. Res. Rev. Mutat. Res. 2016, 768, 1–13. [Google Scholar] [CrossRef]
- Hafez, A.; Squires, R.; Pedracini, A.; Joshi, A.; Seegmiller, R.E.; Oxford, J.T. Col11a1 Regulates Bone Microarchitecture during Embryonic Development. J. Dev. Biol. 2015, 3, 158–176. [Google Scholar] [CrossRef]
- Li, Y.; Lacerda, D.A.; Warman, M.L.; Beier, D.R.; Yoshioka, H.; Ninomiya, Y.; Oxford, J.T.; Morris, N.P.; Andrikopoulos, K.; Ramirez, F. A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell 1995, 80, 423–430. [Google Scholar] [CrossRef]
- Stoffel, W.; Hammels, I.; Jenke, B.; Binczek, E.; Schmidt-Soltau, I.; Brodesser, S.; Odenthal, M.; Thevis, M. Obesity resistance and deregulation of lipogenesis in Δ6-fatty acid desaturase (FADS2) deficiency. EMBO Rep. 2014, 15, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Zhang, Y.; Sampathkumar, A. Cytoskeleton Architecture Regulates Glycolysis Coupling Cellular Metabolism to Mechanical Cues. Trends Biochem. Sci. 2020, 45, 637–638. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yue, M.; Liu, X.; Liu, Y.; Lv, L.; Zhang, P.; Zhou, Y. The PCK2-glycolysis axis assists three-dimensional-stiffness maintaining stem cell osteogenesis. Bioact. Mater. 2022, 18, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Harada, Y.; Yamatoya, K.; Kawano, N.; Kanai, S.; Miyamoto, Y.; Nakamura, A.; Miyado, M.; Hayashi, Y.; Kuroki, Y.; et al. Extra-mitochondrial citrate synthase initiates calcium oscillation and suppresses age-dependent sperm dysfunction. Lab. Investig. 2020, 100, 583–595. [Google Scholar] [CrossRef]
- Chen, Q.; Xin, M.; Wang, L.; Li, L.; Shen, Y.; Geng, Y.; Jiang, H.; Wang, Y.; Zhang, L.; Xu, Y.; et al. Inhibition of LDHA to induce eEF2 release enhances thrombocytopoiesis. Blood 2022, 139, 2958–2971. [Google Scholar] [CrossRef]
- Komrakova, M.; Krischek, C.; Wicke, M.; Sehmisch, S.; Tezval, M.; Rohrberg, M.; Brandsch, T.; Stuermer, K.M.; Stuermer, E.K. Influence of intermittent administration of parathyroid hormone on muscle tissue and bone healing in orchiectomized rats or controls. J. Endocrinol. 2011, 209, 9–19. [Google Scholar] [CrossRef]
- Nian, F.; Qian, Y.; Xu, F.; Yang, M.; Wang, H.; Zhang, Z. LDHA promotes osteoblast differentiation through histone lactylation. Biochem. Biophys. Res. Commun. 2022, 615, 31–35. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Zeng, Q.; Xiao, Z.; Zhang, X.; Xu, M.; He, Y.; Wei, Y.; Deng, X. The Dynamic Counterbalance of RAC1-YAP/OB-Cadherin Coordinates Tissue Spreading with Stem Cell Fate Patterning. Adv. Sci. 2021, 8, 2004000. [Google Scholar] [CrossRef]
- Guo, Y.; Mei, F.; Huang, Y.; Ma, S.; Wei, Y.; Zhang, X.; Xu, M.; He, Y.; Heng, B.C.; Chen, L.; et al. Matrix stiffness modulates tip cell formation through the p-PXN-Rac1-YAP signaling axis. Bioact. Mater. 2021, 7, 364–376. [Google Scholar] [CrossRef]
- Guo, X.; Zheng, H.; Guo, Y.; Heng, B.C.; Yang, Y.; Yao, W.; Jiang, S. A three-dimensional actively spreading bone repair material based on cell spheroids can facilitate the preservation of tooth extraction sockets. Front. Bioeng. Biotechnol. 2023, 11, 1161192. [Google Scholar] [CrossRef]
- Ling, Y.; Zhang, W.; Wang, P.; Xie, W.; Yang, W.; Wang, D.A.; Fan, C. Three-dimensional (3D) hydrogel serves as a platform to identify potential markers of chondrocyte dedifferentiation by combining RNA sequencing. Bioact. Mater. 2021, 6, 2914–2926. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yue, M.; Heng, B.; Liu, Y.; Zhang, P.; Zhou, Y. Metformin can mitigate skeletal dysplasia caused by Pck2 deficiency. Int. J. Oral Sci. 2022, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Gu, L.; Klumpers, D.; Darnell, M.; Bencherif, S.A.; Weaver, J.C.; Huebsch, N.; Lee, H.P.; Lippens, E.; Duda, G.N.; et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2016, 15, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Z.; Liu, H.; Zhu, Y.; Xia, D.; Wang, S.; Gu, R.; Zhang, P.; Liu, Y.; Zhou, Y. Flufenamic Acid Inhibits Adipogenic Differentiation of Mesenchymal Stem Cells by Antagonizing the PI3K/AKT Signaling Pathway. Stem Cells Int. 2020, 2020, 1540905. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhong, D.; Qin, Z.; He, S.; Gong, Y.; Li, W.; Li, X. miR-100-3p inhibits the adipogenic differentiation of hMSCs by targeting PIK3R1 via the PI3K/AKT signaling pathway. Aging 2020, 12, 25090–25100. [Google Scholar] [CrossRef]
- Yang, F.; Liu, X.; Wei, D.; Zhu, Y.; Wang, F.; Liu, X.; Yan, F.; Zhang, X.; Liu, Y. Topical Application of Butyl Flufenamate Ointment Promotes Cranial Defect Healing in Mice by Inducing BMP2 Secretion in Skin Mesenchymal Stem Cells. Cells 2022, 11, 3620. [Google Scholar] [CrossRef]
- Costantini, M.; Idaszek, J.; Szöke, K.; Jaroszewicz, J.; Dentini, M.; Barbetta, A.; Brinchmann, J.E.; Święszkowski, W. 3D bioprinting of BM-MSCs-loaded ECM biomimetic hydrogels for in vitro neocartilage formation. Biofabrication 2016, 8, 035002. [Google Scholar] [CrossRef]


| Primer | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| GAPDH | GGTCACCAGGGCTGCTTTT | GGATCTCGCTCCTGGAAGATG |
| ALP | GACCTCCTCGGAAGACACTC | TGAAGGGCTTCTTGTCTGTG |
| RUNX2 | CCGCCTCAGTGATTTAGGGC | GGGTCTGTAATCTGACTCTGTCC |
| TGFB2 | CAACAGCACCAGGGACTTGC | AACTGGGCAGACAGTTTCGGA |
| COL11A1 | TCCTGGACCACCAGGAAGGAT | GACCAGTCTCACCGGTTGGT |
| COL2A1 | CCCATCTGCCCAACTGACCT | TTTGGTCCTGGTTGCCCACT |
| PPARγ | GAGGAGCCTAAGGTAAGGAG | GTCATTTCGTTAAAGGCTGA |
| C/EBPα | CGCAAGAGCCGAGATAAAGC | CACGGCTCAGCTGTTCCA |
| FADS2 | TGACCGCAAGGTTTACAACAT | AGGCATCCGTTGCATCTTCTC |
| PK | AGAGAGGCAGCCTTCAGACCT | CTGTTTTGTGCCCCGCAAGA |
| CS | TGGGTGTACTGGCACAGCTC | GTGCTCATGGACTTGGGCCT |
| LDHA | GGCTTGAGCTTTGTGGCAGT | GGCTCCTACAGCAAGGACACA |
| PFKM | GAGCACCATGCAGCCAAAAC | GCAGCATTCATACCTTGGGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, M.; Liu, Y.; Zhang, P.; Li, Z.; Zhou, Y. Integrative Analysis Reveals the Diverse Effects of 3D Stiffness upon Stem Cell Fate. Int. J. Mol. Sci. 2023, 24, 9311. https://doi.org/10.3390/ijms24119311
Yue M, Liu Y, Zhang P, Li Z, Zhou Y. Integrative Analysis Reveals the Diverse Effects of 3D Stiffness upon Stem Cell Fate. International Journal of Molecular Sciences. 2023; 24(11):9311. https://doi.org/10.3390/ijms24119311
Chicago/Turabian StyleYue, Muxin, Yunsong Liu, Ping Zhang, Zheng Li, and Yongsheng Zhou. 2023. "Integrative Analysis Reveals the Diverse Effects of 3D Stiffness upon Stem Cell Fate" International Journal of Molecular Sciences 24, no. 11: 9311. https://doi.org/10.3390/ijms24119311
APA StyleYue, M., Liu, Y., Zhang, P., Li, Z., & Zhou, Y. (2023). Integrative Analysis Reveals the Diverse Effects of 3D Stiffness upon Stem Cell Fate. International Journal of Molecular Sciences, 24(11), 9311. https://doi.org/10.3390/ijms24119311







