Physiological and Transcriptomic Analyses Reveal the Effects of Carbon-Ion Beam on Taraxacum kok-saghyz Rodin Adventitious Buds
Abstract
1. Introduction
2. Results
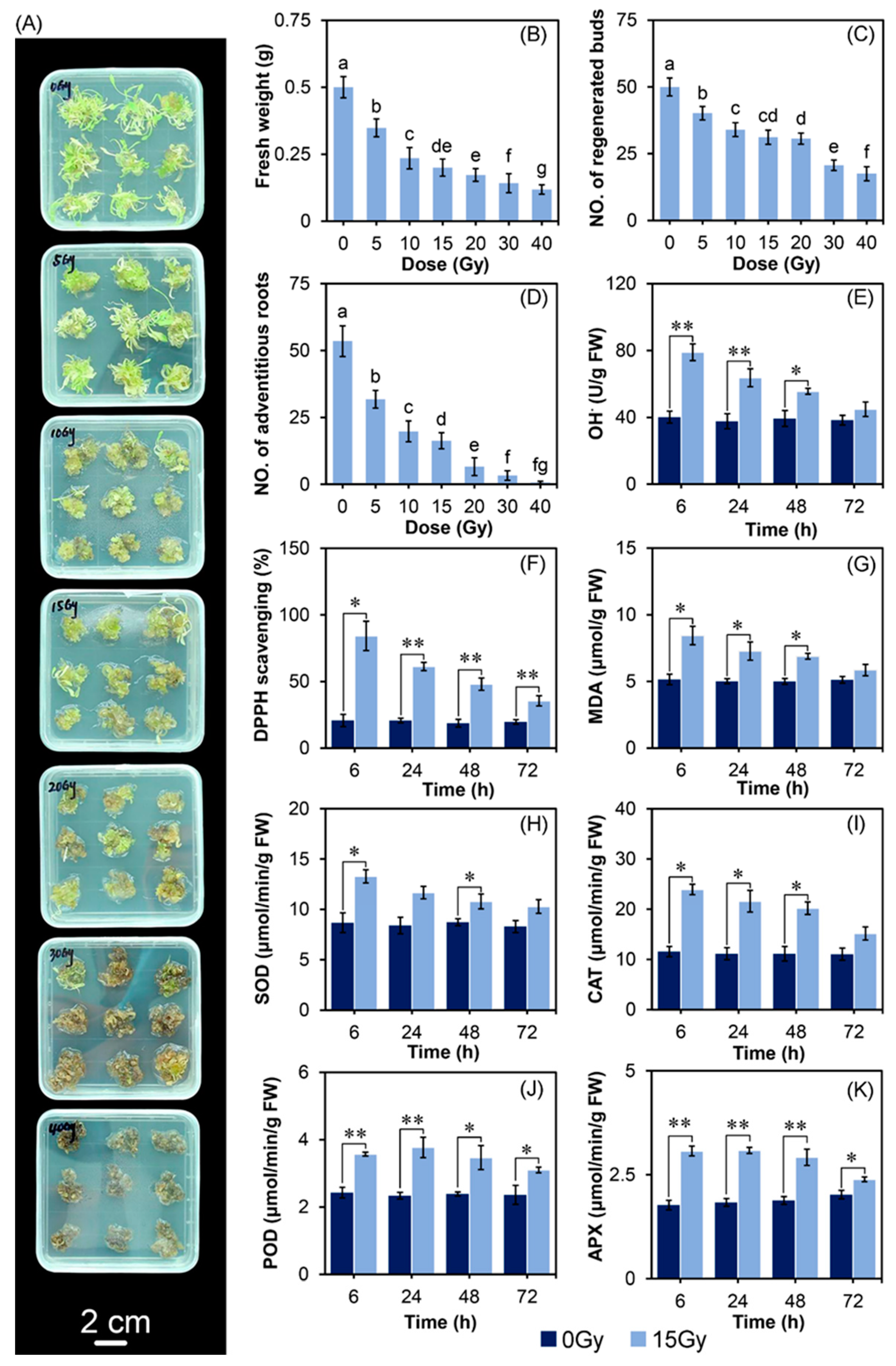
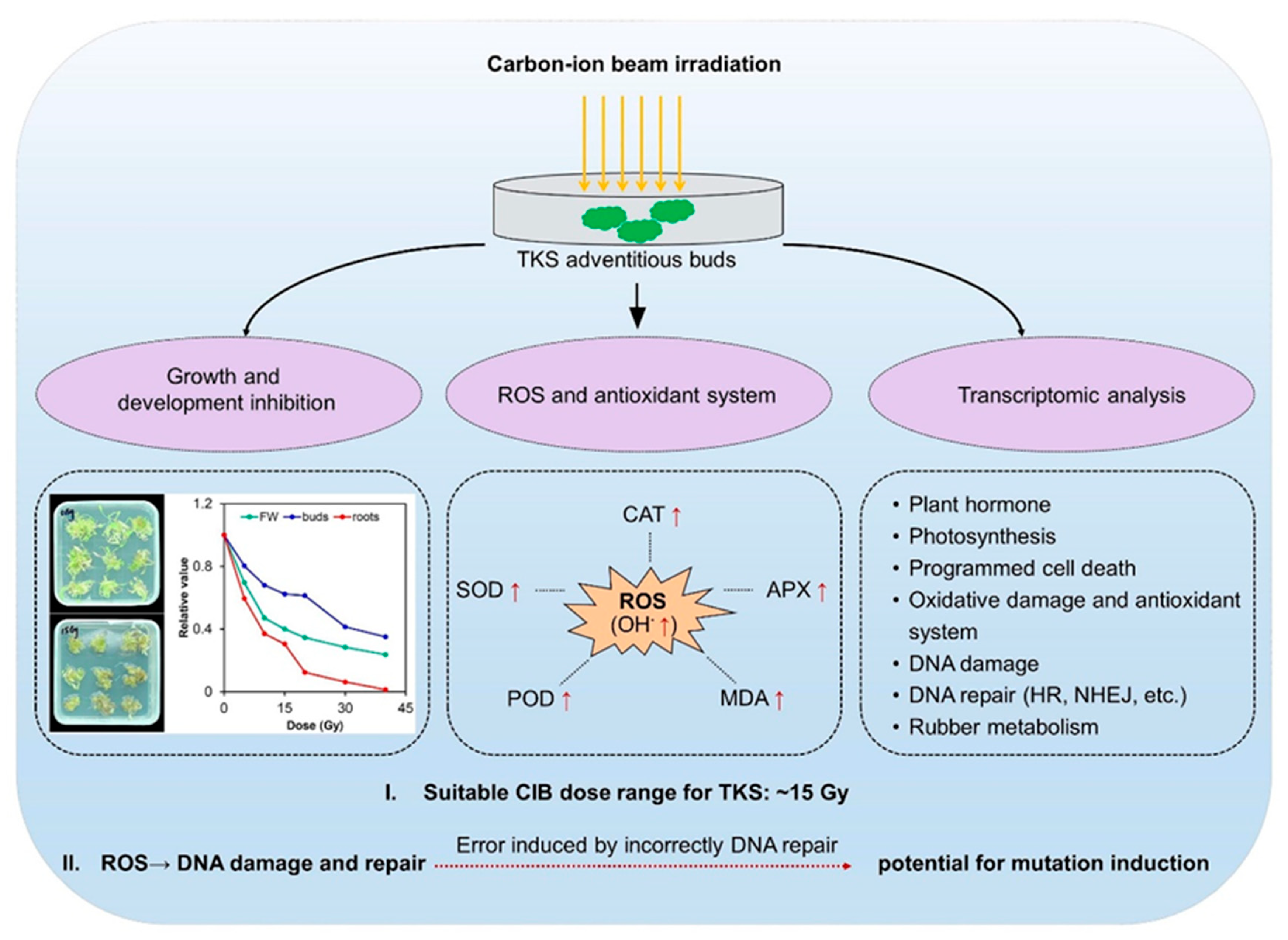
2.1. CIB Irradiation Inhibits the Growth of TKS Adventitious Buds
2.2. The Effect of CIB on Oxidative Damages and Antioxidant System of TKS Adventitious Buds
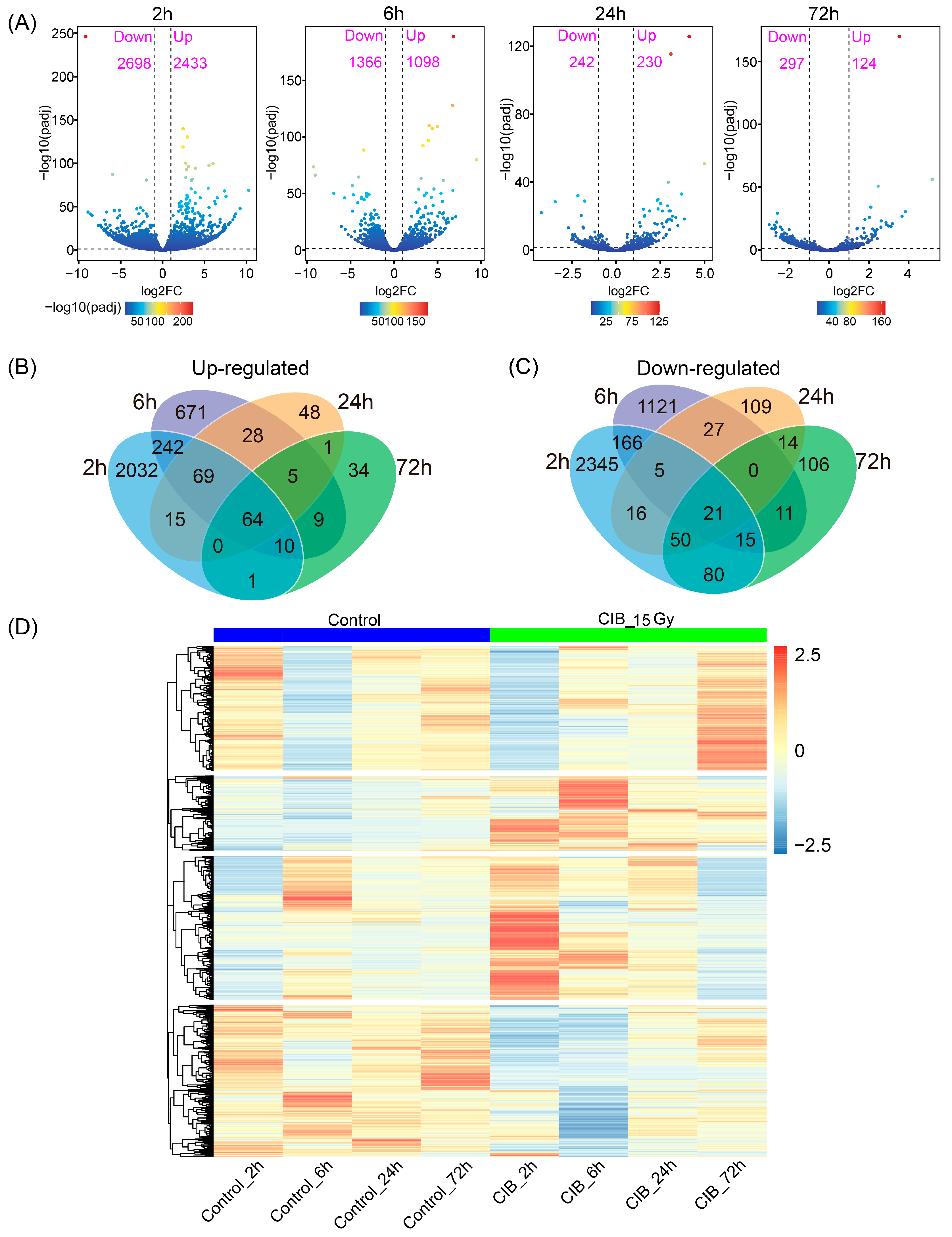
2.3. CIB Triggers Transcriptomic Distinctions in TKS Adventitious Buds
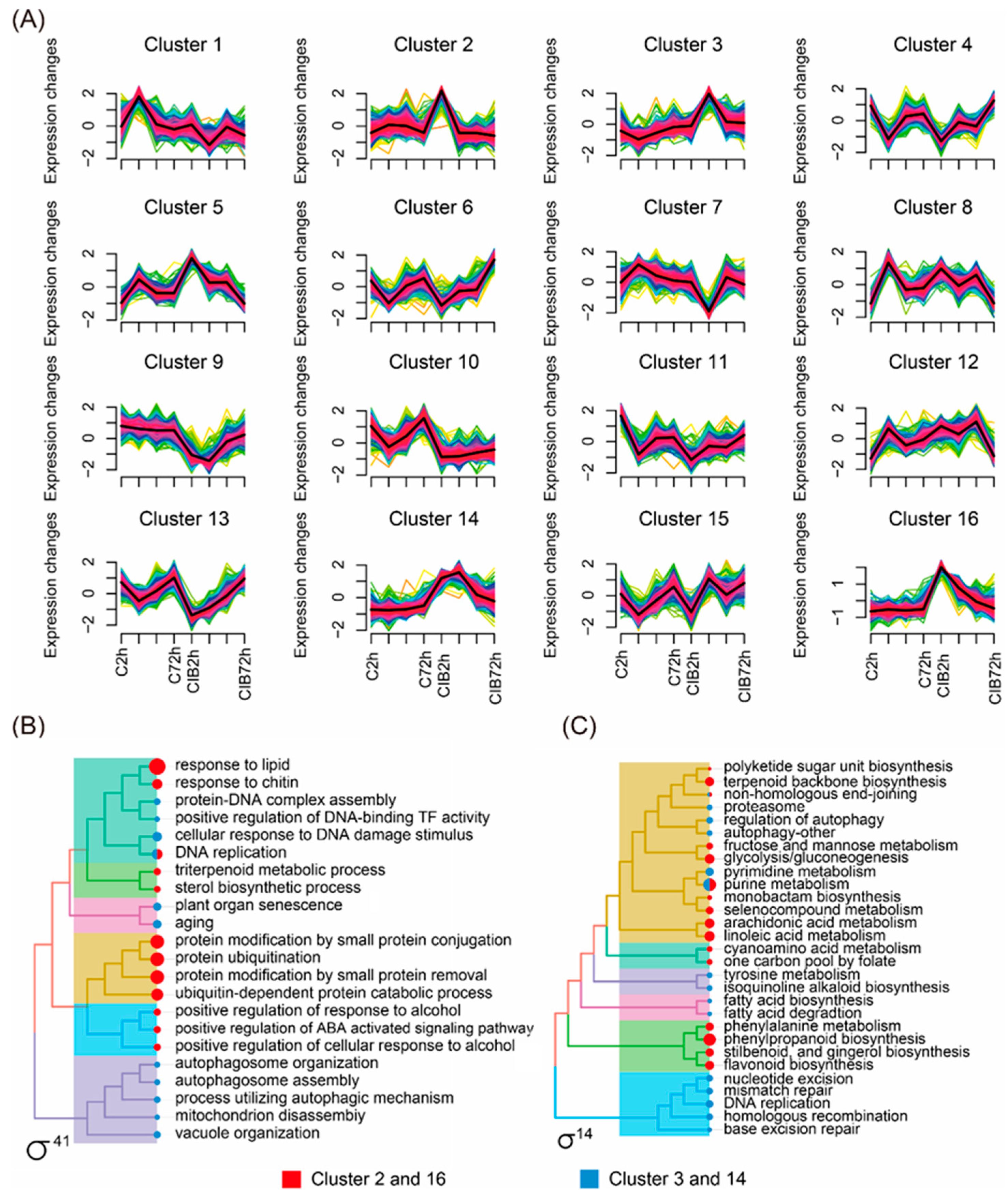
2.4. Time-Series Expression Profile of DEGs Induced by CIB Irradiation
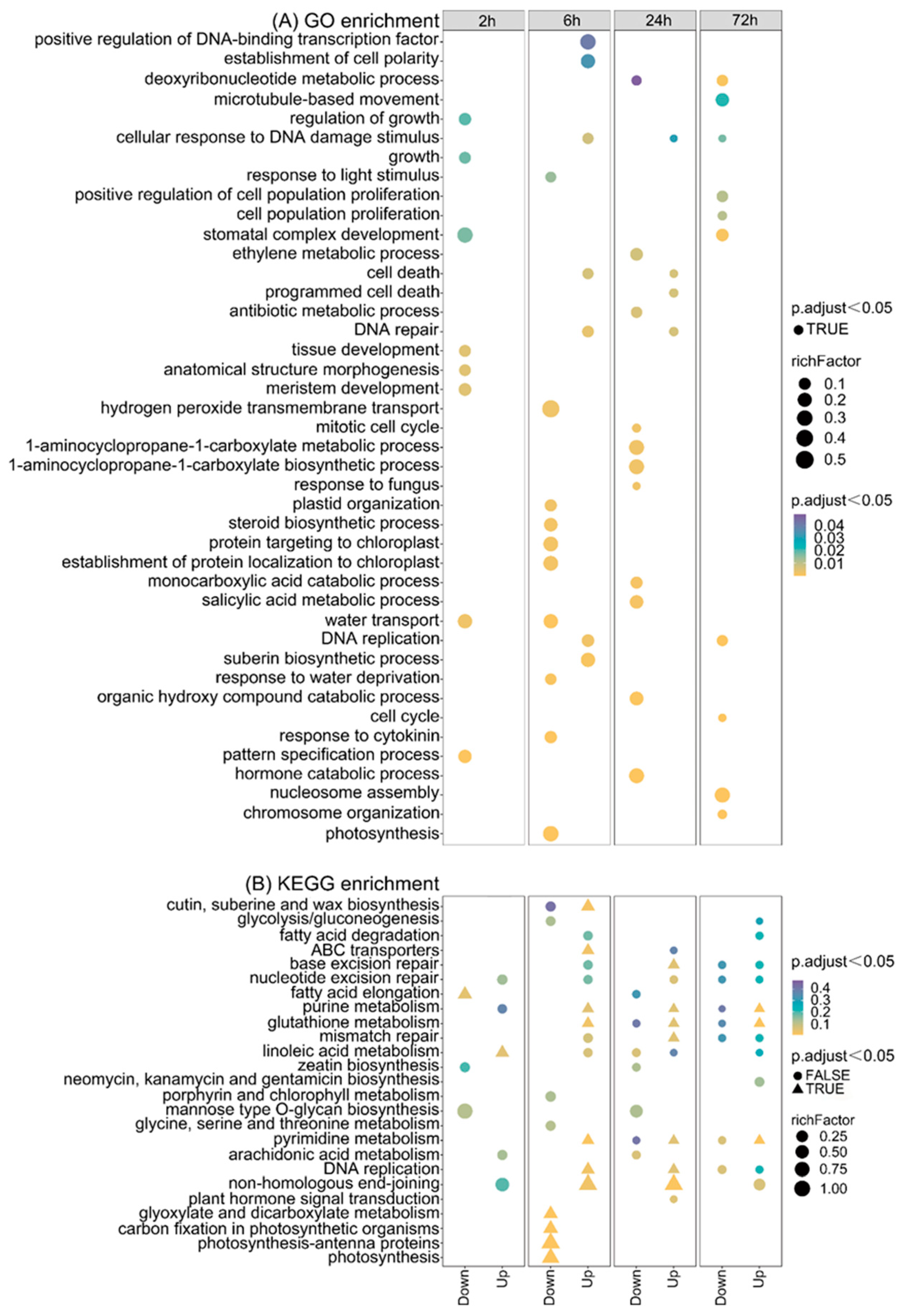
2.5. GO and KEGG Analysis in Different Comparisons after CIB Irradiation
2.6. DNA Replication-/Repair-Related Pathways Are Up-Regulated by CIB in TKS Adventitious Buds
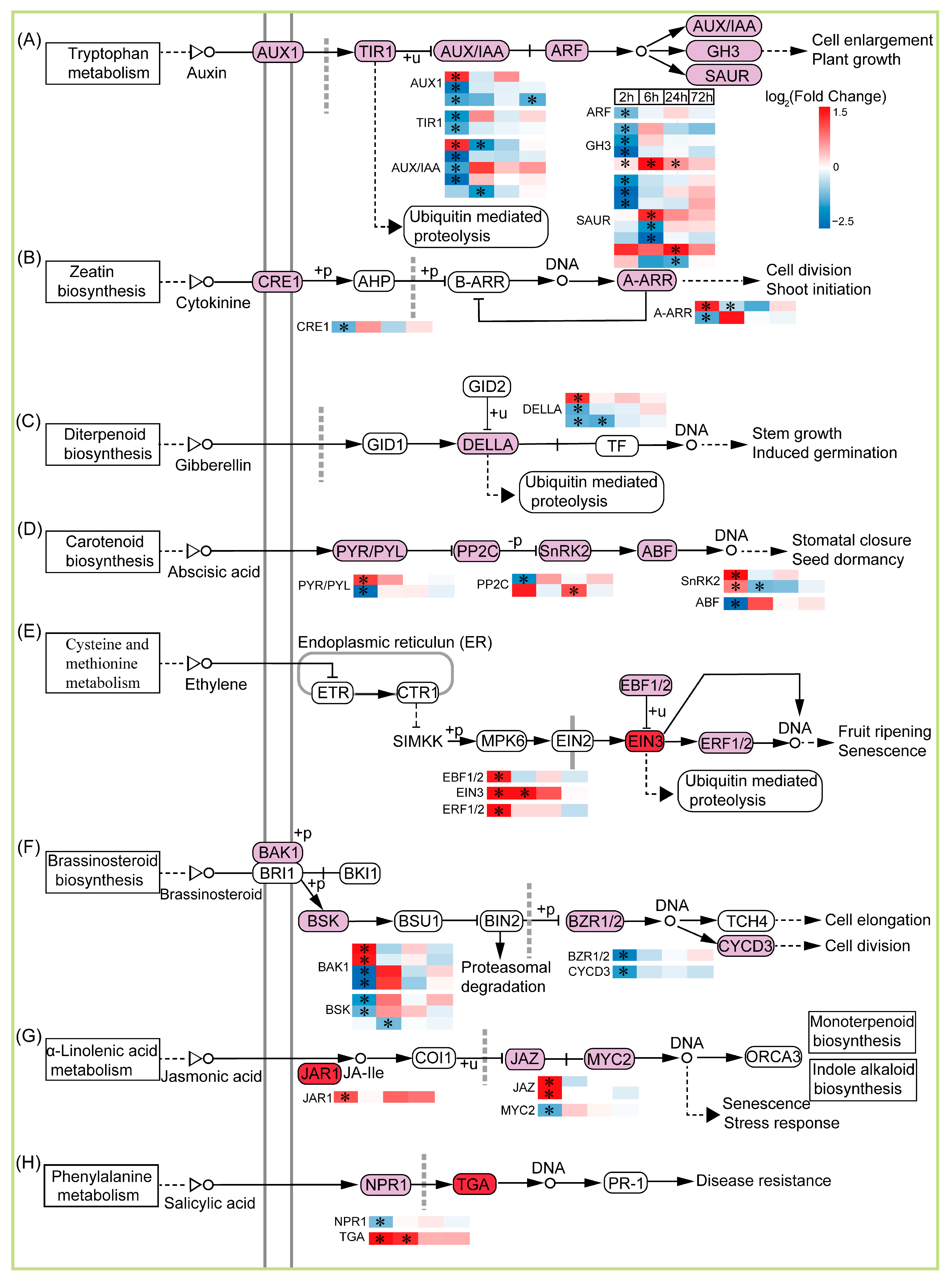
2.7. Plant Hormone signal Transduction Is Involved in TKS Adventitious Buds’ Response to CIB
2.8. Death-Related Pathways Are Up-Regulated by CIB in TKS Adventitious Buds
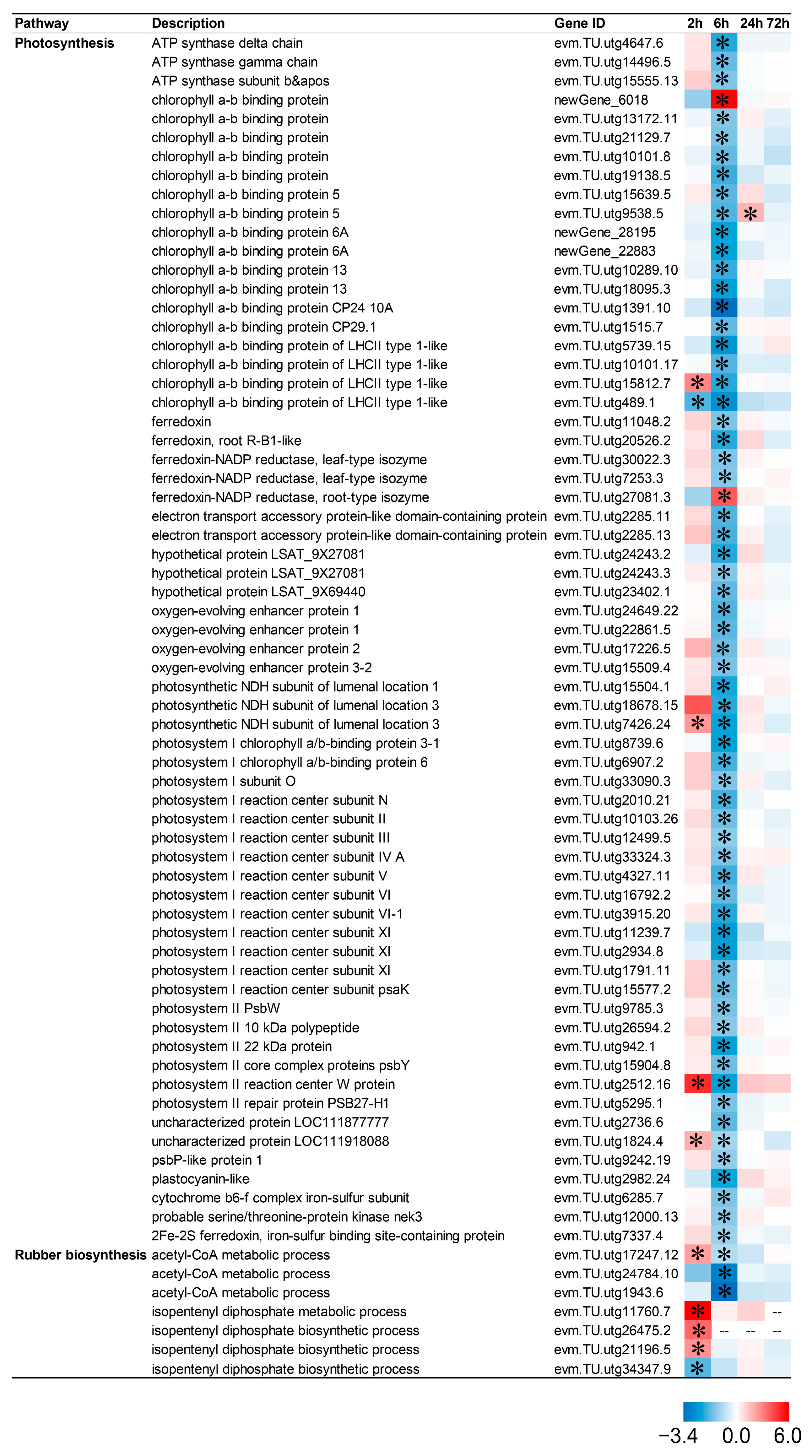
2.9. CIB Down-Regulates Photosynthesis and Up-Regulates Rubber-Metabolism-Related Pathways in TKS Adventitious Buds
3. Discussion
3.1. The Optimum CIB Dose Range for TKS Adventitious Buds
3.2. Transcriptional Map of the CIB Radiation Response in TKS Adventitious Buds
3.3. CIB-Induced Oxidative Damage Response at Physiology and mRNA Levels in TKS Adventitious Buds
3.4. Synergistic Effects of Multiple DNA Repair Pathways Are Induced by CIB in TKS Adventitious Buds
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Irradiation Treatment
4.3. Morphological Observations
4.4. Measurement of OH•, DPPH Radical-Scavenging Activity, and MDA Content
4.5. Determination of Antioxidant Enzyme Activity
4.6. RNA Isolation and Sequencing
4.7. Bioinformatic Analysis of Transcriptome Data
4.8. Statistical Analysis
4.9. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cherian, S.; Ryu, S.B.; Cornish, K. Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol. J. 2019, 17, 2041–2061. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.; Feeney, M.; Ahmadi, M.; Lonoce, C.; Sajari, R.; Di Cola, A.; Frigerio, L. Subcellular localization and interactions among rubber particle proteins from Hevea brasiliensis. J. Exp. Bot. 2017, 68, 5045–5055. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Cadavid, D.A.; Comish, K.; Michel, F.C. Taraxacum kok-saghyz (TK): Compositional analysis of a feedstock for natural rubber and other bioproducts. Ind. Crops Prod. 2017, 107, 624–640. [Google Scholar] [CrossRef]
- Salehi, M.; Bahmankar, M.; Naghavi, M.R.; Cornish, K. Rubber and latex extraction processes for Taraxacum kok-saghyz. Ind. Crops Prod. 2022, 178, 114562. [Google Scholar] [CrossRef]
- Stolze, A.; Wanke, A.; van Deenen, N.; Geyer, R.; Prufer, D.; Gronover, C.S. Development of rubber-enriched dandelion varieties by metabolic engineering of the inulin pathway. Plant Biotechnol. J. 2017, 15, 740–753. [Google Scholar] [CrossRef]
- Kirschner, J.; Stepanek, J.; Cerny, T.; De Heer, P.; van Dijk, P.J. Available ex situ germplasm of the potential rubber crop Taraxacum koksaghyz belongs to a poor rubber producer, T-brevicorniculatum (Compositae-Crepidinae). Genet. Resour. Crop Evol. 2013, 60, 455–471. [Google Scholar] [CrossRef]
- Lin, T.; Xu, X.; Ruan, J.; Liu, S.Z.; Wu, S.G.; Shao, X.J.; Wang, X.B.; Gan, L.; Qin, B.; Yang, Y.S. Genome analysis of Taraxacum kok-saghyz Rodin provides new insights into rubber biosynthesis. Natl. Sci. Rev. 2018, 5, 78–87. [Google Scholar] [CrossRef]
- Ichide, H.; Morita, R.; Shirakawa, Y.; Hayashi, Y.; Abe, T. Targeted exome sequencing of unselected heavy-ion beam-irradiated populations reveals less-biased mutation characteristics in the rice genome. Plant J. 2019, 98, 301–314. [Google Scholar] [CrossRef]
- Mohamad, O.; Sishc, B.J.; Saha, J.; Pompos, A.; Rahimi, A.; Story, M.D.; Davis, A.J.; Kim, D.W.N. Carbon ion radiotherapy: A review of clinical experiences and preclinical research, with an emphasis on DNA damage/repair. Cancers 2017, 9, 66. [Google Scholar] [CrossRef]
- Tanaka, A.; Shikazono, N.; Hase, Y. Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants. J. Radiat. Res. 2010, 51, 223–233. [Google Scholar] [CrossRef]
- Du, Y.; Luo, S.W.; Li, X.; Yang, J.Y.; Cui, T.; Li, W.J.; Yu, L.X.; Feng, H.; Chen, Y.Z.; Mu, J.H.; et al. Identification of substitutions and small insertion-deletions induced by carbon-ion beam irradiation in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1851. [Google Scholar] [CrossRef] [PubMed]
- Okamura, M.; Yasuno, N.; Ohtsuka, M.; Tanaka, A.; Shikazono, N.; Hase, Y. Wide variety of flower-color and-shape mutants regenerated from leaf cultures irradiated with ion beams. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2003, 206, 574–578. [Google Scholar] [CrossRef]
- Shikazono, N.; Yokota, Y.; Kitamura, S.; Suzuki, C.; Watanabe, H.; Tano, S.; Tanaka, A. Mutation rate and novel tt mutants of Arabidopsis thaliana induced by carbon ions. Genetics 2003, 163, 1449–1455. [Google Scholar] [CrossRef]
- Kim, W.J.; Ryu, J.; Im, J.; Kim, S.H.; Kang, S.Y.; Lee, J.H.; Jo, S.H.; Ha, B.K. Molecular characterization of proton beam-induced mutations in soybean using genotyping-by-sequencing. Mol. Genet. Genom. 2018, 293, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- He, J.Y.; Lu, D.; Yu, L.X.; Li, W.J. Pigment analysis of a color-leaf mutant in Wandering Jew (Tradescantia fluminensis) irradiated by carbon ions. Nucl. Sci. Tech. 2011, 22, 77–83. [Google Scholar]
- Yu, L.X.; Li, W.J.; Du, Y.; Chen, G.; Luo, S.W.; Liu, R.Y.; Feng, H.; Zhou, L.B. Flower color mutants induced by carbon ion beam irradiation of geranium (Pelargonium x hortorum, Bailey). Nucl. Sci. Tech. 2016, 27, 112. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, F.; Ma, H.Y.; Li, J.P. Evaluation of the saline-alkaline tolerance of rice (Oryza sativa L.) mutants induced by heavy-ion beam mutagenesis. Biology 2022, 11, 126. [Google Scholar] [CrossRef]
- Takahashi, M.; Kohama, S.; Kondo, K.; Hakata, M.; Hase, Y.; Shikazono, N.; Morikawa, H. Effects of ion beam irradiation on the regeneration and morphology of Ficus thunbergii Maxim. Plant Biotechnol. J. 2005, 22, 63–67. [Google Scholar] [CrossRef]
- Cui, T.; Luo, S.W.; Du, Y.; Yu, L.X.; Yang, J.Y.; Li, W.J.; Chen, X.; Li, X.; Wang, J.; Zhou, L.B. Research of photosynthesis and genomewide resequencing on a yellow-leaf Lotus japonicus mutant induced by carbon ion beam irradiation. Grassl. Sci. 2019, 65, 41–48. [Google Scholar] [CrossRef]
- Shiroshita, Y.; Yuhazu, M.; Hase, Y.; Yamada, T.; Abe, J.; Kanazawa, A. Characterization of chlorophyll-deficient soybean [Glycine max (L.) Merr.] mutants obtained by ion-beam irradiation reveals concomitant reduction in isoflavone levels. Genet. Resour. Crop Evol. 2021, 68, 1213–1223. [Google Scholar] [CrossRef]
- Du, Y.; Luo, S.W.; Zhao, J.; Feng, Z.; Chen, X.; Ren, W.B.; Liu, X.; Wang, Z.Z.; Yu, L.X.; Li, W.J.; et al. Genome and transcriptome-based characterization of high energy carbon-ion beam irradiation induced delayed flower senescence mutant in Lotus japonicus. BMC Plant Biol. 2021, 21, 510. [Google Scholar] [CrossRef] [PubMed]
- Pant, B. Application of plant cell and tissue culture for the production of phytochemicals in medicinal plants. Adv. Exp. Med. Biol. 2014, 808, 25–39. [Google Scholar] [PubMed]
- Loyola-Vargas, V.M.; Ochoa-Alejo, N. An introduction to plant tissue culture: Advances and perspectives. Methods Mol. Biol. 2018, 1815, 3–13. [Google Scholar]
- Dong, Y.H.; Guan, M.J.; Wang, L.X.; Yuan, L.; Sun, X.D.; Liu, S.Q. Transcriptome analysis of low-temperature-induced breaking of garlic aerial bulb dormancy. Int. J. Genom. 2019, 7, 9140572. [Google Scholar] [CrossRef] [PubMed]
- Ishizaka, H. Breeding of fragrant cyclamen by interspecific hybridization and ion-beam irradiation. Breed. Sci. 2018, 68, 25–34. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.W.; Ahn, J.W.; Ryu, J.; Kwon, S.J.; Kang, B.C.; Kim, J.B. Frequency, spectrum, and stability of leaf mutants induced by diverse γ-ray treatments in two Cymbidium hybrids. Plants 2020, 9, 546. [Google Scholar] [CrossRef]
- Hase, Y.; Satoh, K.; Seito, H.; Oono, Y. Genetic consequences of acute/chronic gamma and carbon ion irradiation of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 336. [Google Scholar] [CrossRef]
- Lee, J.W.; Jo, I.H.; Kim, J.U.; Hong, C.E.; Bang, K.H.; Park, Y.D. Determination of mutagenic sensitivity to gamma rays in ginseng (Panax ginseng) dehiscent seeds, roots, and somatic embryos. Hortic. Environ. Biotechnol. 2019, 60, 721–731. [Google Scholar] [CrossRef]
- Perez-Jimenez, M.; Tallon, C.I.; Perez-Tornero, O. Inducing mutations in citrus spp.: Sensitivity of different sources of plant material to gamma radiation. Appl. Radiat. Isot. 2020, 157, 109030. [Google Scholar] [CrossRef]
- Kaul, M.L.H.; Bhan, A.K. Mutagenic effectiveness and efficiency of EMS, DES and gamma-rays in rice. Theor. Appl. Genet. 1977, 50, 241–246. [Google Scholar] [CrossRef]
- Luo, S.W.; Zhou, L.B.; Li, W.J.; Du, Y.; Yu, L.X.; Feng, H.; Mu, J.H.; Chen, Y.Z. Mutagenic effects of carbon ion beam irradiations on dry Lotus japonicus seeds. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2016, 383, 123–128. [Google Scholar] [CrossRef]
- Lei, G.; Zhang, Y.L.; Koppula, P.; Liu, X.G.; Zhang, J.; Lin, S.H.; Ajani, J.A.; Xiao, Q.; Liao, Z.X.; Wang, H.; et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020, 30, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Yang, P.M.; Chuah, Q.Y.; Lee, Y.J.; Hsieh, Y.F.; Peng, C.W.; Chiu, S.J. Autophagy promotes radiation-induced senescence but inhibits bystander effects in human breast cancer cells. Autophagy 2014, 10, 1212–1228. [Google Scholar] [CrossRef]
- Matsumoto, K.; Ueno, M.; Shoji, Y.; Nakanishi, I. Heavy-ion beam-induced reactive oxygen species and redox reactions. Free Radic. Res. 2021, 55, 450–460. [Google Scholar] [CrossRef]
- Scandalios, J.G. Oxidative stress: Molecular perception and transduction of signals triggering antioxidant gene defenses. Braz. J. Med. Biol. Res. 2005, 38, 995–1014. [Google Scholar] [CrossRef]
- Gicquel, M.; Taconnat, L.; Renou, J.P.; Esnault, M.A.; Cabello-Hurtado, F. Kinetic transcriptomic approach revealed metabolic pathways and genotoxic-related changes implied in the Arabidopsis response to ionising radiations. Plant Sci. 2012, 195, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, R.N.; Yin, Y.; Jiao, Z. Antioxidant response of Arabidopsis thaliana seedlings to oxidative stress induced by carbon ion beams irradiation. J. Environ. Radioact. 2018, 195, 1–8. [Google Scholar] [CrossRef]
- Montillet, J.L.; Chamnongpol, S.; Chamnongpol, S.; Rusterucci, C.; Dat, J.; van de Cotte, B.; Agnel, J.P.; Battesti, C.; Inze, D.; Van Breusegem, F.; et al. Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 2005, 138, 1516–1526. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, T.Y.; Lin, Z.K.; Gu, B.J.; Xing, C.H.; Zhao, L.Y.; Dong, H.Z.; Gao, J.Z.; Xie, Z.H.; Zhang, S.L.; et al. A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 2019, 17, 1770–1787. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Grinberg, M.A.; Sukhov, V.; Vodeneev, V. Effect of ionizing radiation on physiological and molecular processes in plants. J. Environ. Radioact. 2019, 202, 8–24. [Google Scholar] [CrossRef]
- Kitamura, S.; Satoh, K.; Oono, Y. Detection and characterization of genome-wide mutations in M1 vegetative cells of gamma-irradiated Arabidopsis. PLoS Genet. 2022, 18, e1009979. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, R.N.; Yin, Y.; Jiao, Z. Role of carbon ion beams irradiation in mitigating cold stress in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2018, 162, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Gu, J.Y.; Irshad, A.; Zhao, L.S.; Guo, H.J.; Xiong, H.C.; Xie, Y.D.; Zhao, S.R.; Ding, Y.P.; Zhou, L.B.; et al. Physiological and differential proteomic analysis at seedling stage by induction of heavy-ion beam radiation in wheat seeds. Front. Genet. 2022, 13, 1779. [Google Scholar] [CrossRef]
- Acuna-Hidalgo, R.; Sengul, H.; Steehouwer, M.; van de Vorst, M.; Vermeulen, S.H.; Kiemeney, L.A.L.M.; Veltman, J.A.; Gilissen, C.; Hoischen, A. Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am. J. Hum. Genet. 2017, 101, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.Z.; Zhang, H.; Zhang, Z.L. Comparing clinical and genetic characteristics of De Novo and inherited COL1A1/COL1A2 variants in a Large chinese cohort of osteogenesis imperfecta. Front. Endocrinol. 2022, 13, 935905. [Google Scholar] [CrossRef]
- Hase, Y.; Satoh, K.; Kitamura, S.; Oono, Y. Physiological status of plant tissue affects the frequency and types of mutations induced by carbon-ion irradiation in Arabidopsis. Sci. Rep. 2018, 8, 1394. [Google Scholar] [CrossRef]
- Hirano, T.; Kazama, Y.; Kunitake, H.; Abe, T. Mutagenic effects of heavy-ion beam irradiation to plant genome. Cytologia 2022, 87, 3–6. [Google Scholar] [CrossRef]
- Dumont, M.; Massot, S.; Doutriaux, M.P.; Gratias, A. Characterization of Brca2-deficient plants excludes the role of NHEJ and SSA in the meiotic chromosomal defect phenotype. PLoS ONE 2011, 6, e26696. [Google Scholar] [CrossRef]
- Du, Y.; Hase, Y.; Satoh, K.; Shikazono, N. Characterization of gamma irradiation-induced mutations in Arabidopsis mutants deficient in non-homologous end joining. J. Radiat. Res. 2020, 61, 639–647. [Google Scholar] [CrossRef]
- Natarajan, A.T.; Palitti, F. DNA repair and chromosomal alterations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2008, 657, 3–7. [Google Scholar] [CrossRef]
- Meschichi, A.; Zhao, L.; Reeck, S.; White, C.; Da Ines, O.; Sicard, A.; Pontvianne, F.; Rosa, S. The plant-specific DDR factor SOG1 increases chromatin mobility in response to DNA damage. EMBO Rep. 2022, 23, e54736. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Shimizu, A.; Nishio, T.; Tsutsumi, N.; Kato, H. Comparison and characterization of mutations induced by gamma-ray and carbon-ion irradiation in rice (Oryza sativa L.) using whole-genome resequencing. G3 Genes Genomes Genet. 2019, 9, 3743–3751. [Google Scholar]
- Permyakova, N.V.; Marenkova, T.V.; Belavin, P.A.; Zagorskaya, A.A.; Sidorchuk, Y.V.; Deineko, E.V. CRISPR/Cas9-mediated targeted DNA integration: Rearrangements at the junction of plant and plasmid DNA. Int. J. Mol. Sci. 2022, 23, 8636. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.T.; Zhao, Y.C.; Wang, B.; Hao, Y.; Wang, Y.X.; Li, Y.Y.; Luo, W.N.; Zong, W.B.; Li, G.S.; Chen, S.F.; et al. Efficient CRISPR/Cas9-based plant genomic fragment deletions by microhomology-mediated end joining. Plant Biotechnol. J. 2020, 18, 2161. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Chaudhary, R.; Deshmukh, R.; Deshmukh, R.; Tiwari, S. Opportunities and challenges with CRISPR-Cas mediated homologous recombination based precise editing in plants and animals. Plant Mol. Biol. 2023, 111, 1–20. [Google Scholar] [CrossRef]
- Park, S.Y.; Vaghchhipawala, Z.; Vasudevan, B.; Lee, L.Y.; Shen, Y.J.; Singer, K.; Waterworth, W.M.; Zhang, Z.Y.J.; West, C.E.; Mysore, K.S.; et al. Agrobacterium T-DNA integration into the plant genome can occur without the activity of key non-homologous end-joining proteins. Plant J. 2015, 81, 934–946. [Google Scholar] [CrossRef]
- Gehrke, F.; Schindele, A.; Puchta, H. Nonhomologous end joining as key to CRISPR/Cas-mediated plant chromosome engineering. Plant Physiol. 2022, 188, 1769–1779. [Google Scholar] [CrossRef]
- Asghar, T.; Jamil, Y.; Iqbal, M.; Abbas, M. Laser light and magnetic field stimulation effect on biochemical, enzymes activities and chlorophyll contents in soybean seeds and seedlings during early growth stages. J. Photochem. Photobiol. B 2016, 165, 283–290. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Plumb-Dhindsa, P.; Thorpe, T. Leaf senescence: Correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Wu, N.; Ren, F.; Wu, X. Assay of hydroxyl radical produced by Co2+/H2O2 reaction using alizarin violet spectrophotometry. J. Instrum. Anal. 2001, 20, 52–54. [Google Scholar]
- Zhang, K.M.; Yu, H.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Xia, X.J. Photoprotective roles of anthocyanins in Begonia semperflorens. Plant Sci. 2010, 179, 202–208. [Google Scholar] [CrossRef]
- Wang, X.; Liu, C.K.; Tu, B.J.; Li, Y.S.; Zhang, Q.Y.; Liu, X.B. Effects of carbon ion beam irradiation on phenotypic variations and biochemical parameters in early generations of soybean plants. Agriculture 2021, 11, 98. [Google Scholar] [CrossRef]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.S.; Arshad, A.; Rajput, L.; Fatima, K.; Ullah, S.; Ahmad, M.; Imran, A. Growth-stimulatory effect of quorum sensing signal molecule N-acyl-homoserine lactone-producing multi-trait Aeromonas spp. on wheat genotypes under salt stress. Front. Microbiol. 2020, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.Z.; Huang, J.J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L.P. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.L.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Du, Y.; Luo, S.; Qu, Y.; Jin, W.; Liu, S.; Wang, Z.; Liu, X.; Feng, Z.; Qin, B.; et al. Physiological and Transcriptomic Analyses Reveal the Effects of Carbon-Ion Beam on Taraxacum kok-saghyz Rodin Adventitious Buds. Int. J. Mol. Sci. 2023, 24, 9287. https://doi.org/10.3390/ijms24119287
Chen X, Du Y, Luo S, Qu Y, Jin W, Liu S, Wang Z, Liu X, Feng Z, Qin B, et al. Physiological and Transcriptomic Analyses Reveal the Effects of Carbon-Ion Beam on Taraxacum kok-saghyz Rodin Adventitious Buds. International Journal of Molecular Sciences. 2023; 24(11):9287. https://doi.org/10.3390/ijms24119287
Chicago/Turabian StyleChen, Xia, Yan Du, Shanwei Luo, Ying Qu, Wenjie Jin, Shizhong Liu, Zhuanzi Wang, Xiao Liu, Zhuo Feng, Bi Qin, and et al. 2023. "Physiological and Transcriptomic Analyses Reveal the Effects of Carbon-Ion Beam on Taraxacum kok-saghyz Rodin Adventitious Buds" International Journal of Molecular Sciences 24, no. 11: 9287. https://doi.org/10.3390/ijms24119287
APA StyleChen, X., Du, Y., Luo, S., Qu, Y., Jin, W., Liu, S., Wang, Z., Liu, X., Feng, Z., Qin, B., & Zhou, L. (2023). Physiological and Transcriptomic Analyses Reveal the Effects of Carbon-Ion Beam on Taraxacum kok-saghyz Rodin Adventitious Buds. International Journal of Molecular Sciences, 24(11), 9287. https://doi.org/10.3390/ijms24119287





