Cinnamaldehyde Restores Ceftriaxone Susceptibility against Multidrug-Resistant Salmonella
Abstract
1. Introduction
2. Results
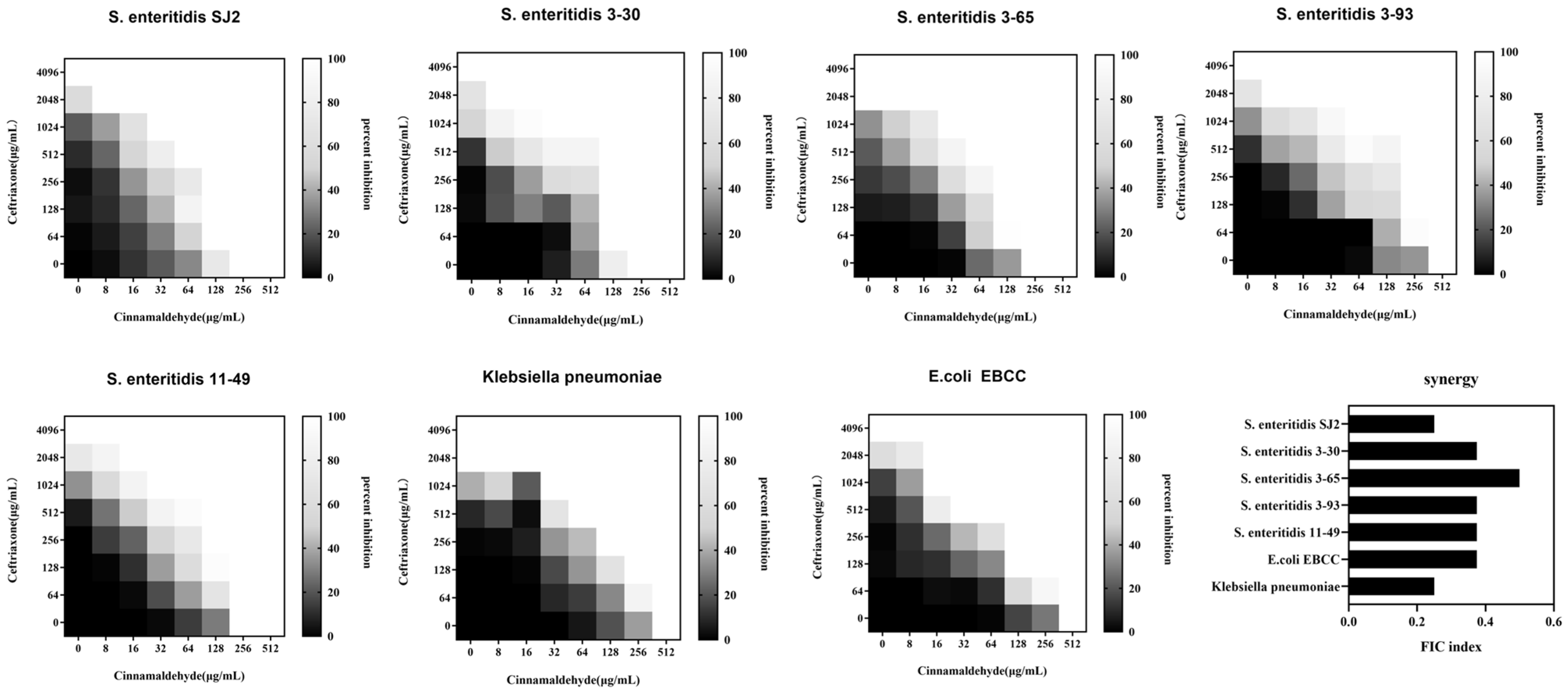
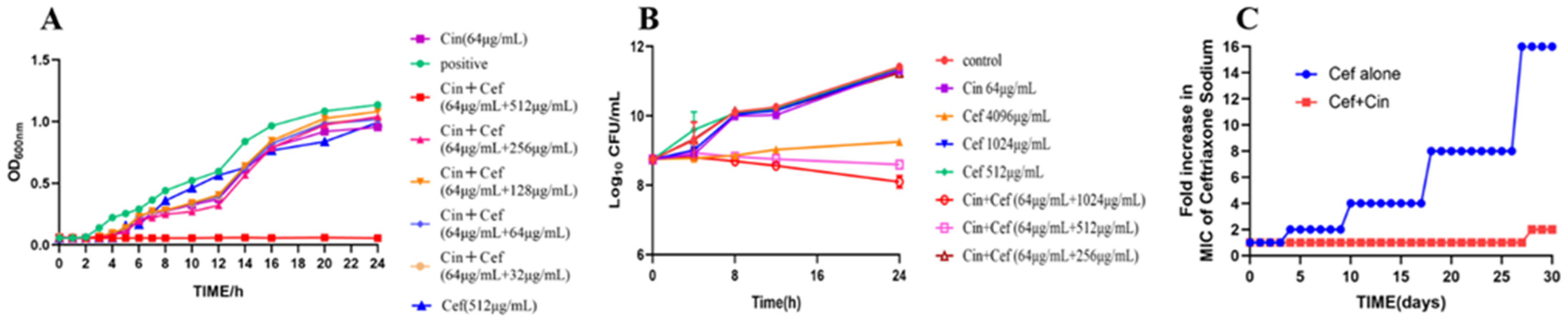
2.1. Cinnamaldehyde Enhances the Effects of Ceftriaxone Sodium against MDR Bacteria
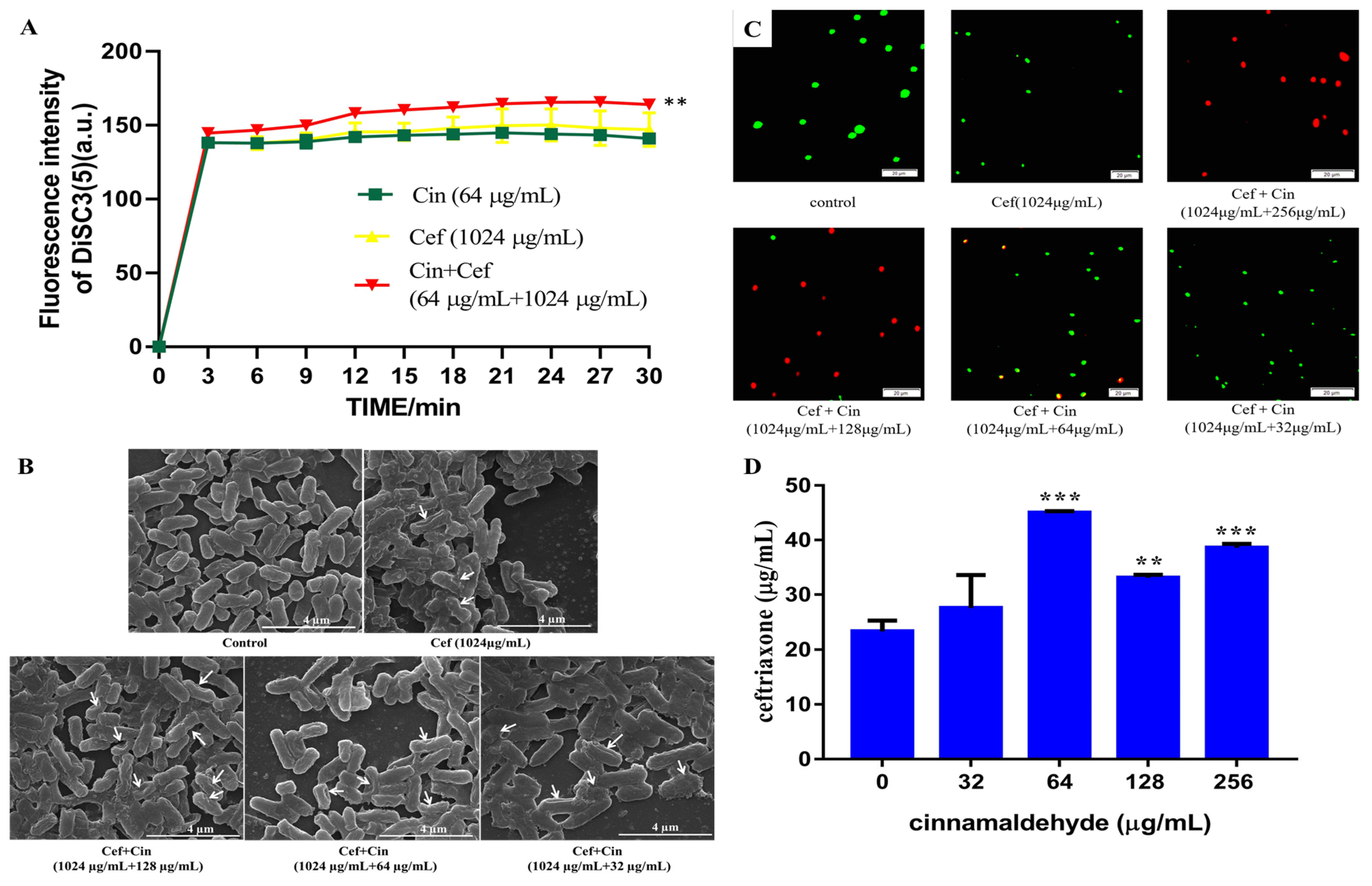
2.2. Cinnamaldehyde Affects the Cell Wall and Cytomembrane of MDR Salmonella
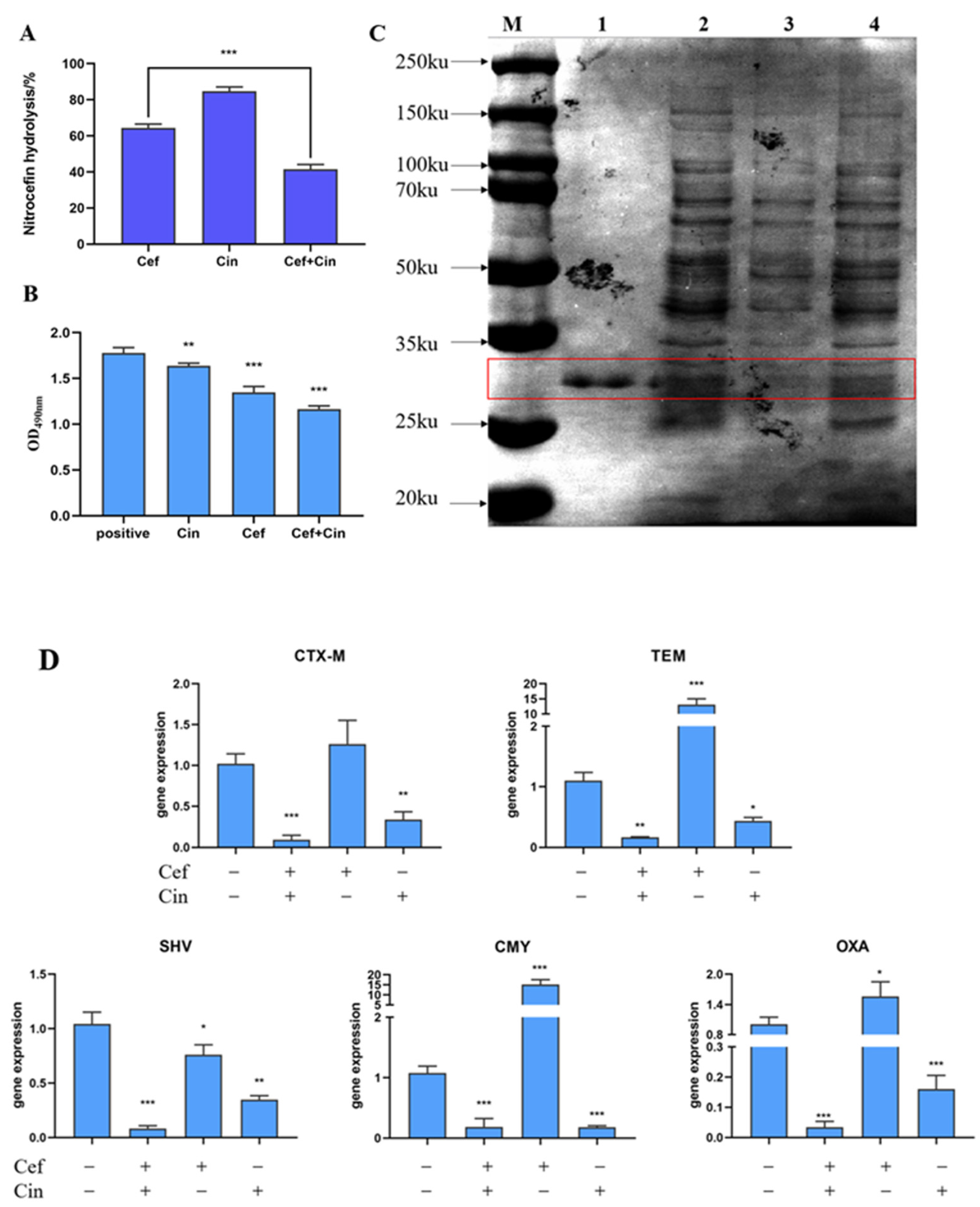
2.3. The Effect of Cinnamaldehyde Combined with Ceftriaxone Sodium on Bacterial β-lactamase and Its Gene Expression
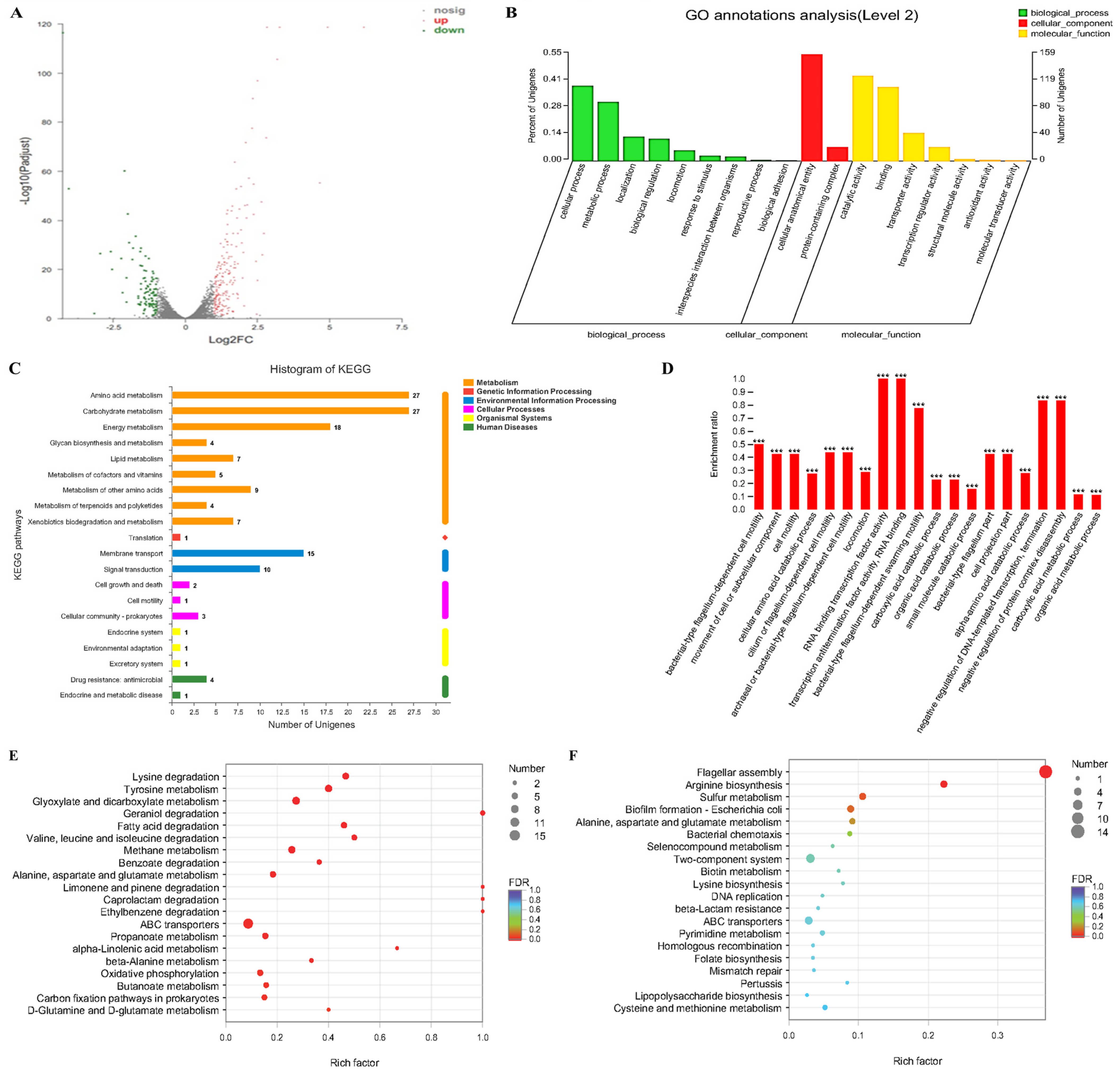
2.4. The Effects of Transcriptome Analyses of Cinnamaldehyde Acting on MDR Salmonella
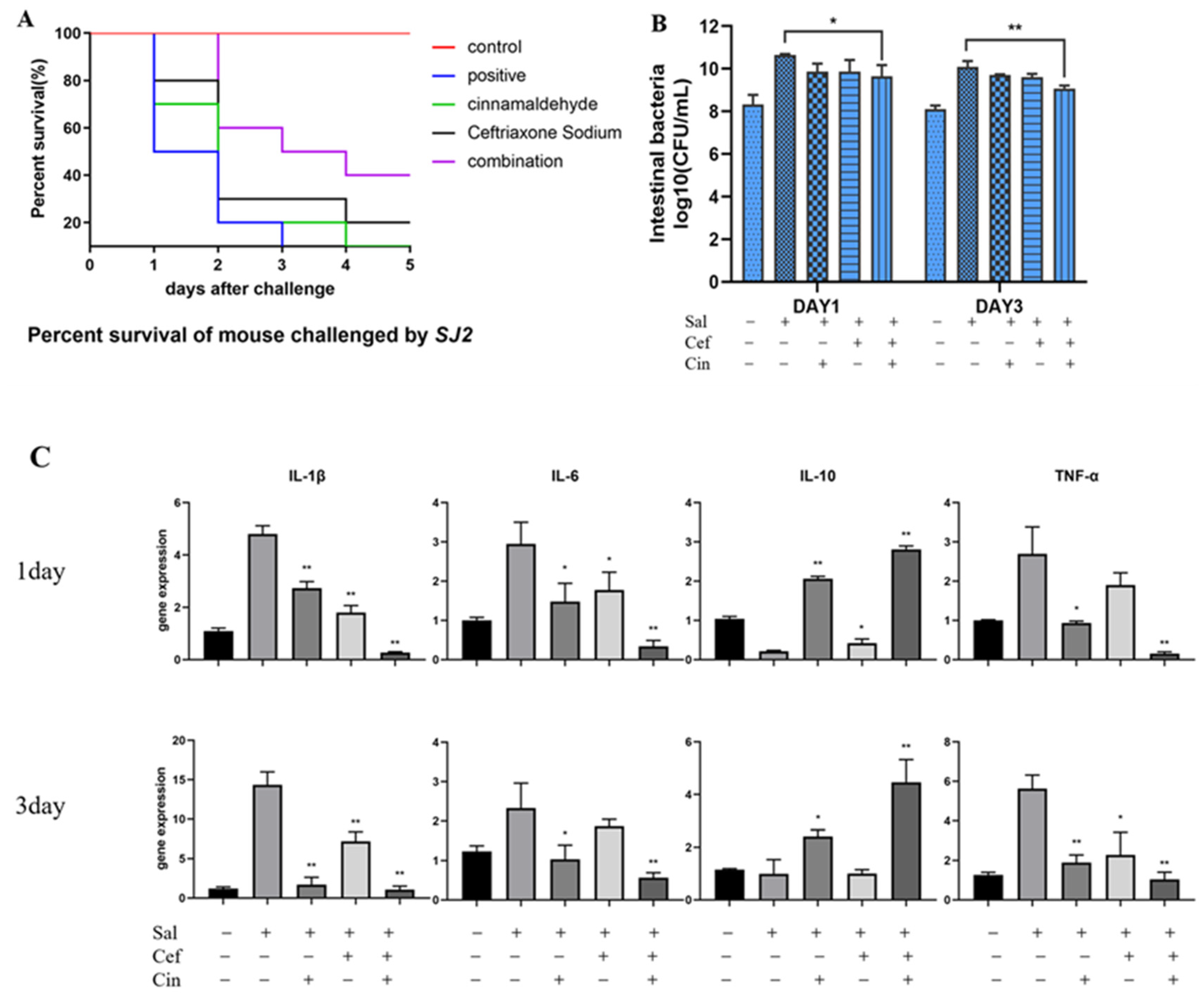
2.5. Cinnamaldehyde Can Reverse the Resistance to Ceftriaxone In Vivo
3. Discussion
4. Materials and Methods
4.1. Bacteria and Reagents
4.2. Minimum Inhibitory Concentration Assay
4.3. Checkerboard Assays
4.4. Bacterial Growth Curves
4.5. Time-Dependent Cytotoxicity Assay
4.6. Analysis of Drug Resistance Development
4.7. Membrane Depolarization Assay
4.8. Scanning Electron Microscopy
4.9. Bacterial Viability Assay
4.10. Antibiotic Accumulation Analysis
4.11. Cephalosporin Hydrolysis Analysis
4.12. SDS-PAGE
4.13. Inhibition of Enzyme Activity
4.14. RT-PCR for Salmonella β-lactamase
4.15. Transcriptomic Analysis
4.16. Animal Experiment
4.17. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Solomon, S.L.; Oliver, K.B. Antibiotic Resistance Threats in the United States: Stepping Back from the Brink. Am. Fam. Physician 2014, 89, 938–941. [Google Scholar] [PubMed]
- Crump, J.A.; Sjölund-Karlsson, M.; Gordon, M.; Parry, C.M. Epidemiology, Clinical Presentation, Laboratory Diagnosis, Antimicrobial Resistance, and Antimicrobial Management of Invasive Salmonella Infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, S.; Fatima, J.; Shakil, S.; Rizvi, S.M.D.; Kamal, M.A. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J. Biol. Sci. 2015, 22, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Upadhaya, S.D.; Kim, I.H. Efficacy of Phytogenic Feed Additive on Performance, Production and Health Status of Monogastric Animals—A Review. Ann. Anim. Sci. 2017, 17, 929–948. [Google Scholar] [CrossRef]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21 st century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef]
- Hammer, K.A.; Carson, C.F.; Riley, T.V. Antifungal effects of Melaleuca alternifolia (tea tree) oil and its components on Candida albicans, Candida glabrata and Saccharomyces cerevisiae. J. Antimicrob. Chemother. 2004, 53, 1081–1085. [Google Scholar] [CrossRef]
- Zhu, L.B.; Olsen, C.; McHugh, T.; Friedman, M.; Jaroni, D.; Ravishankar, S. Apple, Carrot, and Hibiscus Edible Films Containing the Plant Antimicrobials Carvacrol and Cinnamaldehyde Inactivate Salmonella Newport on Organic Leafy Greens in Sealed Plastic Bags. J. Food Sci. 2014, 79, M61–M66. [Google Scholar] [CrossRef]
- Oloketuyi, S.F.; Khan, F. Strategies for Biofilm Inhibition and Virulence Attenuation of Foodborne Pathogen-Escherichia coli O157:H7. Curr. Microbiol. 2017, 74, 1477–1489. [Google Scholar] [CrossRef]
- Shao, P.; Yan, Z.; Chen, H.; Xiao, J. Electrospun poly(vinyl alcohol)/permutite fibrous film loaded with cinnamaldehyde for active food packaging. J. Appl. Polym. Sci. 2018, 135, 46117. [Google Scholar] [CrossRef]
- Lopez, P.; Sánchez, C.; Batlle, R.; Nerin, C. Vapor-phase activities of cinnamon, thyme, and oregano essential oils and key constituents against foodborne microorganisms. J. Agric. Food Chem. 2007, 55, 4348–4356. [Google Scholar] [CrossRef]
- Palaniappan, K.; Holley, R.A. Use of natural antimicrobials to increase antibiotic susceptibility of drug resistant bacteria. Int. J. Food Microbiol. 2010, 140, 164–168. [Google Scholar] [CrossRef]
- Jia, P.; Xue, Y.; Duan, X.; Shao, S. Effect of cinnamaldehyde on biofilm formation and sarA expression by methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 2011, 53, 409–416. [Google Scholar] [CrossRef]
- Liu, Q.; Niu, H.; Zhang, W.; Mu, H.; Sun, C.; Duan, J. Synergy among thymol, eugenol, berberine, cinnamaldehyde and streptomycin against planktonic and biofilm-associated food-borne pathogens. Lett. Appl. Microbiol. 2015, 60, 421–430. [Google Scholar] [CrossRef]
- Dhara, L.; Tripathi, A. Cinnamaldehyde: A compound with antimicrobial and synergistic activity against ESBL-producing quinolone-resistant pathogenic Enterobacteriaceae. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 65–73. [Google Scholar] [CrossRef]
- Shaheen, A.; Tariq, A.; Shehzad, A.; Iqbal, M.; Mirza, O.; Maslov, D.A.; Rahman, M. Transcriptional regulation of drug resistance mechanisms in Salmonella: Where we stand and what we need to know. World J. Microbiol. Biotechnol. 2020, 36, 85. [Google Scholar] [CrossRef]
- Purkait, S.; Bhattacharya, A.; Bag, A.; Chattopadhyay, R.R. Evaluation of antibiofilm efficacy of essential oil components beta-caryophyllene, cinnamaldehyde and eugenol alone and in combination against biofilm formation and preformed biofilms of Listeria monocytogenes and Salmonella typhimurium. Lett. Appl. Microbiol. 2020, 71, 195–202. [Google Scholar] [CrossRef]
- Kimura, M.; Shindo, M.; Moriizumi, T.; Tagawa, N.; Fujinami, A.; Kato, I.; Uchida, Y. Salusin-beta, an Antimicrobially Active Peptide against Gram-Positive Bacteria. Chem. Pharm. Bull. 2014, 62, 586–590. [Google Scholar] [CrossRef]
- Oncul, S.; Cuce, E.M.; Aksu, B.; Garip, A.I. Effect of extremely low frequency electromagnetic fields on bacterial membrane. Int. J. Radiat. Biol. 2016, 92, 42–49. [Google Scholar] [CrossRef]
- Boix-Lemonche, G.; Lekka, M.; Skerlavaj, B. A Rapid Fluorescence-Based Microplate Assay to Investigate the Interaction of Membrane Active Antimicrobial Peptides with Whole Gram-Positive Bacteria. Antibiotics 2020, 9, 92. [Google Scholar] [CrossRef]
- Vicari, G.; Bauer, S.R.; Neuner, E.A.; Lam, S.W. Association Between Colistin Dose and Microbiologic Outcomes in Patients With Multidrug-Resistant Gram-Negative Bacteremia. Clin. Infect. Dis. 2013, 56, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.F.; Drown, B.S.; Riley, A.P.; Garcia, A.; Shirai, T.; Svec, R.L.; Hergenrother, P.J. Predictive compound accumulation rules yield a broad—Spectrum antibiotic. Nature 2017, 545, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Song, M.R.; Liu, Y.; Huang, X.; Ding, S.; Wang, Y.; Shen, J.; Zhu, K. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol. 2020, 5, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Dai, Y.; Chen, H.; He, X.; Ouyang, P.; Huang, X.; Sun, X.; Ai, Y.; Lai, S.; Zhu, L.; et al. Cinnamaldehyde Resist Salmonella Typhimurium Adhesion by Inhibiting Type I Fimbriae. Molecules 2022, 27, 7753. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Nguyen, H.C.B.; Adlanmerini, M.; Hauck, A.K.; Lazar, M.A. Dichotomous engagement of HDAC3 activity governs inflammatory responses. Nature 2020, 584, 286–290. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Su, Y.; Zhang, X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol. Rep. 2018, 39, 2499–2512. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Ohya, S.; Kito, H.; Hatano, N.; Muraki, K. Recent advances in therapeutic strategies that focus on the regulation of ion channel expression. Pharmacol. Ther. 2016, 160, 11–43. [Google Scholar] [CrossRef]
- Liu, Y.; Jia, Y.; Yang, K.; Li, R.; Xiao, X.; Zhu, K.; Wang, Z. Metformin Restores Tetracyclines Susceptibility against Multidrug Resistant Bacteria. Adv. Sci. 2020, 7, 1902227. [Google Scholar] [CrossRef]
- Esposito, S. Parenteral cephalosporin therapy in ambulatory care: Advantages and disadvantages. Drugs 2000, 59 (Suppl. S3), 19–28. [Google Scholar] [CrossRef]
- Esposito, S.; Noviello, S.; Leone, S.; Tice, A.; Seibold, G.; Nathwani, D.; Scaglione, F. Outpatient parenteral antibiotic therapy (OPAT) in different countries: A comparison. Int. J. Antimicrob. Agents 2004, 24, 473–478. [Google Scholar] [CrossRef]
- Zhou, X.J.; Zhang, Z.; Suo, Y.; Cui, Y.; Zhang, F.; Shi, C.; Shi, X. Effect of sublethal concentrations of ceftriaxone on antibiotic susceptibility of multiple antibiotic-resistant Salmonella strains. Fems Microbiol. Lett. 2019, 366, fny283. [Google Scholar] [CrossRef]
- Peterson, S.C.; Lau, T.; Ensom, M.H.H. Combination of Ceftriaxone and Ampicillin for the Treatment of Enterococcal Endocarditis: A Qualitative Systematic Review. Ann. Pharmacother. 2017, 51, 496–503. [Google Scholar] [CrossRef]
- Shreaz, S.; Wani, W.A.; Behbehani, J.M.; Raja, V.; Irshad; Karched, M.; Ali, I.; Siddiqi, W.A.; Hun, L.T. Cinnamaldehyde and its derivatives, a novel class of antifungal agents. Fitoterapia 2016, 112, 116–131. [Google Scholar] [CrossRef]
- Rogiers, G.; Kebede, B.T.; Van Loey, A.; Michiels, C.W. Membrane fatty acid composition as a determinant of Listeria monocytogenes sensitivity to trans-cinnamaldehyde. Res. Microbiol. 2017, 168, 536–546. [Google Scholar] [CrossRef]
- Ghoreishi, F.S.; Roghanian, R.; Emtiazi, G. Inhibition of quorum sensing-controlled virulence factors with natural substances and novel protease, obtained from Halobacillus karajensis. Microb. Pathog. 2020, 149, 104555. [Google Scholar] [CrossRef]
- Shreaz, S.; Bhatia, R.; Khan, N.; Muralidhar, S.; Basir, S.F.; Manzoor, N.; Khan, L.A. Spice oil cinnamaldehyde exhibits potent anticandidal activity against fluconazole resistant clinical isolates. Fitoterapia 2011, 82, 1012–1020. [Google Scholar] [CrossRef]
- Shreaz, S.; Bhatia, R.; Khan, N.; Maurya, I.K.; Ahmad, S.I.; Muralidhar, S.; Manzoor, N.; Khan, L.A. Cinnamic aldehydes affect hydrolytic enzyme secretion and morphogenesis in oral Candida isolates. Microb. Pathog. 2012, 52, 251–258. [Google Scholar] [CrossRef]
- Keelara, S.; Thakur, S.; Patel, J. Biofilm Formation by Environmental Isolates of Salmonella and Their Sensitivity to Natural Antimicrobials. Foodborne Pathog. Dis. 2016, 13, 509–516. [Google Scholar] [CrossRef]
- Kovacs, N.; Jankovics, H.; Vonderviszt, F. Deletion analysis of the flagellum-specific secretion signal in Salmonella flagellin. FEBS Lett. 2018, 592, 3074–3081. [Google Scholar] [CrossRef] [PubMed]
- Sporing, I.; Felgner, S.; Preuße, M.; Eckweiler, D.; Rohde, M.; Häussler, S.; Weiss, S.; Erhardt, M. Regulation of Flagellum Biosynthesis in Response to Cell Envelope Stress in Salmonella enterica Serovar Typhimurium. Mbio 2018, 9, e00736-17. [Google Scholar] [CrossRef] [PubMed]
- Lopatkin, A.J.; Bening, S.C.; Manson, A.L.; Stokes, J.M.; Kohanski, M.A.; Badran, A.H.; Earl, A.M.; Cheney, N.J.; Yang, J.H.; Collins, J.J. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 2021, 371, eaba0862. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, J.M.F.; Lisboa, L.D. Hospital-Acquired Infections Due to Gram-Negative Bacteria. N. Engl. J. Med. 2010, 363, 1482–1483. [Google Scholar]
- Zowawi, H.M.; Harris, P.N.; Roberts, M.J.; Tambyah, P.A.; Schembri, M.A.; Pezzani, M.D.; Williamson, D.A.; Paterson, D.L. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nat. Rev. Urol. 2015, 12, 570–584. [Google Scholar] [CrossRef]
- Kojima, S.; Nikaido, H. Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proc. Natl. Acad. Sci. USA 2013, 110, E2629–E2634. [Google Scholar] [CrossRef]
- Stokes, J.M.; MacNair, C.R.; Ilyas, B.; French, S.; Côté, J.-P.; Bouwman, C.; Farha, M.A.; Sieron, A.O.; Whitfield, C.; Coombes, B.K.; et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017, 2, 17028. [Google Scholar] [CrossRef]
- Lu, M.; Wang, S.; Wang, T.; Hu, S.; Bhayana, B.; Ishii, M.; Kong, Y.; Cai, Y.; Dai, T.; Cui, W.; et al. Bacteria-specific phototoxic reactions triggered by blue light and phytochemical carvacrol. Sci. Transl. Med. 2021, 13, eaba3571. [Google Scholar] [CrossRef]
- Kannappan, A.; Balasubramaniam, B.; Ranjitha, R.; Srinivasan, R.; Packiavathy, I.A.S.V.; Balamurugan, K.; Pandian, S.K.; Ravi, A.V. In vitro and in vivo biofilm inhibitory efficacy of geraniol-cefotaxime combination against Staphylococcus spp. Food Chem. Toxicol. 2019, 125, 322–332. [Google Scholar] [CrossRef]
- Yuan, Z.W.; Ouyang, P.; Gu, K.; Rehman, T.; Zhang, T.; Yin, Z.; Fu, H.; Lin, J.; He, C.; Shu, G.; et al. The antibacterial mechanism of oridonin against methicillin-resistant Staphylococcus aureus (MRSA). Pharm. Biol. 2019, 57, 710–716. [Google Scholar] [CrossRef]
- Hossain, M.A.; Park, H.C.; Lee, K.J.; Park, S.W.; Park, S.C.; Kang, J. In vitro synergistic potentials of novel antibacterial combination therapies against Salmonella enterica serovar Typhimurium. BMC Microbiol. 2020, 20, 118. [Google Scholar] [CrossRef]
- Bendali, F.; Gaillard-Martinie, B.; Hebraud, M.; Sadoun, D. Kinetic of production and mode of action of the Lactobacillus paracasei subsp paracasei anti-listerial bacteriocin, an Algerian isolate. LWT Food Sci. Technol. 2008, 41, 1784–1792. [Google Scholar] [CrossRef]
- Ouyang, P.; He, X.; Yuan, Z.-W.; Yin, Z.-Q.; Fu, H.; Lin, J.; He, C.; Liang, X.; Lv, C.; Shu, G.; et al. Erianin against Staphylococcus aureus Infection via Inhibiting Sortase A. Toxins 2018, 10, 385. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Lozano, F.S.; García, M.I.; García, E.; González, B.; García, M.B.; García, F.J.; García, J.E. Activity of Ertapenem and Ceftriaxone in the eradication of Salmonella in a model of experimental peritonitis in mice. Rev. Esp. Quimioter. 2009, 22, 135–138. [Google Scholar]
| Strain Name | Identification | Strain Source | MIC of Ceftriaxone Sodium (μg/mL) | MIC of Cinnamalde-Hyde (μg/mL) | Antibiotic Resistance a |
|---|---|---|---|---|---|
| ATCC14028 | Salmonella typhimurium | Standard strain | 0.5 | 128 | \ |
| ATCC13036 | Salmonella pullorum | Standard strain | 0.5 | 128 | \ |
| SJ2 | Salmonella enteritidis | Duck | 4096 | 256 | AMP CRO CFZ CTX DOX KAN |
| 3–30 | Salmonella enteritidis | Chicken | 4096 | 256 | AMX CRO CTX |
| 3–65 | Salmonella enteritidis | Chicken | 2048 | 256 | CRO CFZ CTX |
| 3–93 | Salmonella enteritidis | Chicken | 4096 | 512 | AMX CRO CFZ |
| 11–49 | Salmonella enteritidis | Chicken | 4096 | 256 | AMP CRO CTX |
| EBCC | Escherichia coli | Chicken | 1024 | 256 | CRO GEN STR |
| Klebsiella pneumoniae | Klebsiella pneumoniae | Human | 2048 | 512 | AMP AMX CRO CFZ KAN |
| Acinetobacter baumannii | Acinetobacter baumannii | Chicken | 8 | 256 | AMP AMX CRO CTX DOX GEN KAN |
| Primer | Sequence (5′→3′) | Tm (°C) | Product |
|---|---|---|---|
| 16s-f | GCTGCCCTTTGTATTGTC | 56 | 1506 bp |
| 16s-r | AGATGTTGGGTTAAGTCCC | ||
| TEM-f | TCGCCGCATACACTATTCTCAGAATGA | 60 | 800 bp |
| TEM-r | ACGCTCACCGGCTCCAGATTTAT | ||
| SHV-f | GCCTTTATCGGCCTTCACTCAAG | 60 | 898 bp |
| SHV-r | TTAGCGTTGCCAGTGCTCGATCA | ||
| CTX-M-f | ACGCTTTCCAATGTGCAGTA | 60 | 436 bp |
| CTX-M-r | ACGCTTTCCAATGTGCAGTA | ||
| CMY-F | GACAGCCTCTTTCTCCACA | 58 | 1201 bp |
| CMY-R | TGGAACGAAGGCTACGTA | ||
| OXA-F | ATGAAAAACACAATACATATC | 58 | 896 bp |
| OXA-R | CGTATAGGTGTTTCCGTTCT |
| Gene | Primer (5′-3′) | Tm (°C) | Product |
|---|---|---|---|
| β-actin-F | CTACAGCTTCACCACCACAG | 57 | 118 bp |
| β-actin-R | ACCGCTCGTTGCCAATAGTG | ||
| IL-6-F | CTGCAAGAGACTTCCATCCAG | 57 | 132b p |
| IL-6-R | AGTGGTATAGACAGGTCTGTTGG | ||
| IL-1β-F | TGCCACCTTTTGACAGTGATG | 56 | 117 bp |
| IL-1β-R | TGATACTGCCTGCCTGAAGC | ||
| IL-10-F | GAGTGAAGACCAGCAAAGGC | 55 | 168 bp |
| IL-10-R | TTGTCCAGCTGGTCCTT | ||
| TNF-α-F | GCCACCACGCTCTTCTGTCTAC | 58 | 283 bp |
| TNF-α-R | GGGTCTGGGCCATAGAACTGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, L.; Gou, Y.; Dai, Y.; Wang, T.; Gu, K.; Tang, T.; Hussain, S.; Huang, X.; He, C.; Liang, X.; et al. Cinnamaldehyde Restores Ceftriaxone Susceptibility against Multidrug-Resistant Salmonella. Int. J. Mol. Sci. 2023, 24, 9288. https://doi.org/10.3390/ijms24119288
Yin L, Gou Y, Dai Y, Wang T, Gu K, Tang T, Hussain S, Huang X, He C, Liang X, et al. Cinnamaldehyde Restores Ceftriaxone Susceptibility against Multidrug-Resistant Salmonella. International Journal of Molecular Sciences. 2023; 24(11):9288. https://doi.org/10.3390/ijms24119288
Chicago/Turabian StyleYin, Lizi, Yuhong Gou, Yuyun Dai, Tao Wang, Kexin Gu, Ting Tang, Sajjad Hussain, Xiaoli Huang, Changliang He, Xiaoxia Liang, and et al. 2023. "Cinnamaldehyde Restores Ceftriaxone Susceptibility against Multidrug-Resistant Salmonella" International Journal of Molecular Sciences 24, no. 11: 9288. https://doi.org/10.3390/ijms24119288
APA StyleYin, L., Gou, Y., Dai, Y., Wang, T., Gu, K., Tang, T., Hussain, S., Huang, X., He, C., Liang, X., Shu, G., Xu, F., & Ouyang, P. (2023). Cinnamaldehyde Restores Ceftriaxone Susceptibility against Multidrug-Resistant Salmonella. International Journal of Molecular Sciences, 24(11), 9288. https://doi.org/10.3390/ijms24119288






