Role of bZIP Transcription Factors in Plant Salt Stress
Abstract
1. Introduction
2. Structural Characteristics of bZIP Transcription Factors
3. Effects of Salt Stress on Plants
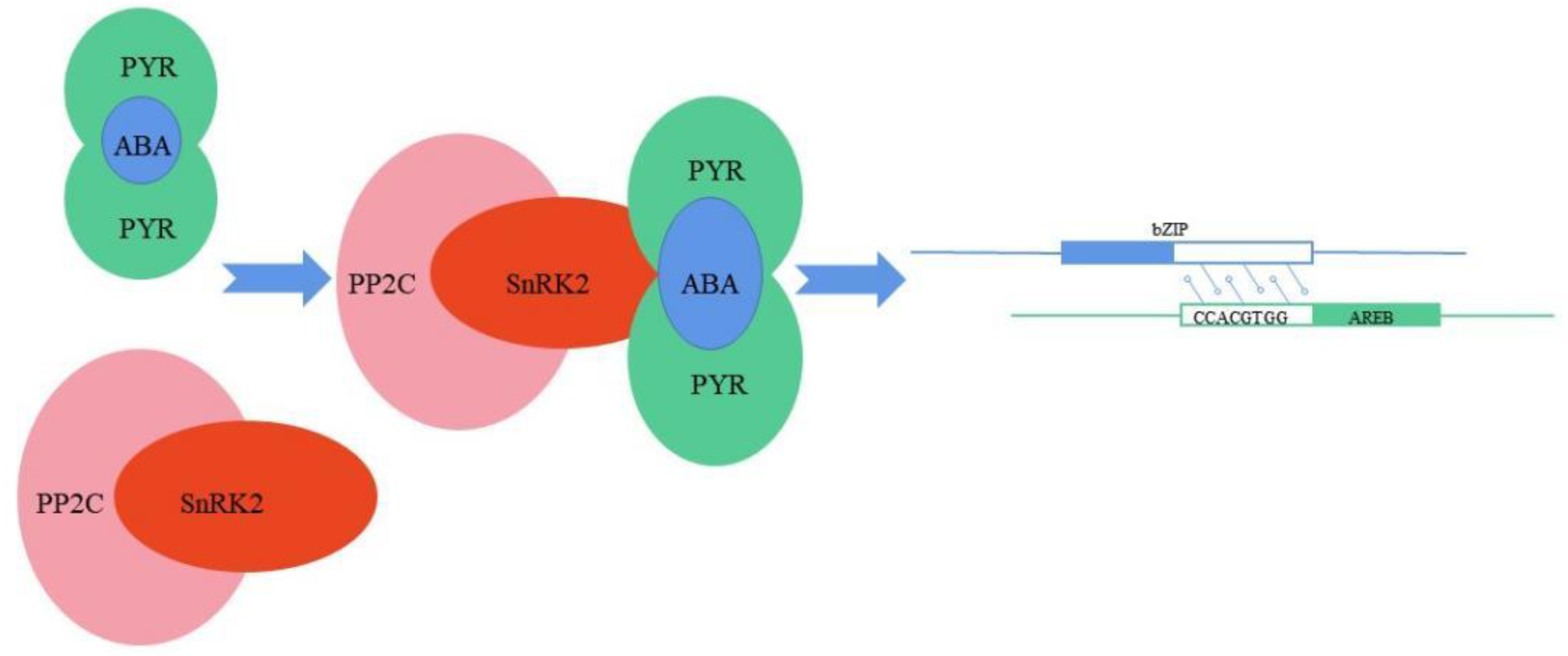
4. Plant bZIP Transcription Factors Interact with Genes in Response to Salt Stress
5. bZIP Transcription Factors Related to Salt Stress in Plants
| Species | bZIPs | References |
|---|---|---|
| Arabidopsis thaliana | AtbZIP1 | [61] |
| AtbZIP17 | [31] | |
| AtbZIP18 | ||
| Oryza sativa | OsbZIP23 | [73] |
| OsbZIP05 | [84] | |
| OsABF1 | [85] | |
| OsABF2 | [57] | |
| Glycine max | GmbZIP44 | [70] |
| GmbZIP62 | ||
| GmbZIP78 | ||
| GmbZIP1 | [86] | |
| GmbZIP132 | [71] | |
| Jatropha curcas | JcbZIP34 | [81] |
| JcbZIP36 | ||
| JcbZIP49 | ||
| JcbZIP50 | ||
| Solanum tuberosum | StbZIP25 | [77] |
| StbZIP65 | [78] | |
| Triticum aestivum | TabZIP15 | [87] |
| Capsicum annuum | CAbZIP1 | [79] |
| Setaria italica | SibZIP35 | [88] |
| SibZIP73 | ||
| SibZIP64 | ||
| SibZIP49 | ||
| SibZIP50 | ||
| SibZIP39 | ||
| SibZIP38 | ||
| SibZIP24 | ||
| Tamarix hispida | ThbZIP1 | [89] |
6. Functional Characterization of bZIP Gene through Gene Editing
7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D.; et al. Radically rethinking agriculture for the 21st century. Science 2010, 327, 833–834. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef]
- Rana, M.; Mark, T. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar]
- Liu, T. Use Model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 2014, 1150, 81–95. [Google Scholar] [PubMed]
- Jia, J.T.; Zhao, P.C.; Cheng, L.Q.; Yuan, G.X.; Yang, W.G.; Liu, S.; Chen, S.Y.; Qi, D.M.; Liu, G.S.; Li, X.X. MADS-box family genes in sheepgrass and their involvement in abiotic stress responses. BMC Plant Biol. 2018, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.; Weijers, D.; Wagner, D. Plant transcription factors-being in the right place with the right company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef]
- Singh, K.B.; Foley, R.C.; Onate-Sanchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Zheng, J.Y.; Liu, F.; Zhu, C.H.; Li, X.F.; Dai, X.Z.; Yang, B.Z.; Zou, X.X.; Ma, Y.Q. Identification, expression, alternative splicing and functional analysis of pepper WRKY gene family in response to biotic and abiotic stresses. PLoS ONE 2019, 14, e0219775. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plants. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S.H. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 2007, 19, 4111–4119. [Google Scholar] [CrossRef]
- Stanković, B.; Vian, A.; Henry-Vian, C.; Davies, E. Molecular cloning and characterization of a tomato cDNA encoding a systemically wound-inducible bZIP DNA-binding protein. Planta 2000, 212, 60–66. [Google Scholar] [CrossRef]
- Mianlengeh, Z.E.; Najafabadi, M.S.; Saidi, A.; Askari, H. Monitoring response of a few bZIP transcription factors in response to osmotic stress in sunflower. Iran. J. Biotechnol. 2018, 16, e1422. [Google Scholar]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, H.E.; Kim, S. Two different domain architectures generate structural and functional diversity among bZIP genes in the Solanaceae family. Front. Plant Sci. 2022, 13, 967546. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Qu, S.C.; Guo, Z.; Du, X.L.; Du, B.B.; Zhang, Z. Biological functions of plant bZIP transcription factors. Acta. Bot. Boreal. Occident Sin. 2011, 31, 1066–1075. [Google Scholar]
- Dröge-Laser, W.; Snoek, B.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 60–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profle of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, R.; Zhang, Z.; Zhao, T.; Zhang, D.; Sofkova, S.; Wu, Y.; Wang, Y. Genome-wide identification and expression analysis of the bZIP gene family in apple (Malus domestica). Tree Genet. Genomes 2016, 12, 1–17. [Google Scholar]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, W.; Li, H.; Wang, Y.; Li, D.; Xue, C.; Liu, Z.; Liu, M.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef]
- Yao, W.; Zhou, B.; Zhang, X.; Zhao, K.; Cheng, Z.; Jiang, T. Transcriptome analysis of transcription factor genes under multiple abiotic stresses in Populus simonii × P. nigra. Gene 2019, 707, 189–197. [Google Scholar] [CrossRef]
- Zhong, X.; Feng, X.; Li, Y.; Guzmán, C.; Lin, N.; Xu, Q.; Zhang, Y.; Tang, H.; Qi, P.; Deng, M.; et al. Genome-wide identification of bZIP transcription factor genes related to starch synthesis in barley (Hordeum vulgare L.). Genome 2021, 64, 1067–1080. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, L.; Wan, L.; Huai, D.; Kang, Y.; Shi, L.; Jiang, H.; Lei, Y.; Liao, B. Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut. BMC Genom. 2019, 20, 51. [Google Scholar] [CrossRef]
- Bensmihen, S.; Rippa, S.; Lambert, G.; Jublot, D.; Pautot, V.; Granier, F. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 2002, 14, 1391–1403. [Google Scholar] [CrossRef]
- Tajima, H.; Iwata, Y.; Iwano, M.; Takayama, S.; Koizumi, N. Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem. Biophys. Res. Commun. 2008, 374, 242–247. [Google Scholar] [CrossRef]
- Büttner, M.; Singh, K.B. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 1997, 94, 5961–5966. [Google Scholar] [CrossRef]
- Chern, M.S.; Bobb, A.J.; Bustos, M.M. The regulator of MAT2 (ROM2) protein binds to early maturation promoters and represses PvALF-activated transcription. Plant Cell 1996, 8, 305–321. [Google Scholar]
- Wani, S.H.; Kumar, V.; Khare, T.; Guddimalli, R.; Parveda, M.; Solymosi, K.; Suprasanna, P.; Kavi, P.B. Engineering salinity tolerance in plants: Progress and prospects. Planta 2020, 251, 76. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Moran, J.F.; Becana, M.; Iturbe-Ormaetxe, I.; Silvia, F.; Robert, V.K.; Pedro, A. Drought induces oxidative stress in pea plants. Planta 1994, 194, 346–352. [Google Scholar] [CrossRef]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Annu. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Annu. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Møller, I.M.; Sweetlove, L.J. ROS signalling–specificity is required. Trends Plant Sci. 2010, 15, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [PubMed]
- Vaahtera, L.; Brosché, M.; Wrzaczek, M.; Kangasjärvi, J. Specificity in ROS signaling and transcript signatures. Antioxid. Redox Signal. 2014, 21, 1422–1441. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Gapper, C.; Dolan, L. Control of plant development by reactive oxygen species. Plant Physiol. 2006, 141, 341–345. [Google Scholar] [CrossRef]
- Mignolet-Spruyt, L.; Xu, E.; Idänheimo, N.; Hoeberichts, F.A.; Mühlenbock, P.; Brosché, M.; Van Breusegem, F.; Kangasjärvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef]
- Onodera, Y.; Suzuki, A.; Wu, C.Y.; Washida, H.; Takaiwa, F. A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J. Biol. Chem. 2001, 276, 14139–14152. [Google Scholar] [CrossRef]
- Lee, S.S.; Yang, S.H.; Berberich, T.; Miyazaki, A.; Kusano, T. Characterization of ATBZIP2, ATBZIP11 and ATBZIP53 from the group S basic region-leucine zipper family in Arabidopsis thaliana. Plant Tissue Cult. Lett. 2006, 23, 249–258. [Google Scholar] [CrossRef]
- Schlögl, P.S.; Nogueira, F.T.; Drummond, R.; Felix, J.M.; De Rosa, V.E., Jr.; Vicentini, R.; Leite, A.; Ulian, E.C.; Menossi, M. Identification of new ABA- and MEJA-activated sugarcane bZIP genes by data mining in the SUCEST database. Plant Cell Rep. 2008, 27, 335–345. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, 15–45. [Google Scholar] [CrossRef]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defense responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Zeevaart, J.; Creelman, R.A. Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 439–473. [Google Scholar] [CrossRef]
- Leung, J.; Giraudat, J. Abscisic acid signal transduction. Annu. Rev. of Plant. Physiol. Plant Mol. Biol. 1998, 49, 199–222. [Google Scholar] [CrossRef]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef]
- Zhang, L.; Xie, J.; Wang, L.; Si, L.; Zheng, S.; Yang, Y.; Yang, H.; Tian, S. Wheat TabZIP8, 9, 13 participate in ABA biosynthesis in NaCl-stressed roots regulated by TaCDPK9-1. Plant Physiol. Biochem. 2020, 151, 650–658. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 1996, 7, 161–167. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D'Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Sun, X.L.; Li, Y.; Cai, H.; Bai, X.; Ji, W.; Ji, Z.J.; Zhu, Y.M. ArabidopsisbZIP1 transcription factor binding to ABRE cis-element regulates abscisic acid signal transduction. Acta Agronom. Sin. 2011, 37, 612–619. [Google Scholar] [CrossRef]
- Qu, G.Z.; Zang, L.; Xilin, H.; Gao, C.; Zheng, T.; Li, K.L. Co-transfer of LEA and bZIP genes from tamarix confers additive salt and osmotic stress tolerance in transgenic tobacco. Plant Mol. Biol. Rep. 2012, 30, 512–518. [Google Scholar] [CrossRef]
- Schubert, U.; Antón, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Vembar, S.S.; Brodsky, J.L. One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008, 9, 944–957. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.L.; Gao, D.S. Endoplasmic reticulum proteins quality control and the unfolded protein response: The regulative mechanism of organisms against stress injuries. Biofactors 2014, 40, 569–585. [Google Scholar] [CrossRef]
- Chen, X.; Shen, J.S.; Prywes, R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 2002, 277, 13045–13052. [Google Scholar] [CrossRef]
- Srivastava, R.; Chen, Y.; Deng, Y.; Brandizzi, F.; Howell, S.H. Elements proximal to and within the transmembrane domain mediate the organelle-to-organelle movement of bZIP28 under ER stress conditions. Plant J. 2012, 70, 1033–1042. [Google Scholar] [CrossRef]
- Srivastava, R.; Deng, Y.; Howell, S.H. Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front. Plant Sci. 2014, 5, 59. [Google Scholar] [CrossRef]
- Srivastava, R.; Deng, Y.; Shah, S.; Rao, A.G.; Howell, S.H. BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 2013, 25, 1416–1429. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Liao, Y.; Zhang, J.S.; Chen, S.Y.; Zhang, W.K. Role of soybean GmbZIP132 under abscisic acid and salt stresses. J. Integr. Plant Biol. 2008, 50, 221–230. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.Y.; Xiong, L.Z. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Piao, W.; Lim, J.H.; Han, S.H.; Kim, Y.S.; An, G.; Paek, N.C. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef]
- Hong, Y.B.; Zhang, H.J.; Huang, L.; Li, D.Y.; Song, F.M. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Li, C.W.; Su, R.C.; Cheng, C.P.; Tsai, Y.C.; Chan, M.T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar] [CrossRef]
- Zhao, P.; Ye, M.H.; Wang, R.Q.; Wang, D.D.; Chen, Q. Systematic identification and functional analysis of potato (Solanum tuberosum L.) bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP65 enhances salt tolerance. Int. J. Biol. Macromol. 2020, 161, 155–167. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Wang, D.; Xu, L.; Li, X.; Guo, Y. Identification and analysis of bZIP family genes in potato and their potential roles in stress responses. Front. Plant Sci. 2021, 12, 637343. [Google Scholar] [CrossRef]
- Lee, B.J.; Park, C.J.; Kim, S.K.; Kim, K.J.; Paek, K.H. In vivo binding of hot pepper bZIP transcription factor CabZIP1 to the G-box region of pathogenesis-related protein 1 promoter. Biochem. Biophys. Res. Commun. 2006, 344, 55–62. [Google Scholar] [CrossRef]
- Ying, S.; Zhang, D.F.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235, 253–266. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, J.; Yuan, W.; Wang, Y.; Hu, P.; Jiao, C.; Xia, H.; Wang, D.; Cai, Q.; Li, J.; et al. Genome-wide characterization of bZIP transcription factors and their expression patterns in response to drought and salinity stress in Jatropha curcas. Int. J. Biol. Macromol. 2021, 181, 1207–1223. [Google Scholar] [CrossRef] [PubMed]
- Gudimella, R.; Singh, P.; Mazumdar, P.; Wong, G.R.; Lau, S.E.; Harikrishna, J.A. Genome-wide regulatory network mapping of miRNA and transcription factors in banana roots. Tropical Plant Biol. 2018, 11, 141–153. [Google Scholar] [CrossRef]
- Kumar, N.; Bharadwaj, C.; Sahu, S.; Shiv, A.; Shrivastava, A.K.; Reddy, S.P.P.; Soren, K.R.; Patil, B.S.; Pal, M.; Soni, A.; et al. Genome-wide identification and functional prediction of salt- stress related long non-coding RNAs (lncRNAs) in chickpea (Cicer arietinum L.). Physiol. Mol. Biol. Plants 2021, 27, 2605–2619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, R.; Wang, H.; Ren, S.; Shi, L.; Huang, S.; Wei, Z.; Guo, B.; Jin, J.; Zhong, Y.; et al. OsWRKY28 positively regulates salinity tolerance by directly activating OsDREB1B expression in rice. Plant Cell Rep. 2023, 42, 223–234. [Google Scholar] [CrossRef]
- Amir Hossain, M.; Lee, Y.; Cho, J.I.; Ahn, C.H.; Lee, S.K.; Jeon, J.S.; Kang, H.; Lee, C.H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 2010, 72, 557–566. [Google Scholar] [CrossRef]
- Gao, S.Q.; Chen, M.; Xu, Z.S.; Zhao, C.P.; Li, L.; Xu, H.J.; Tang, Y.M.; Zhao, X.; Ma, Y.Z. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 2011, 75, 537–553. [Google Scholar] [CrossRef]
- Bi, C.; Yu, Y.; Dong, C.; Yang, Y.; Zhai, Y.; Du, F.; Xia, C.; Ni, Z.; Kong, X.; Zhang, L. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol. J. 2021, 19, 209–211. [Google Scholar] [CrossRef]
- Liu, B.L.; Zhang, L.; Sun, Y. Genome-wide identification of bZIP transcription factors in foxtail millet and their expression analysis under drought and salt stress. Acta Bot. Sin. 2016, 51, 473–487. [Google Scholar]
- Ji, X.; Liu, G.; Liu, Y.; Zheng, L.; Nie, X.; Wang, Y. The bZIP protein from Tamarix hispida, ThbZIP1, is ACGT elements binding factor that enhances abiotic stress signaling in transgenic Arabidopsis. BMC Plant Biol. 2013, 13, 151. [Google Scholar] [CrossRef]
- He, F.; Wei, C.; Zhang, Y.; Long, R.; Li, M.; Wang, Z.; Yang, Q.; Kang, J.; Chen, L. Genome-wide association analysis coupled with transcriptome analysis reveals candidate genes related to salt stress in alfalfa (Medicago sativa L.). Front. Plant Sci. 2002, 12, 826584. [Google Scholar] [CrossRef]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Nakagawa, H.; Ohmiya, K.; Hattori, T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J. 1996, 9, 217–227. [Google Scholar] [CrossRef]
- Kang, J.Y.; Choi, H.I.; Im, M.Y.; Kim, S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 2002, 14, 343–357. [Google Scholar] [CrossRef]
- Hsia, C.C.; McGinnis, W. Evolution of transcription factor function. Curr. Opin. Genet. Dev. 2003, 13, 199–206. [Google Scholar] [CrossRef]
- Wan, C.; Cui, P.; Sun, H.; Guo, W.; Yang, C.; Jin, H.; Fang, B.; Shi, D. Comparative effects of salt and alkali stresses on organic acid accumulation and ionic balance of sea buckthorn (Hippophae rhamnoides L.). Ind. Crop. Prod. 2009, 30, 351–358. [Google Scholar]
- Li, N.; Zhang, Z.H.; Chen, Z.J.; Cao, B.L.; Xu, K. Comparative transcriptome analysis of two contrasting Chinese cabbage (Brassica rapa L.) genotypes reveals that ion homeostasis is a crucial biological pathway involved in the rapid adaptive response to salt stress. Front. Plant Sci. 2021, 12, 215–227. [Google Scholar] [CrossRef]
- Zou, M.J.; Guan, Y.C.; Ren, H.B.; Zhang, F.; Chen, F. Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem. Biophys. Res. Commun. 2007, 360, 307–313. [Google Scholar] [CrossRef]
- Romani, F.; Ribone, P.A.; Capella, M.; Miguel, V.N.; Chan, R.L. A matter of quantity: Common features in the drought response of transgenic plants overexpressing HD-Zip I transcription factors. Plant Sci. 2016, 251, 139–154. [Google Scholar] [CrossRef]
- Kim, S.Y.; Ma, J.; Perret, P.; Li, Z.; Thomas, T.L. Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 2002, 130, 688–697. [Google Scholar] [CrossRef]
- Kagaya, Y.; Hobo, T.; Murata, M.; Ban, A.; Hattori, T. Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 2002, 14, 3177–3189. [Google Scholar] [CrossRef] [PubMed]
- Pei, C.M.; Zhang, Z.Y.; Ma, J. Identification and analysis of salt stress response transcription factors in leaves of salt-tolerant mutant of southern type alfalfa. Seed Plant Breed. 2017, 15, 7. [Google Scholar]


| Species | Number of bZIP Transcription Factor | References |
|---|---|---|
| Arabidopsis | 78 | [9] |
| Zea mays | 58 | [20] |
| Solanum lycopersicum | 69 | [21] |
| Vitis vinifera | 55 | [22] |
| Oryza sativa | 89 | [23] |
| Malus domestica | 114 | [24] |
| Populus simonii | 45 | [25] |
| Ziziphus jujuba | 45 | [26] |
| Poplar | 86 | [27] |
| Hordeum vulgare | 92 | [28] |
| Arachis duranensis | 50 | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. https://doi.org/10.3390/ijms24097893
Liu H, Tang X, Zhang N, Li S, Si H. Role of bZIP Transcription Factors in Plant Salt Stress. International Journal of Molecular Sciences. 2023; 24(9):7893. https://doi.org/10.3390/ijms24097893
Chicago/Turabian StyleLiu, Haotian, Xun Tang, Ning Zhang, Shigui Li, and Huaijun Si. 2023. "Role of bZIP Transcription Factors in Plant Salt Stress" International Journal of Molecular Sciences 24, no. 9: 7893. https://doi.org/10.3390/ijms24097893
APA StyleLiu, H., Tang, X., Zhang, N., Li, S., & Si, H. (2023). Role of bZIP Transcription Factors in Plant Salt Stress. International Journal of Molecular Sciences, 24(9), 7893. https://doi.org/10.3390/ijms24097893







