Updates on the Biofunctionalization of Gold Nanoparticles for the Rapid and Sensitive Multiplatform Diagnosis of SARS-CoV-2 Virus and Its Proteins: From Computational Models to Validation in Human Samples
Abstract
1. Introduction
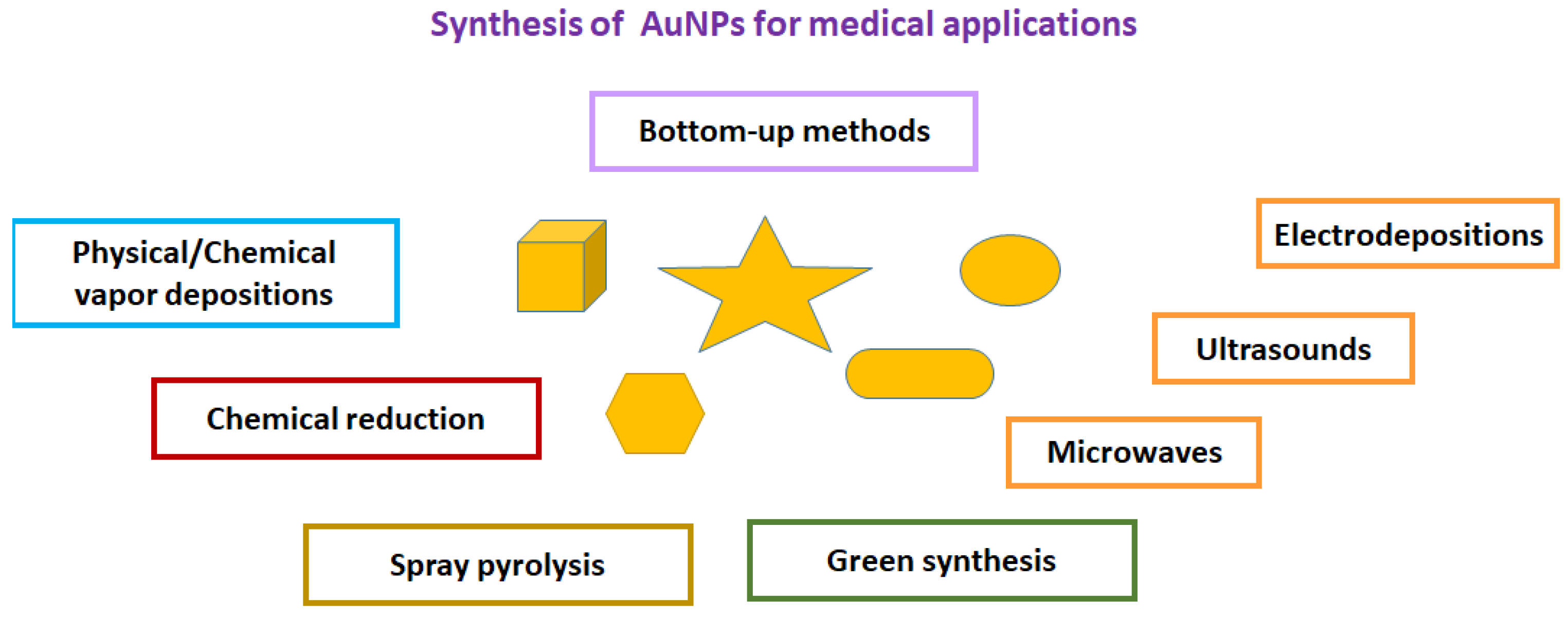
2. Synthesis of Gold Nanoparticles for In-Tube and On-Paper Medical Diagnoses
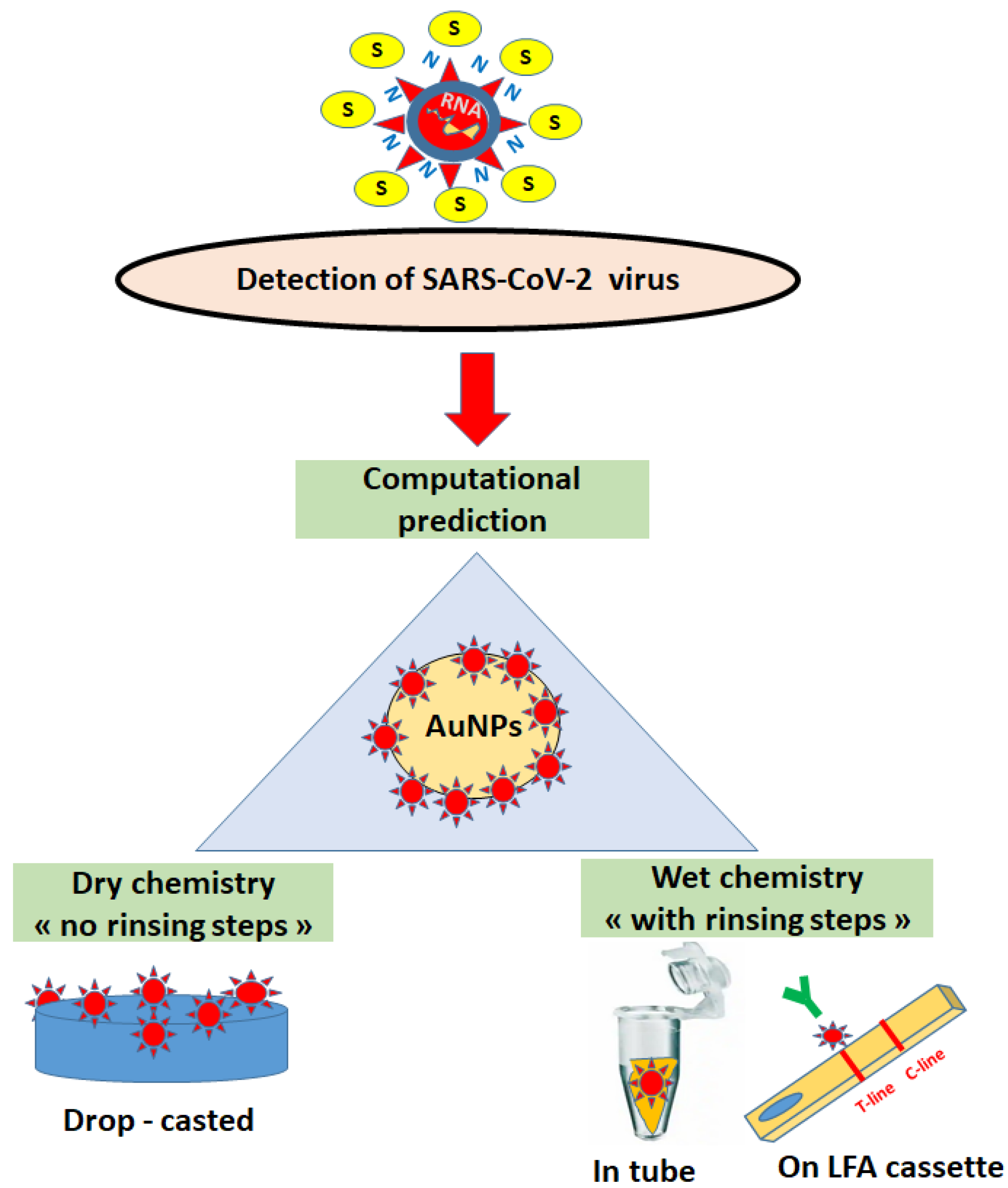
3. Computational Predictions of COVID-19 Infection Using Functionalized Gold Nanoparticles
4. Biofunctionalization of Supports and Viral Detection in Spiked Real Samples
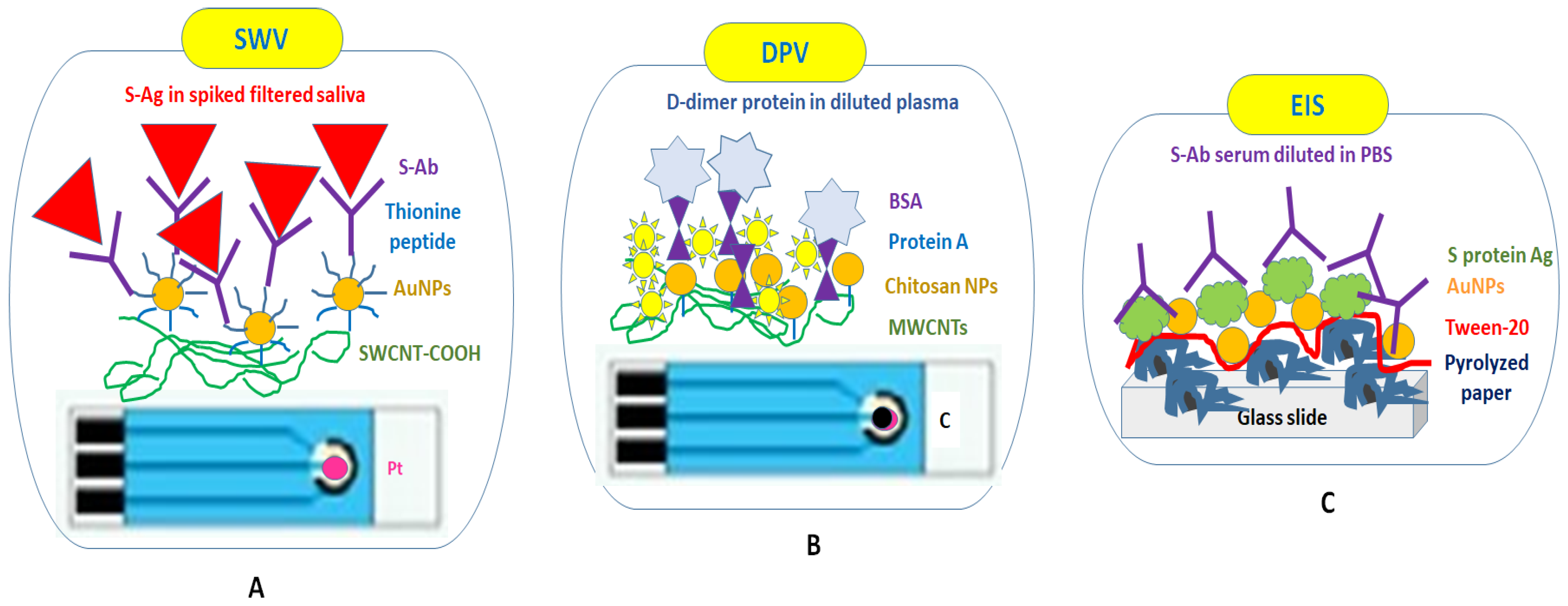
4.1. Using Dry Chemistry Functionalization through the Drop-Cast Approach without Rinsing Steps
| Drop Casting | |||||||
|---|---|---|---|---|---|---|---|
| Support | Method | Antibodies (Ab) | Antigens (Ag) | LOD | Advantages | Disadvantages | Ref |
| SPE-Pt | SWV | Monoclonal anti-S Ab | S protein * | 200 pM in PBS 500 pM in filtered saliva | Presence of SWCNTs | Dried Ab Short incubation time (5 min) between Ab and Ag No rinsing | [99] |
| SPE-C | DPV | D-dimer Ab | D-dimer * | 0.6 μg/L in PBS | Presence of MWCNTs and D-dimer, predictive markers of thromboembolic events important for viral infections Optimized incubation time (15 min) | Dried Ab No rinsing | [100] |
| Pyrolized paper-Pt | EIS | Ab in serum diluted with PBS * | S protein | NA | Presence of Tween-20 | Dried Ab No washing steps | [101] |
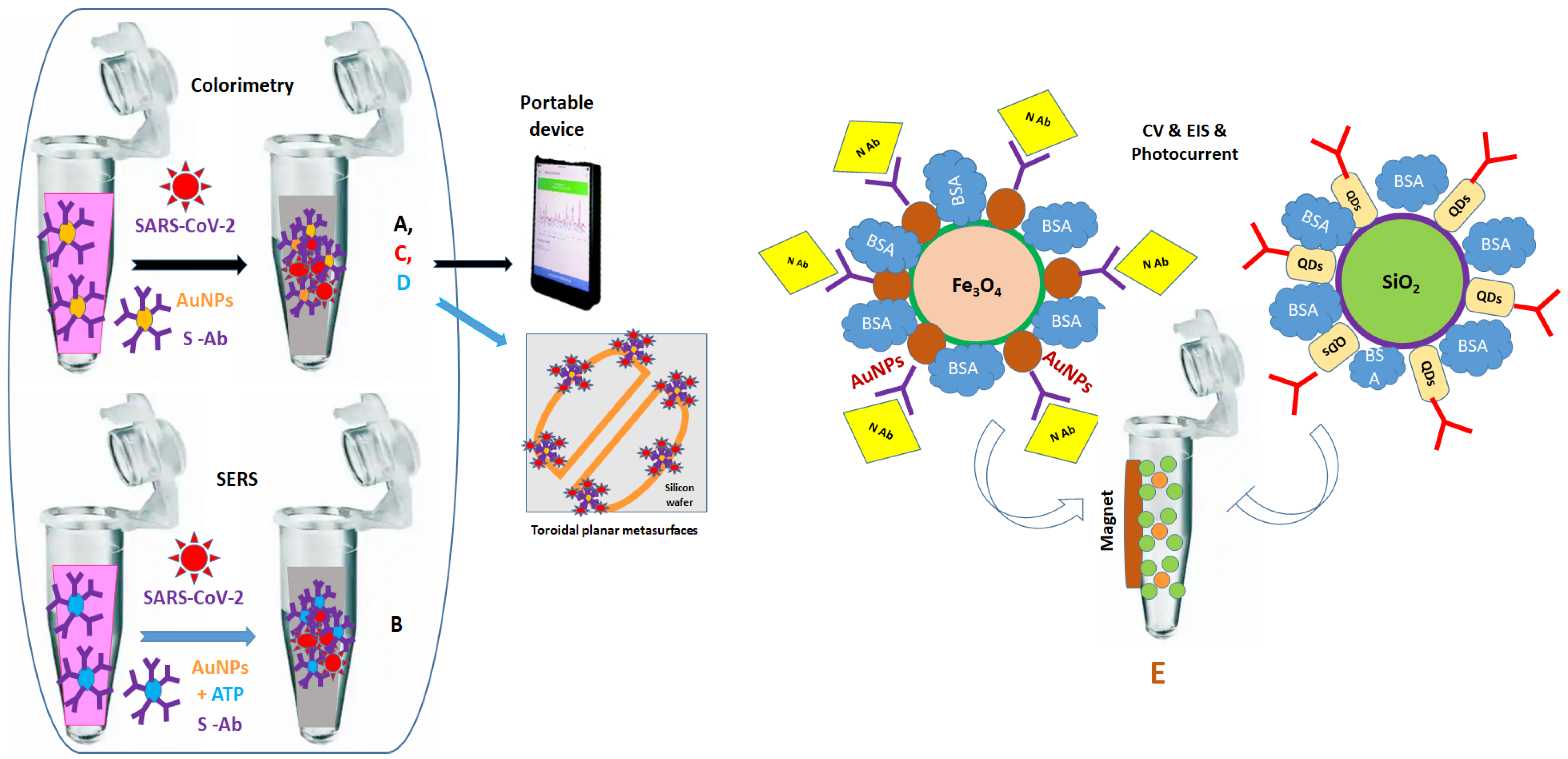
4.2. Using Wet Chemistry for Biofunctionalization in Eppendorf Tubes, including Centrifugation and Rinsing Steps
4.3. Using Wet Chemistry for the Biofunctionalization of Solid Substrates, including Rinsing Steps
| Tubes/Surfaces | |||||||
|---|---|---|---|---|---|---|---|
| Support | Method | Antibodies (Ab) | Antigens (Ag) | LOD | Advantages | Disadvantages | Ref |
| Tubes and glass slide | Colorime-tric and SERS | Monoclonal anti-S Ab | S protein * 1 ng/mL; 4 pg/mL and SARS-CoV-2 * Pseudo-virus 1000 particles/mL: 18 particles/mL | NA | Rapid test (5 min) Presence of 100 ng/mL anti-spike Ab blocked 100% of viral replication | Dried Ag for SERS | [102] |
| Tubes | Colorime-tric | Polyclonal Ab | S protein * and inactivated SARS-CoV-2 * virus in PBS, water river, artificial saliva, human saliva | 2.2 PFU/mL PBS and 0.28 PFU/mL human saliva and 7; 250; 1000; 5000; 6000 PFU mL | Covalent functionalization of AuNPs with MUA and thiol-PEG Optical signal reading with smartphone | Use of polyclonal Ab | [103] |
| Tubes and toroidal planar metasurfaces | Frequency shifts | Monoclonal anti-S Ab | S protein * in PBS (4 fM, 8 fM, 12 fM) | NA | Use of colloidal thiol-AuNPs | Dried Ag on AuNPs No tests with real samples | [104] |
| Tubes and ITO | Photo- current | Two sources of anti-N Ab | N protein * (1 h) | 1.8 pg/mL | Easy NPs washing by holding magnet on tubes | Multiple steps for NPs synthesis | [105] |
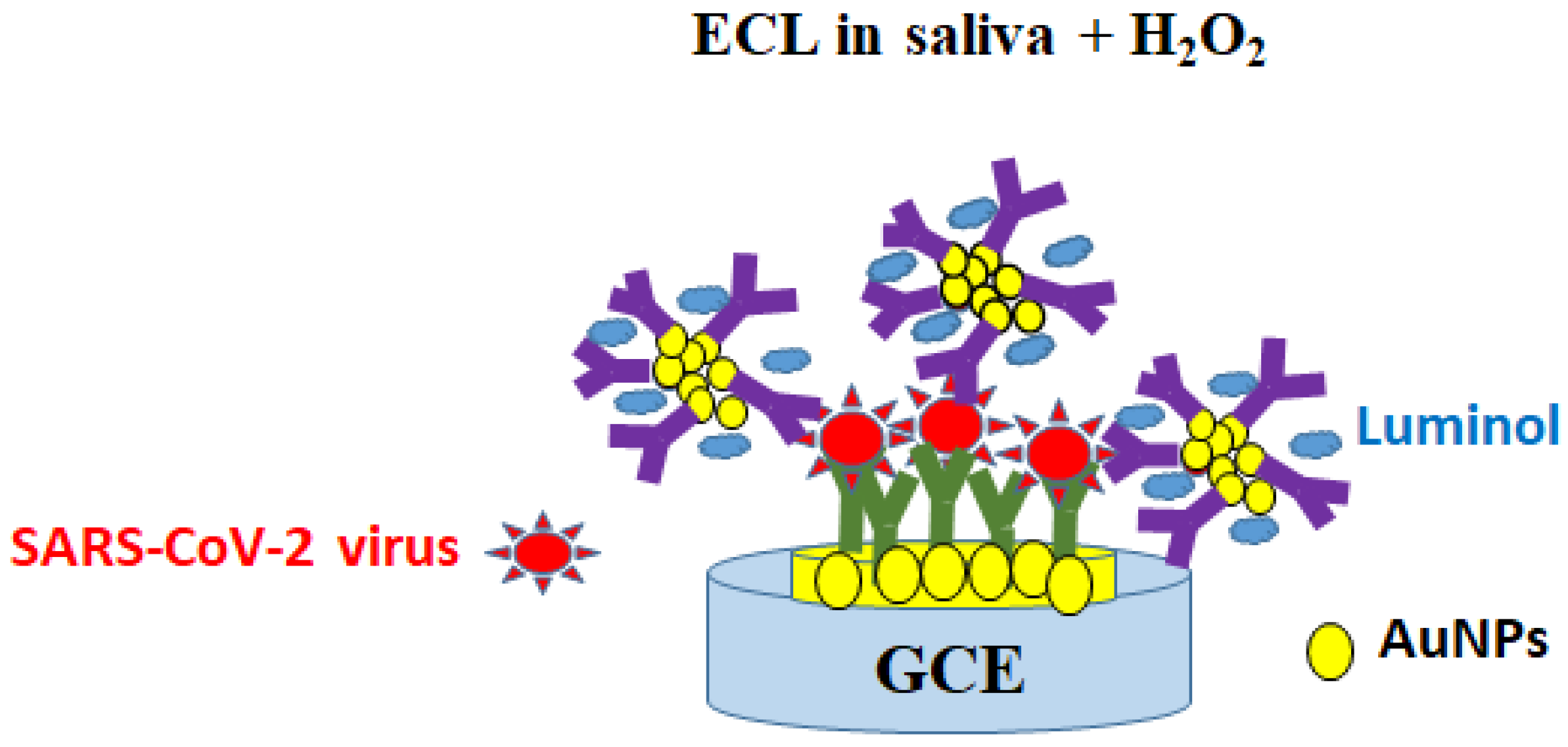
| GCE | ECL | Two sources of polyclonal anti-SARS-CoV-2 Ab | Inactivated SARS-CoV-2 virus * in PBS (10 ng–10 μg/mL) (2 h) | 1.93 ng/mL | Use of active probes (luminol) Optical signal reading with smartphone | Time-consuming protocol (2 days) | [106] |
| IDE | Impedance | SARS-CoV-2 IgG Ab in 10 × serum * (4 μL) | S protein (0.1 mg/mL) | NA | Easy monitoring of positive virus with change in frequency | Multistep preparation of IDE supports | [107] |
4.4. Using Lateral Flow Wet Chemistry for the Biofunctionalization of Metallic and Flexible Paper-Based Substrates including Rinsing Steps
4.4.1. Conventional Protein Antigen/Antibody-Based LFAs: First Generation
Detection of SARS-CoV-2 N protein using nasal swabs
Detection of SARS-CoV-2 RBD Using Nasal/Throat Swabs
Detection of IgA, IgG, and IgM Antibodies in Serum and Nasal Samples
Detection of the SARS-CoV-2 Virus Using Glyco-Based LFA: The Influence of Silver Staining
4.4.2. Pressed Protein Antigen/Antibody LFAs in the Center: Second Generation
4.4.3. SERS Optics for Protein Antigen/Antibody LFAs: Third Generation with Dual- and Tri-Mode Detection Signals
| First LFA | |||||||
|---|---|---|---|---|---|---|---|
| Paper Support | Method | Bioreagents on AuNPs | Antigen (Ag) | Bioreagents on T-Line/C-Line | Advantages | Disadvantages | Ref |
| NC | Colorimetric | Anti-N 20 ng/mL (minimal concentration)/cAuNPs (1:2) | N-protein * (2.14 ng/mL) on nasal swabs | Ab1 2 mg/mL and Ab2 2 mg/mL | Low concentrations and volumes of Ab for conjugation on AuNPs | High optimal concentration of Ab on T/C-lines Local optimization | [110] |
| NC (12 μ pore size) | Colorimetric | Anti- RBD (1 mg/mL)/ cAuNPs (~15 nm, pH 9) | RBD * on nasal swabs in PBS and spiked PBS (LOD 1 ng/mL) | Ab1 1 mg/mL and Ab2 1 mg/mL | 100 μL sample T-line readings with smartphone Signal stability for 21 days | Local optimization | [111] |
| NC CN140 | Colorimetric | RBD (1 mg/mL in PBS):cAuNPs (~20 nm, pH 9) | RBD * in PBS LOD 0.11 ng/mL RBD * in throat swabs | S-IgG (500 μL/Ml in PBS) on T-spot and IgM (1 mg/mL in PBS) on C-spot | Presence of trimethylsilyl cellulose barrier and upper extra sample pad layer before T-line to slow down the flow Improved LOD Target detected on T-spot with no line | Lack of robustness of local optimization: Spacing between barrier/T spot/C spot Homogeneity of spot size | [112] |
| NC + 1 mg/mL BSA | Colorimetric | IgM human (1.5 μg)-cAuNPs (30 nm) | SARS-CoV-2 Ig M * in serum samples (20 μL/80 μL running buffer PBS/BSA/Triton) | SARS-CoV-2 (1 mg/mL) on T-line and goat anti-mouse IgG (2 mg/mL) on C-line | Good selectivity in the presence of other viruses 10x test/sample very low sample volume 10–20 μL) | Laborious strip optimization | [115] |
| NC | Colorimetric | N (70–80 μg/mL) /Au NPs (30 nm) and RBD (60–70 μg/mL)/AuNPs (30 nm) pH 9 | Simultaneous IgM * and IgG * in serum samples | On T-line: goat anti-human IgG (1 mg/mL) and IgM (1.2 mg/mL) On C-line: anti-mouse IgG 2 mg/mL for N and 1.8 mg/mL for RBD | Early detection of SARS-CoV-2 infection (<1 week) Visible T-line | No calibration curve No LOD | [116] |
| NC | Colorimetric | Protein A-cAuNP (pH 8.5–9) | Simultaneous IgM */IgG */IgA * in serum/plasma | N protein (0.9 μg/cm) on T-line and Protein A (0.3 μg/cm) on C-line | No influence from anticoagulants on the color of T-/C-lines No cross-reactivity with other viruses | No calibration curve No LOD | [118] |
| NC Immunopore | Colorimetric | Glycan-AuNPs (35 nm) | Inactivated SARS-CoV-2 * collected from nasal swabs | Spot of SARS-CoV-2 S protein-bearing lentivirus and Lectine on C-line | Sample spot instead of T-line Very low sample volume (2 μL) Improved color visibility due to silver staining step | Need for polymeric coating to reduce nonspecific interaction | [120] |
| NC | Colorimetric and enhanced signal in the presence of HRP/AEC reagents | Anti-S Ab/AuNPs (10 nm) | SARS-CoV-2 S * in saliva Visible to naked eye for: HEP at 3.13 μg/mL in 25 μL, and on HS at 3.13 μg/mL in 50 μL | On T-line: GAG with streptavidin (1 mg/mL) and on C-line: Anti IgG (1 mg/mL) | Signal stability over 47 days at RT Computation data LOD (0.78 μg/mL in 25 μL) 4-fold lower compared with unamplified results | Laborious strip optimization | [121] |
| Second LFA | |||||||
| NC 10 μm pore size (CNPF-SN12) | Colorimetric | Anti-CRP (2 mg/mL)/ AuNPs (20 nm) | CRP protein * in PBS and serum and COVID-19 Ag kit 2-fold increased signal | Anti-CRP Ab (1 mg/mL) on T-line and goat anti-mouse IgG Ab on C-line | Pressure zone between T-line and C-line induces flow delay and signal is enhanced | No tests with samples from SARS-CoV-2 infected patients | [122] |
| Third LFA | |||||||
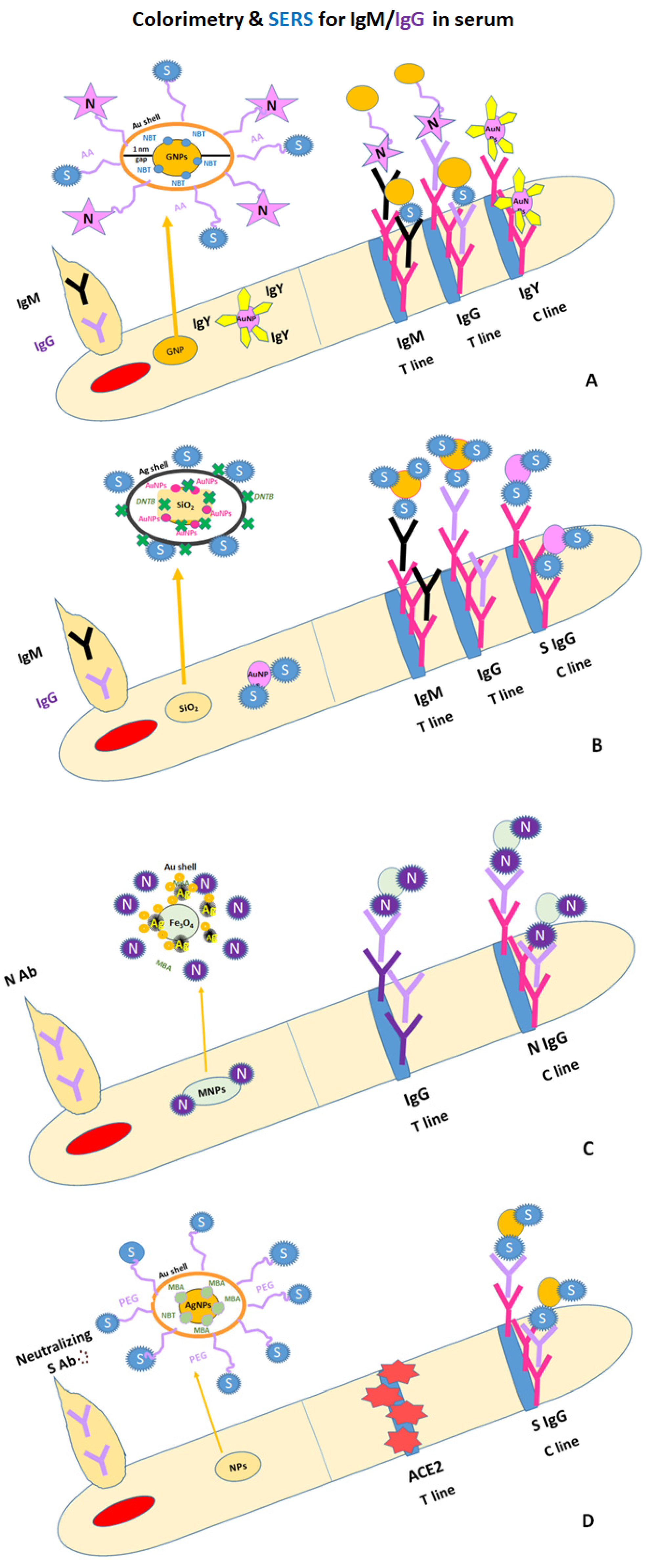
| NC 8 μm pore size (CN140) | Colorimetric and SERS | S-protein/ N-protéiné on Au-GNPs (10 μg/ 1.1 × 10−3 g GNPs (55 nm) + 4-NBT | Simultaneous IgM */IgG * in positive blood/serum samples | Mouse anti-human IgM (1 mg/mL)/ mouse anti-human IgG (1 mg/mL) on two T-lines and polyclonal goat anti-chicken IgY antibody (0.5 mg/mL) on C-line | Two T lines 4-NBT (5 μg/mL) on AuGNPs 100 times more sensitive than colorimetric result LODs: 1 ng/mL for IgM and 0.1 ng/mL for IgG | Instability of 4-NBT on AuNPs No data on storage of Au-GNPs /buffer/tempe-rature | [123] |
| NC ~10 μm pore size (CN140) | Colorimetric and SERS | SARS-CoV-2 S/ nanocomposites (30 μg of S protein/ mL of SERS tags) + DTNB | simultaneous IgM */IgG * in 103×, 104×, 105×, 106× diluted serum and 1% whole blood samples after 25 min immunoreaction | Goat anti-human IgM (0.5 mg/mL)/ goat anti-human IgG (0.6 mg/mL) on two T-lines and SARS-CoV-2 S Ab (0.5 mg/mL) on C-line | DTNB Raman dye SERS signal at 1328 cm–1 for IgM andIgG 800 times more sensitive that LFA based on AuNPs LODSERS 1 pg/mL | No SERS data on individual components of blood/serum samples for patients with different pathologies | [124] |
| NC | Colorimetric and SERS | SARS-CoV-2 N (10 μg/mL)/ trimetallic hybrid MNPs + MBA | anti-SARS-CoV-2 N Ab * in positive serum samples | Mouse anti-SARS-CoV-2 N Ab (1 mg/mL) on T-line and rabbit anti-human Ab (1 mg/mL) on C-line | LOD 10−8 mg/mL MBA Raman dye SERS signal at 1075 cm−1, linear within the range of 10−10 to 10−6 mg /mL Ab * LOD 0.08 pg/mL | No data on the stability of hybrid MNPs with MBA over time and influence of washing, RT and 4 °C in SERS signal evolution | [127] |
| NC CN140 | Colorimetric and Photothermal and SERS | SARS-CoV-2 S protein (10 μg/mL)/ bimetallic Au–Ag HNSs + MBA | Neutralizing SARS-CoV-2 Ab * in spiked serum samples 10x diluted with PBS (20 to 1500 ng/mL) and blood | ACE2 protein on T-line and SARS-CoV-2 S protein Ab on C-line | MBA Raman dye LOD 160 ng/mL LOD 20 ng/mL LOD 20 ng/mL | Missing the SERS signal information used for the calibration curve No data on the stability of NPs with MBA over time and influence of washing, RT and 4 °C in SERS signal evolution | [125] |
| Fourth LFA | |||||||
| NC M170 | Colorimetric | Thiol–DNA probe (4 and 8 μM)/ AuNPs | SARS-CoV-2 N gene * | SARS-CoV-2 N gene complementary on T-line and complementary DNA probe on C-line | 3 sandwich nLFA models 50 bp length of the target influence LOD 5 pM in SSC buffer | Laborious explanation Manually micro- pipetting lines No calibration curves for the three proposed nLFA models | [126] |
| NC HF120 | Colorimetric | Thiol–DNA detection probe/AuNPs (~13 nm) (7 μL) | SARS-CoV-2 RNA * | Biotin–DNA capture probe (0.5 μL of 25 μM) on T-line and biotin–DNA control probe (0.5 μL of 25 μM) on C-line | -Two sugar barriers (10 %) before (2.5 μL) and after (1 μL) the T-line to slow down the flow and improve the test sensitivity by 5-fold | No data on storage and signal stability No data using positive samples | [128] |
| NC | Colorimetric and digital camera and SERS (laser 532 nm) | Two antisense oligonucleotides ASO (25 μM) labeled with FAM (30 μM) and biotin (30 μM) Optimization: ASO1-biotin ASO2-FAM | Inactivated SARS-CoV-2 N gene * (RNA/cDNA) in 10x diluted swab sample and 67250; 3362; 168; 8; 0.42; 0.02; 0.001 copies/μL | Anti-FAM Ab + Av-AuNPs on T-line and anti-Av Ab on C-line | 30 min test after collection of swab Cys-AuNPs as signal enhancer of T-line color for concentrations below 8 copies/μL LOD of 0.02 copies/μL SERS signal at 1617 cm−1 strongly increased Validated in 30 positive nasal/nasopharyngeal samples | Naked-eye color evaluation not possible until 8 copies/μL No calibration curve based on SERS signals Several parameters require optimizations by end users | [129] |
| NC | Colorimetric and smartphone | Anti-FITC Ab/AuNP | SARS-CoV-2 N gene detection in cDNA and SARS-CoV-2 RNA * from swabs tested positive with real-time qRT-PCR (5.6 × 106 to 3.9 × 103 copies/mL) | Streptavidin on T-line and anti-goat IgG Ab on C-line | 5 μL of LAMP-amplified product based on biotin-dUTP/FITC-LF + 20 μL running buffer LOD 3.9 × 103 RNA copies/mL | Laborious protocol Temperature ˃60 °C for active polymerase enzyme and not applicable for kit market | [130] |
| NC | Colorimetric | Anti-FAM Ab/AuNPs | RBD * within the S gene of SARS-CoV-2, (synthetic DNA, 16.7 nM) | SA on T-line and anti-rabbit Ab on C-line | Biotin primer for easy linking to SA-NC FAM probe | Costly protocol No data with real positive samples | [131] |
4.4.4. Nucleic Acid LFAs: Fourth Generation
5. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kamble, P.; Daulatabad, V.; Patil, R.; John, N.A.; John, J. Omicron variant in COVID-19 current pandemic: A reason for apprehension. Horm. Mol. Biol. Clin. Investig. 2023, 44, 89–96. [Google Scholar] [CrossRef]
- Tufa, A.; Gebremariam, T.H.; Manyazewal, T.; Manyazewal, T.; Asrat, Y.; Getinet, T.; Hundie, T.G.; Webb, D.-L.; Hellström, P.M.; Genet, S. Limited value of neutrophil-to-lymphocyte ratio and serum creatinine as point-of-care biomarkers of disease severity and infection mortality in patients hospitalized with COVID-19. PLoS ONE 2022, 17, e0275391. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, N.; Grossegesse, M.; Neumann, M.; Schaade, L.; Nitsche, A. Evaluation of a commercial ELISA as alternative to plaque reduction neutralization test to detect neutralizing antibodies against SARS-CoV-2. Sci. Rep. 2022, 12, 3549. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Abugalyon, Y.; Li, X.J. Multicolorimetric ELISA biosensors on a paper/polymer hybrid analytical device for visualpoint-of-care detection of infection diseases. Anal. Bioanal. Chem. 2021, 413, 4655–4663. [Google Scholar] [CrossRef]
- Perivolaropoulos, C.; Vlacha, V. A reduction of the number of assays and turnaround time by optimizing polymerase chain reaction (PCR) pooled testing for SARS-CoV-2. J. Med. Virol. 2021, 93, 4508–4515. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.E.; Ryu, E.; Kim, J.; Shin, C.J.; Kwon, O.S. Fluorophore-encapsulated nanobeads for on-site, rapid, and sensitive lateral flow assay. Sens. Actuators B Chem. 2023, 381, 133364. [Google Scholar] [CrossRef]
- Jaisankar, A.; Krishnan, S.; Ramalingam, M. Design and development of nanoscale aptasensors for viral diagnostics. Lett. Appl. NanoBioScience 2023, 12, 34. [Google Scholar]
- Hemida, M.G. The next-generation coronavirus diagnosis techniques with particular emphasis on the SARS-CoV-2. J. Med. Virol. 2021, 93, 4219–4241. [Google Scholar] [CrossRef]
- Cardeso, V.M.d.O.; Moreira, B.J.; Comparetti, E.J.; Sampaio, I.; Ferreira, L.M.B.; Lins, P.M.P.; Zucolotto, V. Is nanotechnology helping in the fight against COVID-19. Front. Nanotechnol. 2020, 2, 588915. [Google Scholar] [CrossRef]
- Moabelo, K.L.; Martin, D.R.; Fadaka, A.O.; Sibuyi, N.R.S.; Meyer, M.; Madiehe, A.M. Nanotechnology-based strategies for effective and rapid detection of SARS-CoV-2. Materials 2021, 14, 7851. [Google Scholar] [CrossRef]
- Somu, P.; Mohanty, S.; Chakraborty, S.; Paul, S. Application of nanoscale materials and nanotechnology against viral infection: A special focus on coronaviruses. Adv. Exp. Med. Biol. 2021, 1352, 173–193. [Google Scholar] [PubMed]
- Srivastava, M.; Srivastava, N.; Mishra, P.K.; Malhotra, B.D. Prospects of nanomaterials-enabled biosensors for COVID-19 detection. Sci. Total Environ. 2021, 754, 142363. [Google Scholar] [CrossRef]
- Anh, N.H.; Doan, M.Q.; Dinh, N.X.; Huy, T.Q.; Tri, D.Q.; Ngoc Loan, L.T.; Van Hao, B.; Le, A.-T. Gold nanoparticle-based optical nanosensors for food and health safety monitoring: Recent advances and future perspectives. RSC Adv. 2022, 12, 10950–10988. [Google Scholar] [CrossRef] [PubMed]
- Jazaverin, M.H.; Amani, H.; Pourfatollah, A.A.; Pazoki-Toroudi, H.; Sedighimoghaddam, B. Various methods of gold nanoparticles (GNP) conjugation to antibodies. Sens. Bio-Sens. Res. 2016, 9, 17–22. [Google Scholar]
- Stine, K.J.; Jefferson, K.; Shulga, O.V. Nanoporous gold for enzyme immobilization. Methods Mol. Biol. 2017, 1504, 37–60. [Google Scholar]
- Mateu Ferrando, R.; Lay, L.; Polito, L. Gold nanoparticle-based platforms for vaccine development. Drug Discov. Today Technol. 2020, 38, 57–67. [Google Scholar] [CrossRef]
- Okay, S.; Özcan, O.O.; Karahan, M. Nanoparticles-based delivery platforms for mRNA vaccine development. AIMS Biophys. 2020, 7, 323–338. [Google Scholar] [CrossRef]
- Sengupta, A.; Azharuddin, M.; Al-Otaibi, N.; Hinkula, J. Efficacy and immune response elicited by gold nanoparticle-based nanovaccines against infectious diseases. Vaccines 2022, 10, 505. [Google Scholar] [CrossRef]
- Jafari, A.; Danesh Pouya, F.; Niknam, Z.; Abdollahpour-Alitappeh, M.; Rezaei-Tavirani, M.; Rasmi, Y. Curent advances and challenges in COVID-19 vaccine development: From conventional vaccines to next-generation vaccine platforms. Molec. Biol. Rep. 2022, 49, 4943–4957. [Google Scholar] [CrossRef]
- Moshref Javadi, M.; Taghdisi Hosseinzadeh, M.; Soleimani, N.; Rommasi, F. Evaluating the immunogenity of gold nanoparticles conjugated RBD with Freud’s adjuvant as a potential vaccine against SARS-CoV-2. Microb. Pathog. 2022, 170, 105687. [Google Scholar] [CrossRef]
- Zagorski, K.; Pandey, K.; Rajaiah, R.; Olwenyi, O.A.; Bade, A.N.; Acharya, A.; Johnston, M.; Filliaux, S.; Lyubchenko, Y.L.; Byrareddy, S.N. Modular nanoarray vaccine for SARS-CoV-2. Nanomed. Nanotechnol. Biol. Med. 2022, 46, 102604. [Google Scholar] [CrossRef]
- Mallakpou, S.; Azadi, E.; Hussain, C.M. The latest strategies in the fight against the COVID-19 pandemic: The role of metal and metal oxide nanoparticles. New J. Chem. 2021, 45, 6167–6179. [Google Scholar] [CrossRef]
- Biddram, E.; Esmaelli, Y.; Amini, A.; Sartorious, R.; Tay, F.R.; Shariati, L.; Makvandi, P. Nanobased platforms for diagnosis and treatment of COVID-19: From benchtop to bedside. ACS Biomater. Sci. Eng. 2021, 7, 2150–2176. [Google Scholar] [CrossRef] [PubMed]
- Guarise, C.; Pasquato, L.; De Filippis, V.; Scrimin, P. Gold nanoparticles-based protease assay. Proc. Natl. Acad. Sci. USA 2006, 103, 3978–3982. [Google Scholar] [CrossRef] [PubMed]
- Vidotti, M.; Carvalhal, R.F.; Mendes, R.K.; Ferreira, D.; Kubota, L.T. Biosensors based on gold nanostructures. J. Braz. Chem. Soc. 2011, 22, 3–20. [Google Scholar] [CrossRef]
- Dreaden, E.C.; Alkilany, A.M.; Huang, X.; Murphy, C.J.; El-Sayed, M.A. The golden age: Gold nanoparticles for biomedicine. Chem. Soc. Rev. 2012, 41, 2740–2779. [Google Scholar] [CrossRef]
- Tokmakov, A.A.; Kurotani, A.; Sato, K.-I. Protein pI and intracellular localization. Front. Mol. Biosci. 2021, 8, 775736. [Google Scholar] [CrossRef]
- Hori, K.; Yoshimoto, S.; Yoshino, T.; Zako, T.; Hirao, G.; Fujita, S.; Nakamura, C.; Yamagishi, A.; Kamiya, N. Recent advances in research on biointerfaces: From cell surfaces to artificial interfaces. J. Biosci. Bioeng. 2022, 133, 195–207. [Google Scholar] [CrossRef]
- Hutter, E.; Maysinger, D. Gold-nanoparticles-based biosensors for detection of enzyme activity. Trends Pharmacol. Sci. 2013, 34, 497–507. [Google Scholar] [CrossRef]
- Ramsey, A.V.; Bischoff, A.J.; Francis, M.B. Enzyme activated gold nanoparticles for versatile site-selective bioconjugation. J. Am. Chem. Soc. 2021, 143, 7342–7350. [Google Scholar] [CrossRef]
- Mohammadi, M.; Antoine, D.; Vitt, M.; Dickie, J.M.; Jyoti, S.S.; Wall, J.G.; Johnson, P.A.; Wawrousek, K.E. A fast, ultrasensitive SERS immunoassay to detect SARS-CoV-2 in saliva. Anal. Chim. Acta 2022, 1229, 340290. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, B.; He, M.; Hu, B. A homogeneous nucleic acid assay for simultaneous detection of SARS-CoV-2 and influenza A (H3N2) by single-particle inductively coupled plasma mass spectrometry. Anal. Chim. Acta 2021, 1186, 339134. [Google Scholar] [CrossRef]
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano 2020, 14, 17028–17045. [Google Scholar] [CrossRef] [PubMed]
- Elwakkad, A.; Gamal el Din, A.A.; Saleh, H.A.; Ibrahim, N.E.; Hebishy, M.A.; Mourad, H.H.; El-Kassaby, M.; Abou-Seif, H.S.; Elgattan, G.M. Gold nanoparticles combined baker’s yeast as a successful approach for breast cancer treatment. J. Genet. Eng. Biotechnol. 2023, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- Bao, Q.; Li, G.; Yang, Z.; Liu, J.; Wang, H.; Pang, G.; Guo, Q.; Wei, J.; Cheng, W.; Lin, L. Electrochemical biosensorbased on antibody-modified Au nanoparticles for rapid and senstive analysis of influenza A virus. Ionics 2023, 29, 2021–2029. [Google Scholar] [CrossRef]
- Sinawang, P.D.; Rai, V.; Ionescu, R.E.; Marks, R.S. Electrochemical lateral flow immunosensor for detection and quantification of dengue NS1 protein. Biosens. Bioelectron. 2016, 77, 400–408. [Google Scholar] [CrossRef]
- Karki, M.; Rajak, K.K.; Singh, P.; Fayaz, A.; Kumar, A.; Bhatt, M.; Rai, V.; Einstein, C.; Yadav, A.K.; Singh, R.P. Optimization of lateral flow assay for Canine morbilivirus detection and the application of the strip as sample substitute. J. Immunol. Methods 2023, 514, 113438. [Google Scholar] [CrossRef]
- Shyam, K.U.; Kong, K.-H.; Oh, M.-J.; Kim, T.; Kim, C.-S.; Kim, W.-S. Development of a lateral flow immunochromatographic assay (LFIA) for the detection of hirame novirhabdovirus (HIRRV) in olive flounder (Paralichthys olivaceus). Aquaculture 2023, 568, 739341. [Google Scholar] [CrossRef]
- Martinez-Liu, C.; Machain-Williams, C.; Martinez-Acuna, N.; Lozano-Sepulveda, S.; Galan-Huerta, K.; Arellanos-Soto, D.; Meléndez-villanueva, M.; Avalos-Nolazco, D.; Pérez-Ibarra, K.; Galindo-Rodriguez, S.; et al. Development of a rapid gold nanoparticle-based lateral flow immunoassay for the detection of Dengue virus. Biosensors 2022, 12, 495. [Google Scholar] [CrossRef]
- Najafabad, M.B.; Rastin, S.J.; Taghvaei, F.; Khiyavi, A.A. A review on applications of gold nanoparticles-based biosensors for pathogen detection. Adv. Nat. Sci. Nanosci. Nanotechnol. 2022, 13, 033002. [Google Scholar] [CrossRef]
- Das, A.; Terry, L.R.; Guo, H. A surface-enhanced Raman spectroscopy based smart Petri dish for sensitive and rapid detection of bacterial contamination in shrimp. Food Chem. 2023, 2, 100222. [Google Scholar] [CrossRef]
- Deb, M.; Hunter, R.; Taha, M.; Abdelbary, H.; Anis, H. Rapid detection of bacteria using gold nanoparticles in SERS with three different capping agents: Thioglucose, polyvinylpyrrolidone, and citrate. Spectrochim. Acta Mol. Biomol. Spectrosc. 2022, 280, 121533. [Google Scholar] [CrossRef]
- Peplow, M. Nanotechnology offers alternative ways to fight COVID-19 pandemic with antivirals. Nat. Biotechnol. 2021, 39, 1172–1174. [Google Scholar] [CrossRef]
- Ramaiah, G.B.; Tegegne, A.; Melese, B. Functionality of nanomaterials and its technological aspects-used in preventing, diagnosing and treating COVID-19. Mater. Today Proc. 2021, 47, 2337–2344. [Google Scholar] [CrossRef]
- Sarkar, J.; Das, S.; Aich, S.; Bhattacharyya, P.; Acharya, K. Antiviral potential of nanoparticles for the treatment of Coronavirus infections. J. Trace Elem. Med. Biol. 2022, 72, 126977. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, M.; Vora, A. Nanotechnology-based antiviral therapeutics. Drug Deliv. Transl. Res. 2021, 11, 748–787. [Google Scholar] [CrossRef]
- Barabadi, H.; Noqani, H.; Ashouri, E.; Prasad, A.; Jounaki, K.; Mobaraki, K.; Mohanta, Y.K.; Mostafavi, E. Nanobiotechnological approaches in anticoagulant therapy: The role of bioengineered silver and gold nanomaterials. Talanta 2023, 256, 124279. [Google Scholar] [CrossRef]
- Dheyab, M.A.; Aziz, A.A.; Khaniabadi, P.M.; Jameel, M.S.; Oladzadabbasadi, N.; Rahman, A.A.; Braim, F.S.; Mehrdel, B. Gold nanoparticles-based photothermal therapy for breast cancer. Photodiagn. Photodyn. Ther. 2023, 42, 103312. [Google Scholar] [CrossRef]
- Li, M.; Liu, J.; Shi, L.; Zhou, C.; Zou, M.; Fu, D.; Yuan, Y.; Yao, C.; Zhang, L.; Qin, S.; et al. Gold nanoparticles-embedded ceria with enhanced antioxidant activities for treating inflammatory bowel disease. Bioact. Mater. 2023, 25, 95–106. [Google Scholar] [CrossRef]
- Baker, A.; Wahid, I.; Hassan Baig, M.; Alotaibi, S.S.; Khalid, M.; Uddin, I.; Dong, J.-J.; Khan, M.S. Silk coccon-derived protein bioinspired gold nanoparticles as a formidable anticacer agent. J. Biomed. Nanotechnol. 2021, 17, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, D.Y. Photothermal therapy with gold nanoparticles as an anticancer medication. J. Pharm. Investig. 2017, 47, 19–26. [Google Scholar] [CrossRef]
- Alle, M.; Sharma, G.; Lee, S.-H.; Kim, J.-C. Next-generation engineered nanogold for multimodal cancer therapy and imaging: A clinical perspectives. J. Nanobiotechnol. 2022, 20, 222. [Google Scholar] [CrossRef]
- Diyana Jamaluddin, N.; Ibrahim, N.; Yuziana Mohd Yusof, N.; Ta Goh, C.; Ling Tan, L. Optical reflectometric measurement of SARS-CoV-2 (COVID-19) RNA based on cationic cysteamine-capped gold nanoparticles. Opt. Laser Technol. 2023, 157, 108763. [Google Scholar] [CrossRef]
- Luo, X.; Yue, W.; Zhang, S.; Liu, H.; Chen, Z.; Qiao, L.; Wu, C.; Li, P.; He, Y. SARS-CoV-2 proteins monitored by long-range surface plasmon field-enhanced Raman scattering with hybrid bowtie nanoperture arrays and nanocavities. Lab Chip 2022, 23, 388–399. [Google Scholar] [CrossRef]
- Luo, Y.; Zhou, M.; Fan, C.; Song, Y.; Wang, L.; Xu, T.; Zhang, X. Active enrichment of nanoparticles for ultra-trace point-of-care COVID-19 detection. Anal. Chem. 2023, 95, 5316–5322. [Google Scholar] [CrossRef] [PubMed]
- Behrouzi, K.; Lin, L. Gold nanoparticle based plasmonic sensing for the detection of SARS-CoV-2 nucleocapsid proteins. Biosens. Biolectron. 2022, 195, 113669. [Google Scholar] [CrossRef]
- Oliveira, B.B.; Ferreira, D.; Fernandes, A.R.; Baptista, P.V. Engineering gold nanoparticles for molecular diagnosis and biosensing. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15, e1836. [Google Scholar]
- Zeng, J.; Duarte, P.A.; Ma, Y.; Savchenko, O.; Shoute, L.; Khaniani, Y.; Babiuk, S.; Zhuo, R.; Abdelrasoul, G.N.; Charlton, C.; et al. An impedimetric biosensor for COVID-19 serology test and modification of sensor performance via diaelectrophoresis force. Biosens. Bioelectron. 2022, 213, 114476. [Google Scholar] [CrossRef] [PubMed]
- Khaniani, Y.; Ma, Y.; Ghadiri, M.; Zeng, J.; Wishart, D.; Babiuk, S.; Charlton, C.; Kanji, J.N.; Chen, J. A gold nanoparticle-protein G electrochemical affinity biosensor for the detection of SARS-CoV-2 antibodies: A surface modification approach. Sci. Rep. 2022, 12, 12850. [Google Scholar] [CrossRef] [PubMed]
- Brazaca, L.C.; Imamura, A.H.; Gomes, N.O.; Almeida, M.B.; Scheidt, D.T.; Raymundo-Pereira, P.A.; Oliveira, O.N., Jr.; Janegitz, B.C.; Machado, S.A.S.; Carriho, E. Electrochemical immunosensors using electrodeposited gold nanostructures for detecting the S proteins from SARS-CoV and SARS-CoV-2. Anal. Bioanal. Chem. 2022, 414, 5507–5517. [Google Scholar] [CrossRef]
- Zeng, R.; Qiu, M.; Wan, Q.; Huang, Z.; Liu, X.; Tang, D.; Lnopp, D. Smartphone-based electrochemical immunoassay for point-of-care detection of SARS-CoV-2 nucleocapsid protein. Anal. Chem. 2022, 94, 15155–15161. [Google Scholar] [CrossRef]
- Mandal, D.; Indaleeb, M.M.; Younan, A.; Banerjee, S. Piezoelectric point-of-care biosensor for the detection of SARS-CoV-2 (COVID-19) antibodies. Sens. Bio-Sens. Res. 2022, 37, 100510. [Google Scholar] [CrossRef]
- Peng, Y.-C.; Cheng, C.-H.; Yatsud, H.; Liu, S.-H.; Liu, S.-J.; Kogai, T.; Kuo, C.-Y.; Wang, R.Y.L. Novel rapid test to detect anti-SARS-CoV-2 N protein IgG based on shear horizontal surface acoustic wave (SH-SAW). Diagnostics 2021, 11, 1838. [Google Scholar] [CrossRef]
- Iravani, S. Nano- and biosensors for the detection of SARS-CoV-2: Challenges and opportunities. Adv. Mater. 2020, 1, 3092–3103. [Google Scholar] [CrossRef]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef] [PubMed]
- Tessaro, L.; Aquino, A.; Panzenhagen, P.; Ochioni, A.C.; Mutz, Y.S.; Raymundo-Pereira, P.A.; Vieira, I.R.S.; Belem, N.K.R.; Conte-Junior, C.A. Development and application of an SPR nanobiosensor based on AuNPs for the detection of SARS-CoV-2 on food surfaces. Biosensors 2022, 12, 1101. [Google Scholar] [CrossRef] [PubMed]
- Paladhi, A.G.; Manohar, M.; Pal, K.; Vallinayagam, S.; Packirisamy, A.S.B.; Bashreer, V.A.; Sai Nandhini, R.; Ukhurebor, K. Novel electrochemical biosensor key significance of smart intelligence IoMT & IoHT) of COVID-19 virus control management. Process Biochem. 2022, 122, 105–109. [Google Scholar] [PubMed]
- Lee, A.S.; Kim, S.M.; Kim, K.R.; Park, C.; Lee, D.-G.; Heo, H.R.; Cha, H.J.; Kim, C.S. A colorimetric lateral flow immunoassay based on oriented antibody immobilization for sensitive detection of SARS-CoV-2. Sens. Actuators B Chem. 2023, 379, 133245. [Google Scholar] [CrossRef] [PubMed]
- Fox, T.; Geppert, J.; Dinnes, J.; Scandrett, K.; Bigio, J.; Sulis, G.; Hettiarachchi, D.; Mathangasinghe, Y.; Weeratunga, P.; Wickramasinghe, D.; et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2022, 11, CD013652. [Google Scholar]
- Shaw, A.M.; Hyde, C.; Merrick, B.; James-Pemberton, P.; Squires, B.K.; Olkhov, R.V.; Batra, R.; Patel, A.; Bisnauthsing, K.; Nebbia, G.; et al. Real-world evaluation of a novel technology for quantitative simultaneous antibody detection against multiple SARS-CoV-2 antigens in a cohort of patients presenting with COVID-19 syndrome. Analyst 2020, 145, 5638–5646. [Google Scholar] [CrossRef] [PubMed]
- Guglielmi, G. Rapid coronavirus tests: A guide for the perplexed. Nature 2021, 590, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Koller, G.; Morrel, A.P.; Galao, R.P.; Pickering, S.; MacMahon, E.; Johnson, J.; Ignatyev, K.; Neil, S.J.D.; Elsharkawy, S.; Fleck, R.; et al. More than the eye can see: Shedding new light on SARS-CoV-2 lateral flow device-based immunoassays. ACS Appl. Mater. Interfaces 2021, 13, 25694–25700. [Google Scholar] [CrossRef]
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Bio/Technology 1993, 11, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Nyaruaba, R.; Mwaliko, C.; Dobnik, D.; Neuzil, P.; Amoth, P.; Mwau, M.; Yu, J.; Yang, H.; Wei, H. Digital PCR applications in the SARS-CoV-2/COVID-19 era: A roadmap for future outbreaks. Clin. Microbiol. Rev. 2022, 35, e00168-21. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.; Sharma, P.; Hassan, M.R.; Singh, S.; Thakur, D.; Narang, J. Gold nanomaterials—the golden approach from synthesis to applications. Mater. Sci. Energy Technol. 2022, 5, 375–390. [Google Scholar] [CrossRef]
- Fu, X.; Cai, J.; Zhang, X.; Li, W.-D.; Ge, H.; Hu, Y. Top-down fabrication of shape-controlled, monodisperse nanoparticles for biomedical applications. Adv. Drug Deliv. Rev. 2018, 132, 169–187. [Google Scholar] [CrossRef]
- Aryal, S.; Park, H.; Leary, J.F.; Key, J. Top-down fabrication-based nano/microparticles for molecular imaging and drug delivery. Int. J. Nanomed. 2019, 14, 6631–6644. [Google Scholar] [CrossRef]
- Kumar, S.; Bhushan, P. Bhattacharya, Fabrication of nanostructures with bottom-up approach and their utility in diagnosis, therapeutics, and others. In Environmental, Chemical and Medical Sensors; Energy, Environment, and Sustainability; Springer: Singapore, 2018; pp. 167–198. [Google Scholar]
- Ji, X.; Song, X.; Li, J.; Bai, Y.; Yang, W.; Peng, X. Size control of gold nanocrystals in citrate reduction: The third role of citrate. J. Am. Chem. Soc. 2007, 129, 13939–13948. [Google Scholar] [CrossRef]
- Borse, V.; Konwar, A.N. Synthesis and characterization of gold nanoparticles as a sensing tool for the lateral flow immunoassay development. Sens. Inter. 2020, 1, 100051. [Google Scholar] [CrossRef]
- Feng, M.; Chen, J.; Xun, J.; Dai, R.; Zhao, W.; Lu, H.; Xu, J.; Chen, L.; Sui, G. Development of a sensitive immunochromatographic method using lanthanide fluorescent microsphere for rapid serodiagnosis of COVID-19. ACS Sens. 2020, 5, 2331–2337. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.Y.; Wu, Y.H.; Xiong, Q.R.; Xu, H.Y.; Xiong, Y.H.; Liu, K.; Jin, Y.; Lai, W.H. Advantages of fluorescent microspheres compared with colloidal gold as a label in immunochromatographic lateral flow assays. Biosens. Bioelectron. 2014, 54, 262–265. [Google Scholar] [CrossRef]
- Thangavelu, R.M.; Kadirvel, N.; Balasubramaniam, P.; Viswanathan, R. Ultrasensitive nano-gold labelled, duplex lateral flow immunochromatographic assay for early detection of sugarcane mosaic viruses. Sci. Rep. 2022, 12, 4144. [Google Scholar] [CrossRef] [PubMed]
- Prabhabar, A.; Bansal, I.; Jaiswar, A.; Roy, N.; Verma, D. A simple cost-effective microfluidic platform for rapid synthesis of diverse metal nanoparticles: A novel approach towards fighting SARS-CoV-2. Mater. Today Proc. 2023, 80, 1852–1857. [Google Scholar]
- Marques, A.C.; Pinheiro, T.; Morais, M.; Martins, C.; Andrade, A.F.; Martins, R.; Sales, M.G.F.; Fortunto, E. Bottom-up microwave-assisted seed-mediated syhthesis of gold nanopartciles onto nanocellulose to boos stability and high performane for SERS applications. Appl. Surf. Sci. 2021, 561, 150060. [Google Scholar] [CrossRef]
- Vinnacombe-Wilson, G.A.; Conti, Y.; Jonas, S.J.; Weiss, P.S.; Mihi, A.; Scarabelli, L. Surface lattice plasmon resonaces by direct in situ substrate growth of gold nanoparticles in ordered arrays. Adv. Matter. 2022, 34, 2205330. [Google Scholar] [CrossRef]
- Minopoli, A.; Scardapane, E.; Acunzo, A.; Della Ventura, B.; Veotta, R. Antonio Minpopoli Analysis of the optical response of a SARS-CoV-2-directed colorimetric immunosensor. AIP Adv. 2021, 11, 065319. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 antibodiesreceptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Vermma, N.; Badhe, Y.; Gupta, R.; Maparu, A.K.; Rai, B. Peptide mediated colorimetric detection of SARS-CoV-2 using gold nanoparticles: A molecular dynamics simulation study. J. Mol. Model. 2022, 28, 202. [Google Scholar] [CrossRef]
- Sun, D.; Sun, M.; Zhang, J.; Lin, X.; Zhang, Y.; Lin, F.; Zhang, P.; Yang, C.; Song, J. Computational tools for aptamer identification and optimization. TrAC Trends Anal. Chem. 2022, 157, 116767. [Google Scholar] [CrossRef]
- Saad, Y.; Gazzah, M.H.; Mougin, K.; Selmi, M.; Belmabrouk, H. Sensitive detection of SARS-CoV-2 using a novel plasmonic fiber optic biosensor design. Plasmonics 2022, 17, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Baidya, S.; Hassan, A.M. SARS-CoV-2 detection using colorimetric plasmonic sensors: A proof-of-concept computational study. IEEE Trans. Nanobiosci. 2023, 22, 71–77. [Google Scholar] [CrossRef]
- Baidya, S.; Hassan, A.M. A computational study of COVID-19 detection using colorimetric plasmonic sensors. In Proceedings of the 2021 IEEE International Symposium on Antennas and Propagation and North American Radio Science Meeting, APS/URSI 2021—Proceedings, Singapore, 4–10 December 2021; pp. 1731–1732. [Google Scholar]
- Valagiannopoulos, C.; Sihvola, A. Maximal interaction of electromagnetic radiation with corona virions. Phys. Rev. B 2021, 103, 014114. [Google Scholar] [CrossRef]
- Mehranfar, A.; Izadyar, M. Theoretical design of functionalized gold nanoparticles as antiviral agents against severe acute respiratoty syndrome coronavirus 2 (SARS-CoV-2). J. Phys. Chem. Lett. 2020, 11, 10284–10289. [Google Scholar] [CrossRef]
- De, P.; Kumar, V.; Kar, S.; Roy, K.; Leszczynski, J. Repurposing FDA approved drugs as possible anti-SARS-CoV-2 medications using ligand-based computational approaches: Sum of ranking difference-based model selection. Struc. Chem. 2022, 33, 1741–1753. [Google Scholar] [CrossRef]
- Kaliyaraj Selva Kumar, A.; Zhang, Y.; Li, D.; Compton, R.G. A mini-review: How reliable is the drop casting technique? Electrochem. Commun. 2020, 121, 106867. [Google Scholar] [CrossRef]
- Farsaeivahid, N.; Grenier, C.; Nazarian, S.; Wang, M.L. A rapid label-free disposable electrochemical salivary point-of-care sensor for SARS-CoV-2 detection and quatification. Sensors 2023, 23, 433. [Google Scholar] [CrossRef] [PubMed]
- Tortolini, C.; Gigli, V.; Angeloni, A.; Galantini, L.; Tasca, F.; Antiochia, R. Disposable voltammetric immunosensor for D-dimer detection as early biomarker of thromboembolic disease and of COVID-19 prognosis. Biosensors 2023, 13, 43. [Google Scholar] [CrossRef]
- Nicoliche, C.Y.N.; Pascon, A.M.; Bezerra, I.R.S.; de Castro, A.C.H.; Martos, G.R.; Bettini, J.; Alves, A.W.; Santhiago, M.; Lima, R.S. In situ nanocoating on porous pyrolyzed paper enables antibiofouling and sensitive electrochemical analyses in biological fluids. ACS Appl. Mater. 2022, 14, 2522–2533. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, A.; Gao, Y.; Patibandla, S.; Mitra, D.; McCandless, M.G.; Fassero, L.A.; Gates, K.; Tandon, R.; Chandra Ray, P. The rapid diagnosis and effective inhibition of coronavirus using spike antibody attached gold nanoparticles. Nanoscale Adv. 2021, 3, 1588–1596. [Google Scholar] [CrossRef]
- Materon, E.M.; Gomez, F.R.; Almeidia, M.B.; Azevedo, R.B.; Goncalves, D. Colorimetric detection of SARS-CoV-2 using plasmonic biosensors and smartphones. ACS Appl. Mater. Interfaces 2022, 14, 44538–54527. [Google Scholar] [CrossRef]
- Ahmadivand, A.; Gerislioglu, B.; Ramezani, Z.; Kaushik, A.; Manickam, P.; Ghoreishi, S.A. Functionalized terahertz plasmonic metasensors: Femtomolar-level detection of SARS-CoV-2 spike proteins. Biosens. Bioelectron. 2021, 177, 112971. [Google Scholar] [CrossRef]
- Guo, A.; Pei, F.; Hu, W.; Xia, M.; Mu, X.; Tong, Z.; Wang, F.; Liu, B. CdTe QDs-sensitized TiO2 nanocomposite for magnetic-assisted photoelectrochemical immunoassay of SARS-CoV-2 nucleocapsid protein. Bioelectrochemistry 2023, 150, 108358. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, M.; Sobhanie, E.; Salehnia, F.; Xu, G.; Rabbani, H.; Sheikholeslami, M.N.; Firoozbakhtian, A.; Sadeghi, N.; Farajollah, M.H.; Reza Ganjali, M.; et al. Development of sandwich electrochemi-luminescence immunosensor for COVID-19 diagnosis by SARS-CoV-2 spike protein detection based on Au@BSA-luminol nanocomposites. Bioelectrochemistry 2022, 147, 108161. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Jiang, T.; Ma, Y.; TO, D.; Wang, S.; Chen, J. A portable, low-cost and high-throughput electrochemical impedance spectroscopy device for point-of-care biomarker detection. Biosens. Bioelectron. X 2023, 13, 100301. [Google Scholar] [CrossRef]
- Khlebtsov, B.N.; Tumskys, R.S.; Burov, A.M.; Pylaev, T.E.; Khlebtsov, N.G. Quantifying the numbers of gold nanoparticles in the test zone of lateral flow immunoassay strips. ACS Appl. Nano Mater. 2019, 2, 5020–5028. [Google Scholar] [CrossRef]
- Foncea, P.; Mondschein, S.; Olivares, M. Replacing quarantine of COVID-19 contacts with periodic testing is also effective in mitigating the risk of transmission. Sci. Rep. 2022, 12, 3620. [Google Scholar] [CrossRef]
- Singh, S.; Singha, T.; Maheshwari, R.; Yadav, P.; Kumar, A.; Bhoj, H.; Sharma, B.; Verma, A.; Gupta, A.; Singh, H.; et al. Development of colloidal gold nanoparticle based lateral-flow assay for rapid detection of SARS-CoV-2 showing enhanced sensitivity and specificity. J. Appl. Biol. Biotechnol. 2023, 11, 253–258. [Google Scholar] [CrossRef]
- Prakashan, D.; Shrikrishna, N.S.; Byakodi, M.; Nagamani, K.; Gandhi, S. Gold nanoparticle conjugate-based lateral flow immunoassay (LFIA) for rapid detection of RBD antigen of SARS-CoV-2 in clinical samples using a smartphone-based application. J. Med. Virol. 2023, 95, e28416. [Google Scholar] [CrossRef]
- Srithong, P.; Chaiyo, S.; Pasomsub, E.; Rengpipat, S.; Chailapakul, O.; Praphairaksit, N. A novel delayed lateral flow immunoassay for enhanced detection of SARS-CoV-2 spike NC. Microchim. Acta 2022, 189, 386. [Google Scholar] [CrossRef]
- Higgins, V.; Fabros, A.; Wang, X.Y.; Bhandari, M.; Daghfai, D.J.; Kulasingam, V. Anti-SARS-CoV-2 IgM improves clinical sensitivity early in disease course. Clin. Biochem. 2021, 90, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nicol, T.; Lefeuvre, C.; Serri, O.; Pivert, A.; Joubaud, F.; Dubée, V.; Kouatchet, A.; Ducancelle, A.; Lunel-Fabiani, F.; Le Guillou-Guilemette, H. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech). J. Clin. Virol. 2020, 129, 104511. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wen, T.; Shi, F.-J.; Zeng, X.-Y.; Jiao, Y.-J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticles based lateral-flow assay. ACS Omega 2020, 5, 12550–12556. [Google Scholar] [CrossRef] [PubMed]
- Salem, R.; Elshamy, A.M.; Kamel, N.; Younes, S.; Marie, O.M.; Waly, F.R.; El-Kholy, A.A.; Elmenofy, W. A gold nanoparticles-based lateral flow assay utilizing baculovirus expressed recombinant nucelocapsid and receptor binding domain proteins for serodetection of IgG and IgM against SARS-CoV-2. Biotechnol. Lett. 2022, 4, 1507–1517. [Google Scholar] [CrossRef]
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef]
- Ngo, H.T.; Van Tran, S.; Nguyen, H.D.; Truong, P.Q. Humoral immune response in COVID-19 patients and novel design of lateral flow assay strip for simultaneous rapid detection of IgA/IgM/IgG antibodies against the SARS-CoV-2 virus. J. Appl. Biol. Biotechnol. 2023, 11, 102–113. [Google Scholar] [CrossRef]
- Mehdipour, A.R.; Hummer, G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl. Acad. Sci. USA 2021, 118, e2100425118. [Google Scholar] [CrossRef]
- Baker, A.N.; Richards, S.-J.; Pandey, S.; Guy, C.S.; Ahmad, A.; Hasan, M.; Biggs, C.I.; Georgiou, P.G.; Zwetsloot, A.J.; Straube, A.; et al. Glycan-based flow-through device for the detection of SARS-CoV-2. ACS Sens. 2021, 6, 3696–3705. [Google Scholar] [CrossRef]
- Kim, S.H.; Kearns, F.L.; Rosenfeld, M.A.; Casalino, L.; Papanikolas, M.J.; Simmerling, C.; Amaro, R.E.; Freeman, R. GlycoGrip: Cell surface-inspired universal sensor for betacoronaviruses. ACS Cent. Sci. 2022, 8, 22–42. [Google Scholar] [CrossRef]
- Park, S.B.; Shin, J.H. Pressed lateral flow assay strips for flow delay-induced signal enhancement in lateral flow assay strips. Biochip J. 2022, 16, 480–489. [Google Scholar] [CrossRef]
- Chen, S.; Meng, L.; Wang, L.; Huang, X.; Ali, S.; Chen, X.; Yu, M.; Yi, M.; Li, L.; Chen, X.; et al. SERS-based lateral flow immunoassay for sensitive and simultaneous detection of anti-SARS-CoV-2 IgM and IgG antibodies by using gap-enhanced Raman nanotags. Sens. Actuators B Chem. 2021, 348, 130706. [Google Scholar] [CrossRef]
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators B Chem. 2021, 329, 129196. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Liang, P.; Ren, J.; Zhu, J.; Yang, X.; Bian, H.; Li, J.; Cui, X.; Fu, C.; Xing, J.; et al. Gold-silver alloy hollow nanoshells-based lateral flow immunoassay for colorimetric, photothermal and SERS tri-mode detection of SARS-CoV-2 neutralizing antibody. Anal. Chim. Acta 2023, 1255, 341102. [Google Scholar] [CrossRef]
- Dighe, K.; Moitra, P.; Alafeef, M.; Gunaseelan, N.; Pan, D. A rapid RNA extraction-free lateral flow assay for molecular point-of-care detection of SARS-CoV-2 augmented by chemical probes. Biosens. Bioelectron. 2022, 200, 113900. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, P.; Zhao, T.; Guo, G.; Zhu, J.; Wen, C.; Zeng, J. Colorimetric and Raman dual-mode lateral flow immunoassay detection of SARS-CoV-2 N protein antibody based on Ag nanoparticles with ultrathin Au shell assembled onto Fe3O4 nanoparticles. Anal. Bioanal. Chem. 2023, 415, 545–554. [Google Scholar] [CrossRef]
- Tang, R.; Alam, N.; Li, M.; Xie, M.; Ni, Y. Dissolvable sugar barriers to enhance the sensitivity of nitrocellulose lateral flow assay for COVID-19 nucleic acid. Carbohydr. Polym. 2021, 268, 118259. [Google Scholar] [CrossRef] [PubMed]
- Çam Derin, D.; Gültekin, E.; Içen Taskin, I.; Yakupogullan, Y. Development of nucleic acid based lateral flow assays for SARS-CoV-2 detection. J. Biosci. Bioeng. 2023, 135, 87–92. [Google Scholar] [CrossRef]
- Agarwal, S.; Warmt, C.; Henkel, J.; Schrick, L.; Nitschne, A.; Bier, F.F. Lateral flow-based nucleic acid detection of SARS-CoV-2 using enzymatic incorporation of biotin-labeled dUTP for POCT use. Anal. Bioanal. Chem. 2022, 414, 3177–3186. [Google Scholar] [CrossRef]
- Lee, H.N.; Lee, J.; Kang, Y.K.; Lee, J.H.; Yang, S.; Chung, H.J. A lateral flow assay for nucleic acid detection based on rolling circle amplification using capture ligand-modified oligonucleotides. Biochip J. 2022, 16, 441–450. [Google Scholar] [CrossRef]
- Fabre, M.; Ruiz-Martinez, S.; Monserrat Cantera, M.E.; Cortizo Garrido, A.; Beunza Fabra, Z.; Peran, M.; Benito, R.; Mateo, P.; Paules, C.; Oros, D. SARS-CoV-2 immunochromatographic IgM/IgG rapid test in pregnancy: A false friend? Anal. Clin. Biochem. 2021, 58, 149–152. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.-H. Current advances in paper-based biosensor technologies for rapid COVID-19 diagnosis. Biochip J. 2022, 16, 376–396. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Trinh, H.D.; Lee, Y.; Kang, T.; Chen, L.; Yoon, S.; Choo, J. SERS-ELISA using silica-encapsulated Au core-satellite nanotags for sensitive detection of SARS-CoV-2. Sens. Actuators B Chem. 2023, 382, 133521. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Suzuki, H.; Maekawa, M.; Hariyama, T. Combination of the Nanosuit method and gold/platinum particle-based lateral flow assay for quantitative and highly sensitive diagnosis using a desktop scanning electron microscope. J. Pharm. Biomed. Anal. 2021, 196, 113924. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Qin, R.; Jiang, M.; Yang, Z.; Wen, W.; Li, J. Clinical applications of detecting IgG, IgM or IgA antibody for the diagnosis of COVID-19: A meta-analysis and systematic review. Int. J. Infect. Dis. 2021, 104, 415–422. [Google Scholar] [CrossRef]
- Nuccetelli, M.; Pieri, M.; Gisone, F.; Sarubbi, S.; Ciotti, M.; Andreoni, M.; Bernardini, S. Evaluation of a new simultaneous anti-SARS-CoV-2 IgA, IgM and IgG screening automated assay based on native inactivated virus. Int. Immunopharmacol. 2021, 92, 107330. [Google Scholar] [CrossRef]
- Malla, B.; Thakali, O.; Shrestha, S.; Segawa, T.; Kitajima, M.; Haramoto, E. Application of a high-throughput quantitative PCR system for simultaneous monitoring of SARS-CoV-2 variants and other viruses in wastewater. Sci. Total Environ. 2022, 853, 158659. [Google Scholar] [CrossRef]
- Sena-Torralba, A.; Alvarez-Diduk, R.; Parolo, C.; Piper, A.; Merkoci, A. Toward next generation lateral flow assays: Integration of nanomaterials. Chem. Rev. 2022, 122, 14881–14910. [Google Scholar] [CrossRef]
- Kamat, S.; Kumari, M.; Jayabaskaran, C. Nano-engineered tools in the diagnosis, therapeutics, prevention, and mitigation of SARS-CoV-2. J. Control Release 2021, 338, 813–836. [Google Scholar] [CrossRef]
- Tyagi, P.K.; Tyagi, S.; Mishra, M.; Dashora, K. Prevention, diagnosis and treatment of COVID-19: A nanotechnological perspective. Curr. Nanosci. 2021, 17, 418–422. [Google Scholar] [CrossRef]
- Ramakrishnan, S.G.; Robert, B.; Salim, A.; Ananthan, P.; Sivaramakrishnan, M.; Subramaniam, S.; Natesan, S.; Suresh, R.; Rajeshkumar, G.; Maran, J.P.; et al. Nanotechnology based solutions to combat zootic viruses with special attention to SARS, MERS, and COVID-19: Detection, protection and medication. Microb. Pathog. 2021, 159, 1051133. [Google Scholar] [CrossRef]
- Ionescu, R.E.; Marks, R.S.; Gheber, L.A. Nanolithography using protease etching of protein surfaces. Nano Lett. 2003, 3, 1639–1642. [Google Scholar] [CrossRef]
- Ionescu, R.E.; Marks, R.S.; Gheber, L.A. Manufacturing of nanochannels with controlled dimensions using protease nanolithography. Nano Lett. 2005, 5, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Taha, B.A.; Al Mashhadany, Y.; Al-Jubouri, Q.; Rashid, A.R.B.A.; Luo, Y.; Chen, Z.; Rustagi, S.; Chaudhary, V.; Arsad, N. Next-generation nanophotonic-enabled biosensors for intelligent diagnosis of SARS-CoV-2 variants. Sci. Total Environ. 2023, 880, 163333. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Wang, B.; Huang, H.; Yan, K.; Li, W.; Wang, S.; Liu, F. Portable high-throughput multimodal immunoassay platform for rapid on-site COVID-19 diagnosis. Anal. Chim. Acta 2023, 1238, 340634. [Google Scholar] [CrossRef]
- Fatima, M.; Sadaf Zaidi, N.U.S.; Amraiz, D.; Afzal, F. In vitro antiviral activity of Cinnamomum cassia and its nanoparticles against H7N3 influenza a virus. J. Microbiol. Biotechnol. 2016, 26, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, S.; Rabiee, N.; Fatahi, Y.; Hooshmad, S.E.; Bagherzadeh, M.; Rabiee, M.; Jajarmi, V.; Dinarvand, R.; Habibzadeh, S.; Saeb, M.R.; et al. Green chemistry and coronavirus. Sustain. Chem. Pharm. 2021, 21, 100415. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ionescu, R.E. Updates on the Biofunctionalization of Gold Nanoparticles for the Rapid and Sensitive Multiplatform Diagnosis of SARS-CoV-2 Virus and Its Proteins: From Computational Models to Validation in Human Samples. Int. J. Mol. Sci. 2023, 24, 9249. https://doi.org/10.3390/ijms24119249
Ionescu RE. Updates on the Biofunctionalization of Gold Nanoparticles for the Rapid and Sensitive Multiplatform Diagnosis of SARS-CoV-2 Virus and Its Proteins: From Computational Models to Validation in Human Samples. International Journal of Molecular Sciences. 2023; 24(11):9249. https://doi.org/10.3390/ijms24119249
Chicago/Turabian StyleIonescu, Rodica Elena. 2023. "Updates on the Biofunctionalization of Gold Nanoparticles for the Rapid and Sensitive Multiplatform Diagnosis of SARS-CoV-2 Virus and Its Proteins: From Computational Models to Validation in Human Samples" International Journal of Molecular Sciences 24, no. 11: 9249. https://doi.org/10.3390/ijms24119249
APA StyleIonescu, R. E. (2023). Updates on the Biofunctionalization of Gold Nanoparticles for the Rapid and Sensitive Multiplatform Diagnosis of SARS-CoV-2 Virus and Its Proteins: From Computational Models to Validation in Human Samples. International Journal of Molecular Sciences, 24(11), 9249. https://doi.org/10.3390/ijms24119249








