Antibacterial Activity and Mechanisms of TroHepc2-22, a Derived Peptide of Hepcidin2 from Golden Pompano (Trachinotus ovatus)
Abstract
1. Introduction
2. Results
2.1. Peptides Synthesis and Structure Analysis
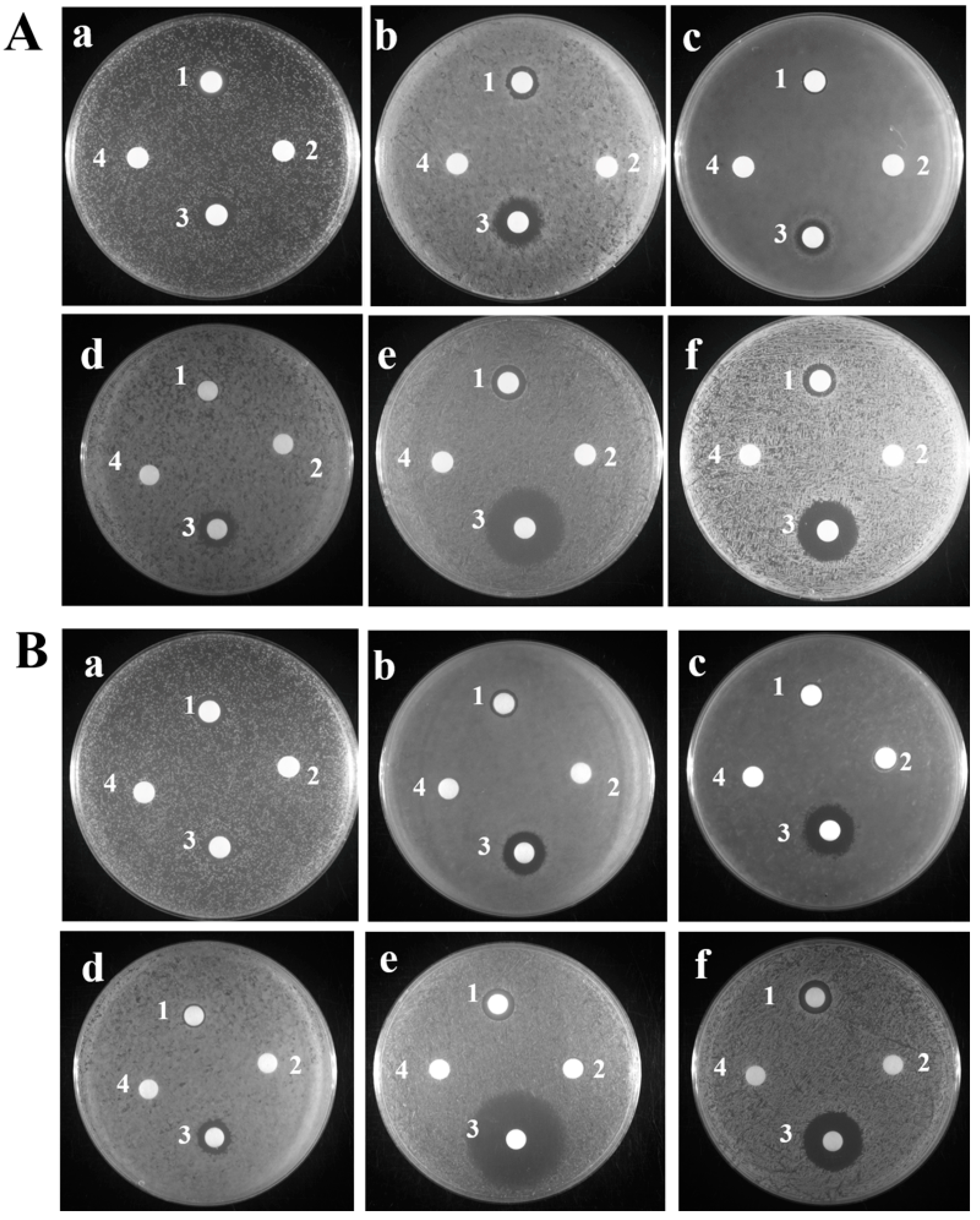
2.2. Antibacterial Activity of TroHepc2-22
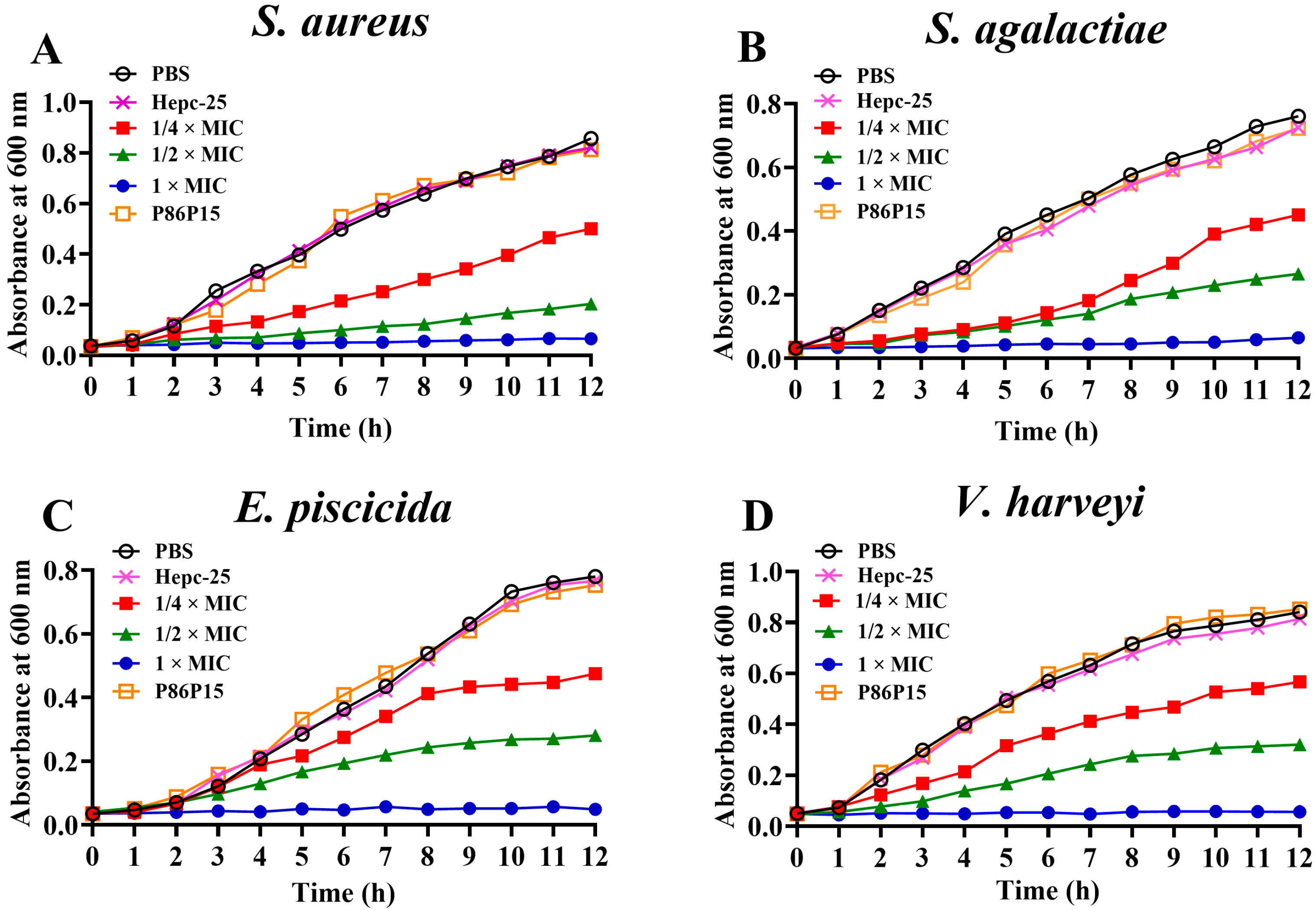
2.3. Effects of TroHepc2-22 on Bacterial Growth
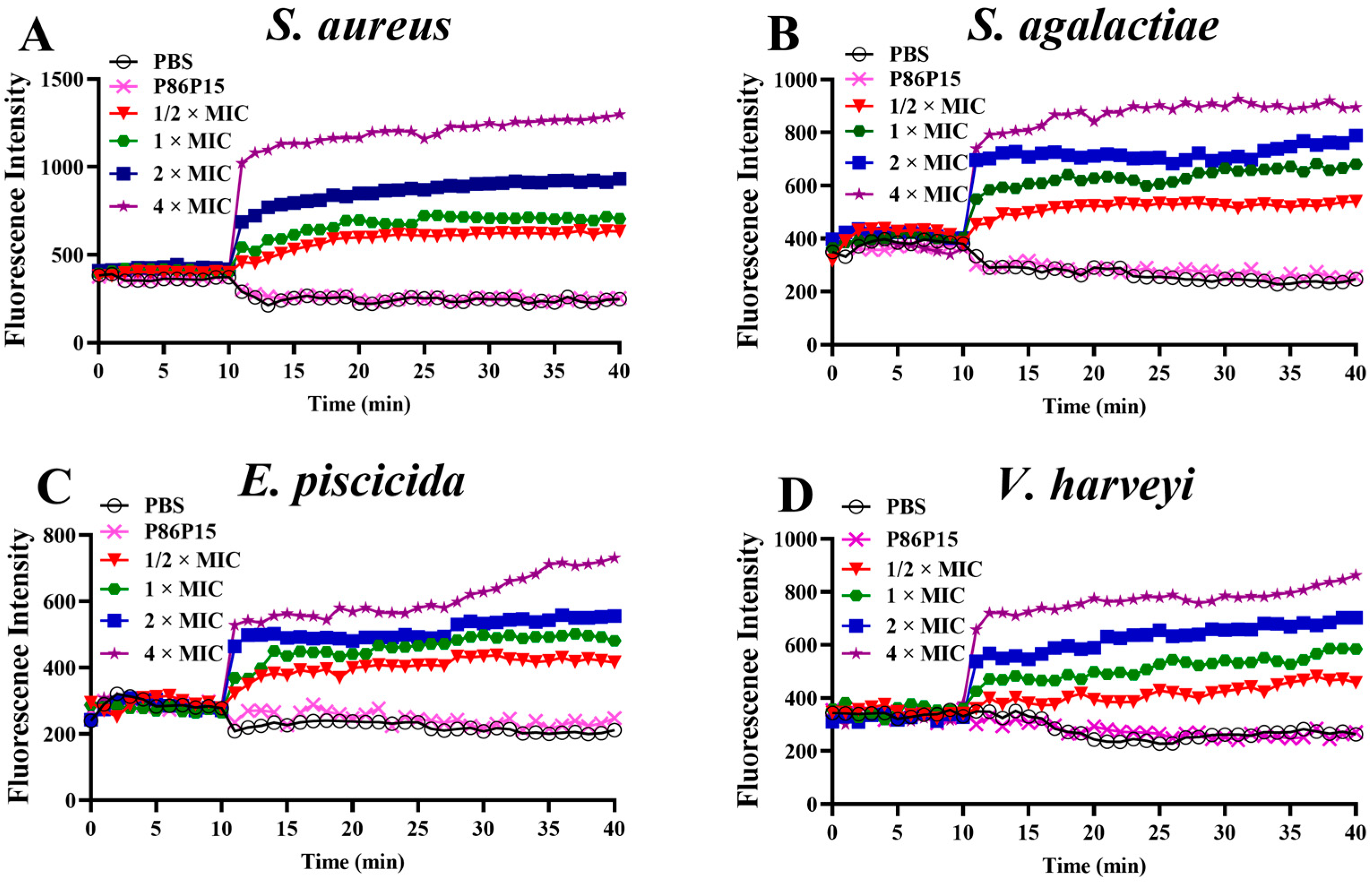
2.4. Bacterial Membrane Depolarization Induced by TroHepc2-22
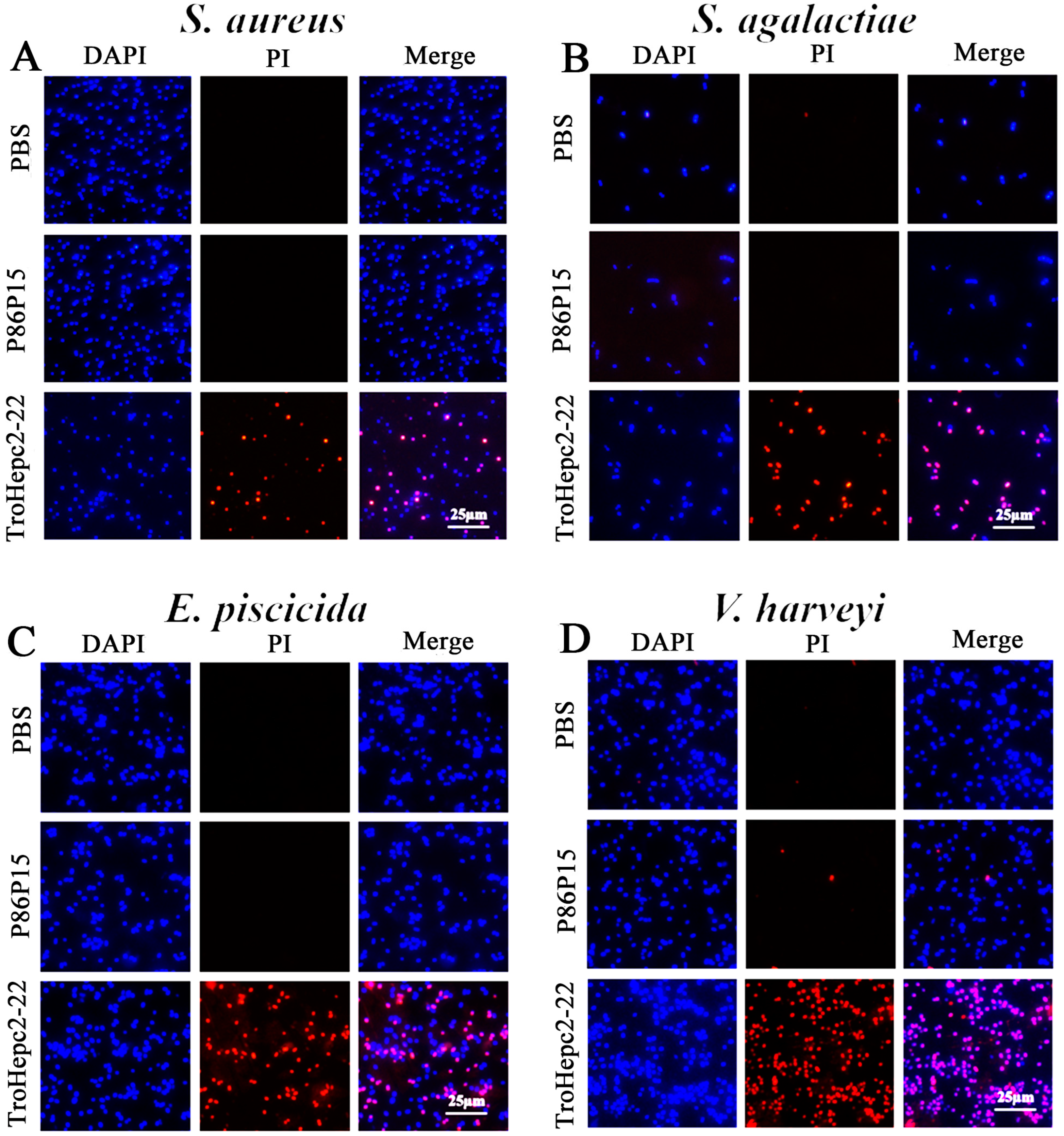
2.5. PI Staining Analysis
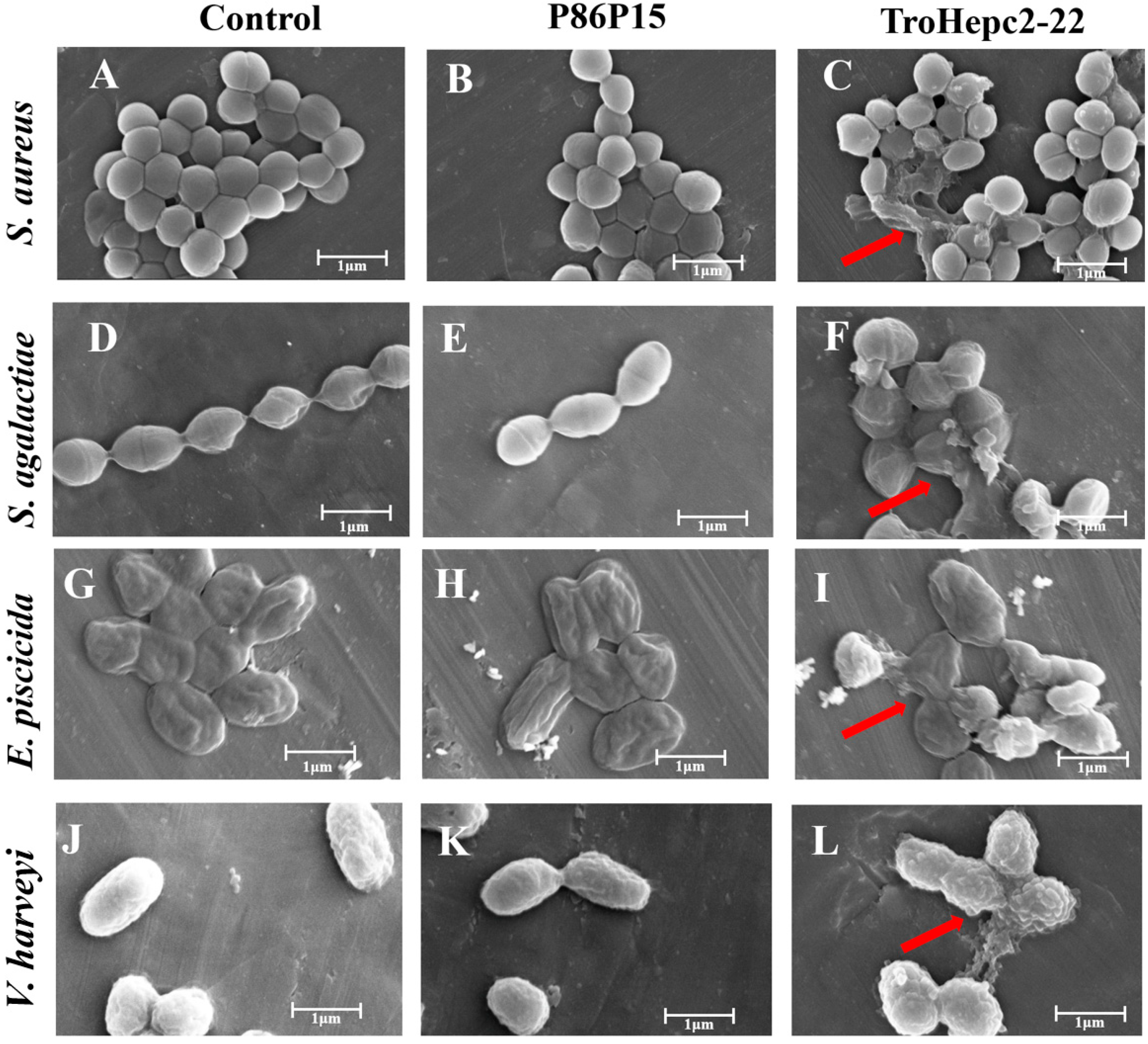
2.6. Scanning Electron Microscopy (SEM) Observation
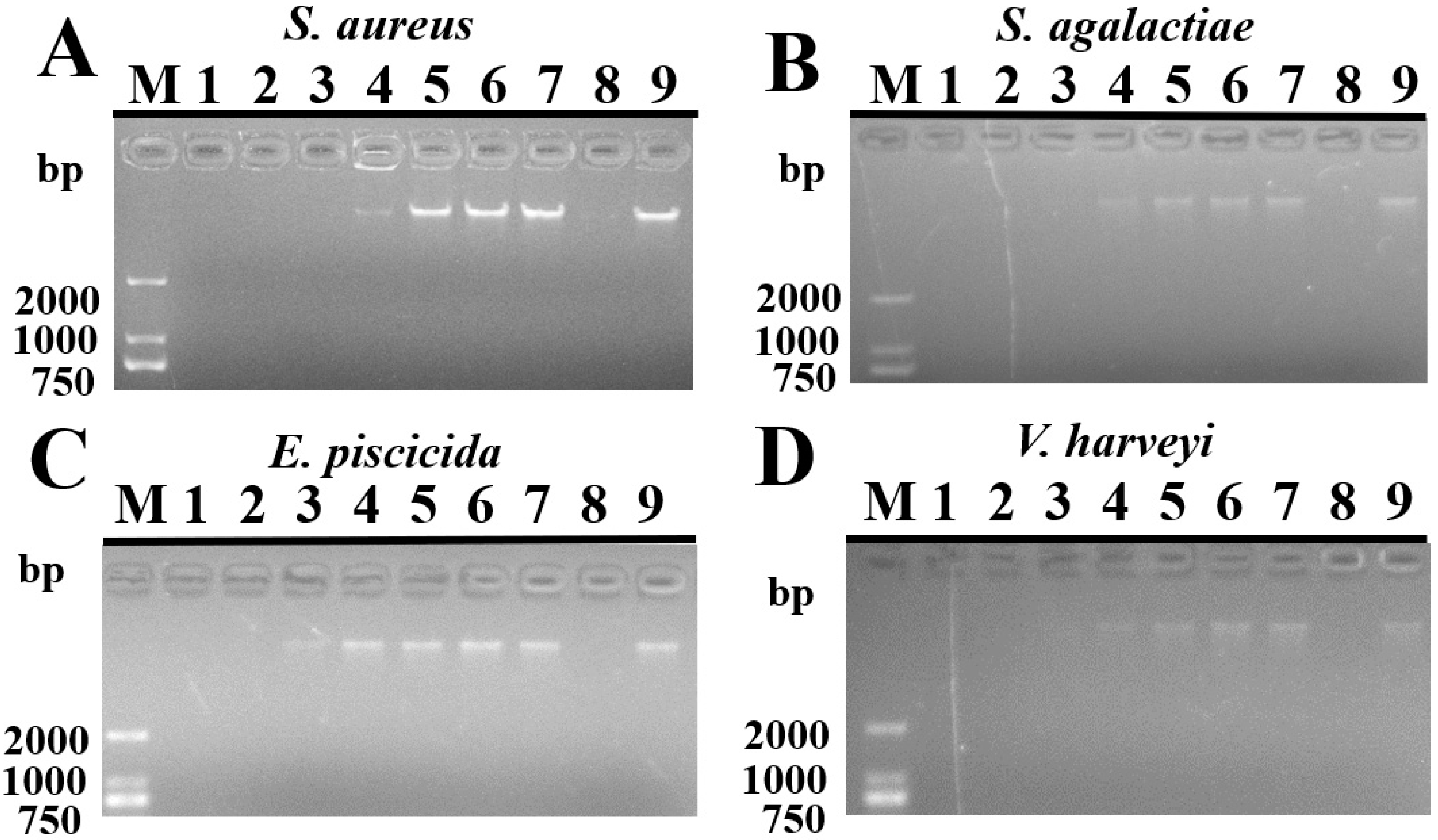
2.7. Binding Activity of TroHepc2-22 on Bacterial Genomic DNA
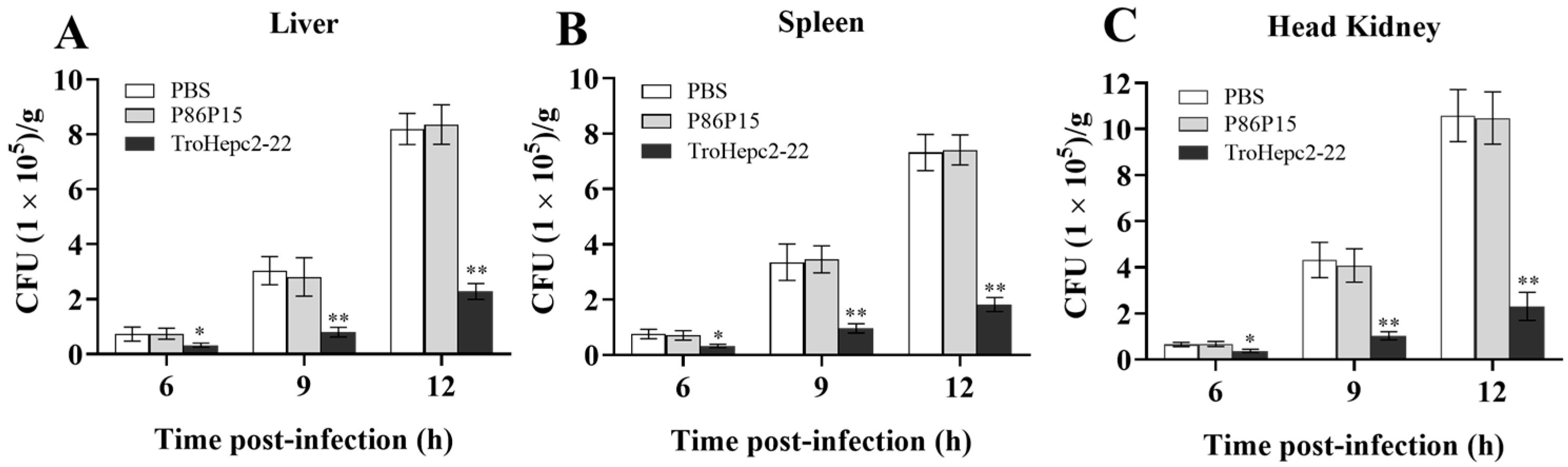
2.8. Effects of TroHepc2-22 on Bacterial Infection
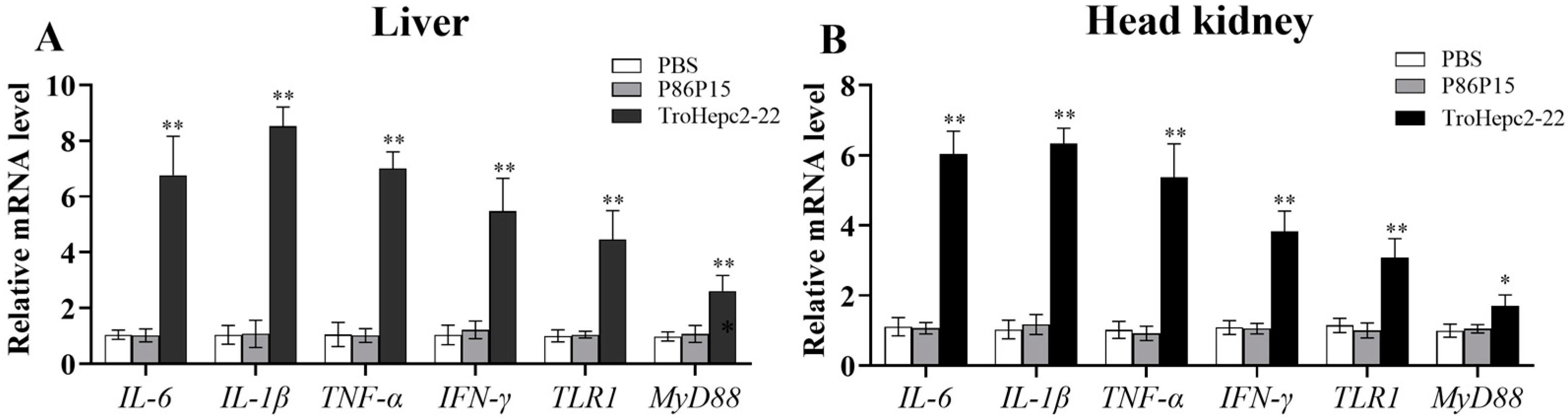
2.9. Trohepc2-22 Regulates Immune-Related Gene Expressions against V. harveyi Infection
3. Discussion
4. Materials and Methods
4.1. Fish, Bacterial Strains and Culture Conditions
4.2. Peptides Synthesis and Structure Analysis
4.3. Antibacterial Activity of TroHepc2-22
4.3.1. Inhibitory Zone Assay
4.3.2. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) Assay
4.3.3. Growth Curve Assay
4.4. Antibacterial Mechanisms of TroHepc2-22
4.4.1. Bacterial Membrane Depolarization Assay
4.4.2. Propidium Iodide (PI) Staining Assay
4.4.3. Scanning Electron Microscopy (SEM) Visualization
4.4.4. Gel Retardation Assay
4.5. In Vivo Study on Pathogens Infection
4.6. Expression of Immune-Related Genes Induced by TroHepc2-22
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lafferty, K.D.; Harvell, C.D.; Conrad, J.M.; Friedman, C.S.; Kent, M.L.; Kuris, A.M.; Powell, E.N.; Rondeau, D.; Saksida, S.M. Infectious diseases affect marine fisheries and aquaculture economics. Ann. Rev. Mar. Sci. 2015, 7, 471–496. [Google Scholar] [CrossRef] [PubMed]
- Muniesa, A.; Basurco, B.; Aguilera, C.; Furones, D.; Reverté, C.; Sanjuan-Vilaplana, A.; Jansen, M.D.; Brun, E.; Tavornpanich, S. Mapping the knowledge of the main diseases affecting sea bass and sea bream in Mediterranean. Transbound. Emerg. Dis. 2020, 67, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Limbu, S.M.; Chen, L.Q.; Zhang, M.L.; Du, Z.Y. A global analysis on the systemic effects of antibiotics in cultured fish and their potential human health risk: A review. Rev. Aquac. 2021, 13, 1015–1059. [Google Scholar] [CrossRef]
- Mrozik, W.; Vinitnanthara, T.S.; Thongsamer, T.; Pansuk, N.; Pattanachan, P.; Thayanukul, P.; Acharya, K.; Baluja, M.Q.; Hazlerigg, C.; Robson, A.F.; et al. The food-water quality nexus in periurban aquacultures downstream of Bangkok. Thail. Sci. Total Environ. 2019, 695, 133923. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef]
- Mo, W.Y.; Chen, Z.; Leung, H.M.; Leung, A.O. Application of veterinary antibiotics in China’s aquaculture industry and their potential human health risks. Environ. Sci. Pollut. Res. Int. 2017, 24, 8978–8989. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Hancock, S.J.; Phan, M.D.; Peters, K.M.; Forde, B.M.; Chong, T.M.; Yin, W.F.; Chan, K.G.; Paterson, D.L.; Walsh, T.R.; Beatson, S.A.; et al. Identification of Inca/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob. Agents Chemother. 2017, 61, e01740-16. [Google Scholar] [CrossRef]
- Hancock, R.E. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef]
- Devine, D.A.; Hancock, R.E. Cationic peptides: Distribution and mechanisms of resistance. Curr. Pharm. Des. 2002, 8, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.; Belanger, C.R.; Haney, E.F.; Hancock, R.E.W. 10-Host Defense (Antimicrobial) Peptides. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Koutsopoulos, S., Ed.; Woodhead Publishing: Sawston, UK, 2018; pp. 253–285. [Google Scholar] [CrossRef]
- Shabir, U.; Ali, S.; Magray, A.R.; Ganai, B.A.; Firdous, P.; Hassan, T.; Nazir, R. Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microb. Pathog. 2018, 114, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.J.; Desbois, A.P.; Dyrynda, E.A. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar. Drugs 2010, 8, 1213–1262. [Google Scholar] [CrossRef] [PubMed]
- Maisetta, G.; Petruzzelli, R.; Brancatisano, F.L.; Esin, S.; Vitali, A.; Campa, M.; Batoni, G. Antimicrobial activity of human hepcidin 20 and 25 against clinically relevant bacterial strains: Effect of copper and acidic pH. Peptides 2010, 31, 1995–2002. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, E.; Tuttle, M.S.; Powelson, J.; Vaughn, M.B.; Donovan, A.; Ward, D.M.; Ganz, T.; Kaplan, J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004, 306, 2090–2093. [Google Scholar] [CrossRef]
- Hocquellet, A.; le Senechal, C.; Garbay, B. Importance of the disulfide bridges in the antibacterial activity of human hepcidin. Peptides 2012, 36, 303–307. [Google Scholar] [CrossRef]
- Park, C.H.; Valore, E.V.; Waring, A.J.; Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 2001, 276, 7806–7810. [Google Scholar] [CrossRef]
- Hunter, H.N.; Fulton, D.B.; Ganz, T.; Vogel, H.J. The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J. Biol. Chem. 2002, 277, 37597–37603. [Google Scholar] [CrossRef]
- Krijt, J.; Cmejla, R.; Sýkora, V.; Vokurka, M.; Vyoral, D.; Necas, E. Different expression pattern of hepcidin genes in the liver and pancreas of C57BL/6N and DBA/2N mice. J. Hepatol. 2004, 40, 891–896. [Google Scholar] [CrossRef]
- Iizuka, D.; Yoshioka, S.; Kawai, H.; Okazaki, E.; Kiriyama, K.; Izumi, S.; Nishimura, M.; Shimada, Y.; Kamiya, K.; Suzuki, F. Hepcidin-2 in mouse urine as a candidate radiation-responsive molecule. J. Radiat. Res. 2016, 57, 142–149. [Google Scholar] [CrossRef]
- Hilton, K.B.; Lambert, L.A. Molecular evolution and characterization of hepcidin gene products in vertebrates. Gene 2008, 415, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Muncaster, S.; Kraakman, K.; Gibbons, O.; Mensink, K.; Forlenza, M.; Jacobson, G.; Bird, S. Antimicrobial peptides within the Yellowtail Kingfish (Seriola lalandi). Dev. Comp. Immunol. 2018, 80, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.G.; Liu, S.S.; Sun, B.; Wang, X.L.; Wang, N.; Chen, S.L. Iron-metabolic function and potential antibacterial role of Hepcidin and its correlated genes (Ferroportin 1 and Transferrin Receptor) in turbot (Scophthalmus maximus). Fish Shellfish Immunol. 2013, 34, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, W.; Xu, Y.W.; Chen, R.Y.; Xu, Q. Sequence analysis of hepcidin in barbel steed (Hemibarbus labeo): QSHLS motif confers hepcidin iron-regulatory activity but limits its antibacterial activity. Dev. Comp. Immunol. 2021, 114, 103845. [Google Scholar] [CrossRef]
- Nemeth, E.; Preza, G.C.; Jung, C.L.; Kaplan, J.; Waring, A.J.; Ganz, T. The N-terminus of hepcidin is essential for its interaction with ferroportin: Structure-function study. Blood 2006, 107, 328–333. [Google Scholar] [CrossRef]
- Liu, D.; Gan, Z.S.; Ma, W.; Xiong, H.T.; Li, Y.Q.; Wang, Y.Z.; Du, H.H. Synthetic Porcine Hepcidin Exhibits Different Roles in Escherichia coli and Salmonella Infections. Antimicrob. Agents Chemother. 2017, 61, e02638-16. [Google Scholar] [CrossRef]
- Vyoral, D.; Petrak, J. Therapeutic potential of hepcidin-the master regulator of iron metabolism. Pharmacol. Res. 2017, 115, 242–254. [Google Scholar] [CrossRef]
- Katzenback, B.A. Antimicrobial Peptides as Mediators of Innate Immunity in Teleosts. Biology 2015, 4, 607–639. [Google Scholar] [CrossRef]
- Neves, J.V.; Ramos, M.F.; Moreira, A.C.; Silva, T.; Gomes, M.S.; Rodrigues, P.N.S. Hamp1 but not Hamp2 regulates ferroportin in fish with two functionally distinct hepcidin types. Sci. Rep. 2017, 7, 14793. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Hussain, B.R.A.; Pande, A. Antimicrobial peptides of fish: Innocuous alternatives to antibiotics. Rev. Aquac. 2020, 12, 85–106. [Google Scholar] [CrossRef]
- Gui, L.; Zhang, P.; Zhang, Q.; Zhang, J. Two hepcidins from spotted scat (Scatophagus argus) possess antibacterial and antiviral functions in vitro. Fish Shellfish Immunol. 2016, 50, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Kung, C.W.; Chen, J.Y. Antiviral activity by fish antimicrobial peptides of epinecidin-1 and hepcidin 1-5 against nervous necrosis virus in medaka. Peptides 2010, 31, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.G.; Wei, J.G.; Xu, D.; Cui, H.C.; Yan, Y.; Ou-Yang, Z.L.; Huang, X.H.; Huang, Y.H.; Qin, Q.W. Molecular cloning and characterization of two novel hepcidins from orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol. 2011, 30, 559–568. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, L.P.; Li, M.F.; Sun, L. Turbot (Scophthalmus maximus) hepcidin-1 and hepcidin-2 possess antimicrobial activity and promote resistance against bacterial and viral infection. Fish Shellfish Immunol. 2014, 38, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Lauth, X.; Babon, J.J.; Stannard, J.A.; Singh, S.; Nizet, V.; Carlberg, J.M.; Ostland, V.E.; Pennington, M.W.; Norton, R.S.; Westerman, M.E. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J. Biol. Chem. 2005, 280, 9272–9282. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.J.; Cai, J.J.; Cai, L.; Qu, H.D.; Yang, M.; Zhang, M. Cloning and expression of a hepcidin gene from a marine fish (Pseudosciaena crocea) and the antimicrobial activity of its synthetic peptide. Peptides 2019, 30, 638–646. [Google Scholar] [CrossRef]
- Xie, J.; Obiefuna, V.; Hodgkinson, J.W.; McAllister, M.; Belosevic, M. Teleost antimicrobial peptide hepcidin contributes to host defense of goldfish (Carassius auratus L.) against Trypanosoma carassii. Dev. Comp. Immunol. 2019, 94, 11–15. [Google Scholar] [CrossRef]
- Solstad, T.; Larsen, A.N.; Seppola, M.; Jørgensen, T.Ø. Identification, cloning and expression analysis of a hepcidin cDNA of the Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. 2008, 25, 298–310. [Google Scholar] [CrossRef]
- Shike, H.; Shimizu, C.; Lauth, X.; Burns, J.C. Organization and expression analysis of the zebrafish hepcidin gene, an antimicrobial peptide gene conserved among vertebrates. Dev. Comp. Immunol. 2004, 28, 747–754. [Google Scholar] [CrossRef]
- Huang, P.H.; Chen, J.Y.; Kuo, C.M. Three different hepcidins from tilapia, Oreochromis mossambicus: Analysis of their expressions and biological functions. Mol. Immunol. 2007, 44, 1922–1934. [Google Scholar] [CrossRef]
- Chang, W.T.; Pan, C.Y.; Rajanbabu, V.; Cheng, C.W.; Chen, J.Y. Tilapia (Oreochromis mossambicus) antimicrobial peptide, hepcidin 1-5, shows antitumor activity in cancer cells. Peptides 2011, 32, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Xu, M.Y.; Ji, X.S.; Yu, G.C.; Liu, Y. Cloning, characterization, and expression analysis of hepcidin gene from red sea bream (Chrysophrys major). Antimicrob. Agents Chemother. 2005, 49, 1608–1612. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.N.; Vázquez-Dorado, S.; Neves, J.V.; Wilson, J.M. Dual function of fish hepcidin: Response to experimental iron overload and bacterial infection in sea bass (Dicentrarchus labrax). Dev. Comp. Immunol. 2006, 30, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.V.; Caldas, C.; Vieira, I.; Ramos, M.F.; Rodrigues, P.N. Multiple Hepcidins in a teleost fish, Dicentrarchus labrax: Different hepcidins for different roles. J. Immunol. 2015, 195, 2696–2709. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Ma, L.; Yu, Y.; Yu, H.; Mohammed, S.; Chu, G.; Mu, L.; Zhang, Q. Identification and characterization of a hepcidin from half-smooth tongue sole Cynoglossus semilaevis. Fish Shellfish Immunol. 2012, 33, 213–219. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, Q.; Ji, R.; Zou, W.; Guo, G. Isolation and characterization of a hepcidin peptide from the head kidney of large yellow croaker, Pseudosciaena crocea. Fish Shellfish Immunol. 2009, 26, 864–870. [Google Scholar] [CrossRef]
- Qu, H.D.; Chen, B.; Peng, H.; Wang, K.J. Molecular cloning, recombinant expression, and antimicrobial activity of EC-hepcidin3, a new four-cysteine hepcidin isoform from Epinephelus coioides. Biosci. Biotechnol. Biochem. 2013, 77, 103–110. [Google Scholar] [CrossRef]
- Yin, F.; Sun, P.; Tang, B.; Dan, X.; Li, A. Immunological, ionic and biochemical responses in blood serum of the marine fish Trachinotus ovatus to poly-infection by Cryptocaryon irritans. Exp. Parasitol. 2015, 154, 113–117. [Google Scholar] [CrossRef]
- Wang, J.; Gatlin, D.M.; Li, L.H.; Wang, Y.; Jin, N.; Lin, H.Z.; Zhou, C.P.; Huang, Z.; Yu, W.; Guo, Y.J. Dietary chromium polynicotinate improves growth performance and feed utilization of juvenile golden pompano (Trachinotus ovatus) with starch as the carbohydrate. Aquaculture 2019, 505, 405–411. [Google Scholar] [CrossRef]
- Ning, L.J.; Gao, L.L.; Zhou, W.; Liu, S.; Chen, X.Y.; Pan, Q. Beneficial effects of dietary mulberry leaf along with multi-enzyme premix on the growth, immune response and disease resistance of golden pompano. Trachinotus Ovatus. Aquac. 2021, 535, 736396. [Google Scholar] [CrossRef]
- Guo, S.; Mo, Z.Q.; Wang, Z.; Xu, J.; Li, Y.W.; Dan, X.M.; Li, A.X. Isolation and pathogenicity of Streptococcus iniae in offshore cage-cultured Trachinotus ovatus in China. Aquaculture 2018, 492, 247–252. [Google Scholar] [CrossRef]
- Han, F.F.; Gao, Y.H.; Luan, C.; Xie, Y.G.; Liu, Y.F.; Wang, Y.Z. Comparing bacterial membrane interactions and antimicrobial activity of porcine lactoferricin-derived peptides. J. Dairy Sci. 2013, 96, 3471–3487. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Chino, M.; Pane, K.; Pistorio, V.; De Santis, A.; Pizzo, E.; D’Errico, G.; Pavone, V.; Lombardi, A.; Del Vecchio, P.; et al. Exploring the role of unnatural amino acids in antimicrobial peptides. Sci. Rep. 2018, 8, 8888. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, C.A.; Santana, P.A.; Salinas-Parra, N.; Beltrán, D.; Guzmán, F.; Vega, B.; Acosta, F.; Mercado, L. Immune Modulation ability of Hepcidin from teleost fish. Animals 2022, 12, 1586. [Google Scholar] [CrossRef]
- Álvarez, C.A.; Guzmán, F.; Cárdenas, C.; Marshall, S.H.; Mercado, L. Antimicrobial activity of trout hepcidin. Fish Shellfish Immunol. 2014, 41, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nie, L.; Chen, J. Mudskipper (Boleophthalmus pectinirostris) Hepcidin-1 and Hepcidin-2 present different gene expression profile and antibacterial activity and possess distinct protective effect against Edwardsiella tarda infection. Probiotics Antimicrob. Proteins 2018, 10, 176–185. [Google Scholar] [CrossRef]
- Hu, Y.; Kurobe, T.; Liu, X.; Zhang, Y.A.; Su, J.; Yuan, G. Hamp type-1 promotes antimicrobial defense via direct microbial killing and regulating iron metabolism in grass carp (Ctenopharyngodon idella). Biomolecules 2020, 10, 825. [Google Scholar] [CrossRef]
- Jordan, J.B.; Poppe, L.; Haniu, M.; Arvedson, T.; Syed, R.; Li, V.; Kohno, H.; Kim, H.; Schnier, P.D.; Harvey, T.S.; et al. Hepcidin revisited, disulfide connectivity, dynamics, and structure. J. Biol. Chem. 2009, 284, 24155–24167. [Google Scholar] [CrossRef]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.Y. Antimicrobial peptides: New hope in the war against multidrug resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef]
- Dong, G.L.; Song, Y.S.; Kim, D.H.; Seo, M.Y.; Kang, J.H.; Lee, Y. Antifungal mechanism of a cysteine-rich antimicrobial peptide, Ib-AMP1, from impatiens balsamina against Candida albicans. Biotechnol. Lett. 1999, 21, 1047–1050. [Google Scholar] [CrossRef]
- Álvarez, C.A.; Acosta, F.; Montero, D.; Guzmán, F.; Torres, E.; Vega, B.; Mercado, L. Synthetic hepcidin from fish: Uptake and protection against Vibrio anguillarum in sea bass (Dicentrarchus labrax). Fish Shellfish Immunol. 2016, 55, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Chamorro, C.; Boerman, M.A.; Arnusch, C.J.; Breukink, E.; Pieters, R.J. Enhancing membrane disruption by targeting and multivalent presentation of antimicrobial peptides. Biochim. Biophys. Acta 2012, 1818, 2171–2174. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zheng, L.B.; Mao, Y.; Wang, J.; Lin, L.S.; Su, Y.Q.; Li, Y. The antibacterial activity and mechanism analysis of piscidin 5 like from Larimichthys crocea. Dev. Comp. Immunol. 2019, 92, 43–49. [Google Scholar] [CrossRef]
- Lin, M.C.; Hui, C.F.; Chen, J.Y.; Wu, J.L. The antimicrobial peptide, shrimp anti-lipopolysaccharide factor (SALF), inhibits proinflammatory cytokine expressions through the MAPK and NF-κB pathways in Trichomonas vaginalis adherent to HeLa cells. Peptides 2012, 38, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef]
- Huang, T.; Gu, W.; Wang, B.; Zhang, Y.; Cui, L.; Yao, Z.; Zhao, C.; Xu, G. Identification and expression of the hepcidin gene from brown trout (Salmo trutta) and functional analysis of its synthetic peptide. Fish Shellfish Immunol. 2019, 87, 243–253. [Google Scholar] [CrossRef]
- van der Does, Y.; Limper, M.; Jie, K.E.; Schuit, S.C.E.; Jansen, H.; Pernot, N.; van Rosmalen, J.; Poley, M.J.; Ramakers, C.; Patka, P.; et al. Procalcitonin-guided antibiotic therapy in patients with fever in a general emergency department population: A multicentre non-inferiority randomized clinical trial (HiTEMP study). Clin. Microbiol. Infect. 2018, 24, 1282–1289. [Google Scholar] [CrossRef]
- Zhang, D.; He, Y.; Ye, Y.; Ma, Y.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Little antimicrobial peptides with big therapeutic roles. Protein Pept. Lett. 2019, 26, 564–578. [Google Scholar] [CrossRef]
- Wei, X.L.; Sarath-Babu, V.; Lin, L.; Hu, Y.Z.; Zhang, Y.L.; Liu, X.L.; Su, J.G.; Li, J.; Zhao, L.J.; Yuan, G.L. Hepcidin protects grass carp (Ctenopharyngodon idellus) against Flavobacterium columnare infection via regulating iron distribution and immune gene expression. Fish Shellfish Immunol. 2018, 75, 274–283. [Google Scholar] [CrossRef]
- Barroso, C.; Carvalho, P.; Nunes, M.; Gonçalves, J.F.M.; Rodrigues, P.N.S.; Neves, J.V. The era of antimicrobial peptides: Use of hepcidins to prevent or treat bacterial infections and iron disorders. Front. Immunol. 2021, 12, 754437. [Google Scholar] [CrossRef]
- Shahmiri, M.; Enciso, M.; Mechler, A. Controls and constrains of the membrane disrupting action of Aurein 1.2. Sci. Rep. 2015, 5, 16378. [Google Scholar] [CrossRef] [PubMed]
- AlMatar, M.; Makky, E.A.; Yakıcı, G.; Var, I.; Kayar, B.; Köksal, F. Antimicrobial peptides as an alternative to anti-tuberculosis drugs. Pharmacol. Res. 2018, 128, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.Y.; Peng, K.C.; Lin, C.H.; Chen, J.Y. Transgenic expression of tilapia hepcidin 1-5 and shrimp chelonianin in zebrafish and their resistance to bacterial pathogens. Fish Shellfish Immunol. 2011, 31, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Rebl, A.; Rebl, H.; Korytář, T.; Goldammer, T.; Seyfert, H.M. The proximal promoter of a novel interleukin-8-encoding gene in rainbow trout (Oncorhynchus mykiss) is strongly induced by CEBPA, but not NF-κB p65. Dev. Comp. Immunol. 2014, 46, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Ronza, P.; Bermúdez, R.; Losada, A.P.; Sitjà-Bobadilla, A.; Pardo, B.G.; Quiroga, M.I. Immunohistochemical detection and gene expression of TNFα in turbot (Scophthalmus maximus) enteromyxosis. Fish Shellfish Immunol. 2015, 47, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Kim, Y.J. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 2007, 76, 447–480. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Rajanbabu, V.; Chen, J.Y. The antimicrobial peptide, tilapia hepcidin 2-3, and PMA differentially regulate the protein kinase C isoforms, TNF-α and COX-2, in mouse RAW264.7 macrophages. Peptides 2011, 32, 333–341. [Google Scholar] [CrossRef]
- Hsieh, J.C.; Pan, C.Y.; Chen, J.Y. Tilapia hepcidin (TH)2-3 as a transgene in transgenic fish enhances resistance to Vibrio vulnificus infection and causes variations in immune-related genes after infection by different bacterial species. Fish Shellfish Immunol. 2010, 29, 430–439. [Google Scholar] [CrossRef]
- Shen, B.; Wei, K.; Yang, J.J.; Jing, F.; Zhang, J.S. Molecular characterization and functional analyses of a hepcidin gene from Bostrychus sinensis. Aquaculture 2021, 54, 737114. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, Y.H.; Xiao, Z.Z.; Sun, Y.; Sun, L. Construction and analysis of experimental DNA vaccines against megalocytivirus. Fish Shellfish Immunol. 2012, 33, 1192–1198. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, G.; Hao, D.; Wang, C.; Zhang, M. Antimicrobial and Immunoregulatory Activities of TS40, a Derived Peptide of a TFPI-2 Homologue from Black Rockfish (Sebastes schlegelii). Mar. Drugs 2022, 20, 353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cao, Z.J.; Diao, Q.Y.; Zhou, Y.C.; Ao, J.; Liu, C.S.; Sun, Y. Antimicrobial activity and mechanisms of a derived antimicrobial peptide TroNKL-27 from golden pompano (Trachinotus ovatus) NK-lysin. Fish Shellfish Immunol. 2022, 126, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Lin, H.; Ramesh-Pavase, T.; Mi, N.; Sui, J. Extraction, identification, modification, and antibacterial activity of histone from immature testis of Atlantic salmon. Mar. Drugs 2020, 18, 133. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.M.; Lei, Y.; Cao, Z.J.; Zhou, Y.C.; Sun, Y. TroCCL4, a CC chemokine of Trachinotus ovatus, is involved in the antimicrobial immune response. Fish Shellfish Immunol. 2019, 86, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.Y.; Kim, M.K.; Mereuta, L.; Seo, C.H.; Luchian, T.; Park, Y. Mechanism of action of antimicrobial peptide P5 truncations against Pseudomonas aeruginosa and Staphylococcus aureus. AMB Express 2019, 9, 122. [Google Scholar] [CrossRef]
- Xie, Z.; Wei, H.; Meng, J.; Cheng, T.; Song, Y.; Wang, M.; Zhang, Y. The analogs of temporin-gha exhibit a broader spectrum of antimicrobial activity and a stronger antibiofilm potential against Staphylococcus aureus. Molecules 2019, 24, 4173. [Google Scholar] [CrossRef]
- Zhang, T.; Jiang, S.; Sun, L. A fish galectin-8 possesses direct bactericidal activity. Int. J. Mol. Sci. 2020, 22, 376. [Google Scholar] [CrossRef]
- Li, H.; Sun, Y.; Sun, L. A Teleost CXCL10 Is Both an Immunoregulator and an Antimicrobial. Front. Immunol. 2022, 13, 917697. [Google Scholar] [CrossRef]
- Diao, Q.Y.; Du, H.H.; Zhao, N.; Wu, Y.; Du, X.Y.; Sun, Y.; Zhou, Y.C. Cathepsin C (CTSC) contributes to the antibacterial immunity in golden pompano (Trachinotus ovatus). Fish Shellfish Immunol. 2022, 128, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.S.; Hu, X.C.; Diao, Q.Y.; Zhang, P.P.; Wu, Y.; Cao, Z.J.; Zhou, Y.C.; Liu, C.S.; Sun, Y. Macrophage migration inhibitory factor (MIF) of golden pompano (Trachinotus ovatus) is involved in the antibacterial immune response. Dev. Comp. Immunol. 2022, 133, 104445. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Qiu, R.; Shen, Y.; Zhou, Y.C.; Cao, Z.J.; Sun, Y. Molecular characterization and antibacterial immunity functional analysis of liver-expressed antimicrobial peptide 2 (LEAP-2) gene in golden pompano (Trachinotus ovatus). Fish Shellfish Immunol. 2020, 106, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Zhang, X.Q.; Huang, S.; Cao, Z.J.; Qin, Q.W.; Hu, W.T.; Sun, Y.; Zhou, Y.C. Selection of reference genes for quantitative real-time RT-PCR on gene expression in Golden Pompano (Trachinotus ovatus). Pol. J. Vet. Sci. 2017, 20, 583–594. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, Y.C.; Cao, Z.J.; Chen, X.J.; Du, H.H.; Sun, Y. Interferon regulatory factor 7 contributes to the host response during Vibrio harveyi infection in the golden pompano Trachinotus ovatus. Dev. Comp. Immunol. 2021, 117, 103959. [Google Scholar] [CrossRef]
| Physical and Chemical Parameters | TroHepc2-22 |
|---|---|
| Molecular formula | C100H166N32O23S8 |
| Total atomic number | 329 |
| Net electric charge | +4 |
| Amino acid residue | 22 |
| Molecular weight | 2441.10 |
| Constant electric point | 8.78 |
| Coefficient of fat | 66.36 |
| Coefficient of instability | −2.91 |
| Hydrophilicity index | 1.018 |
| Hydrophobic index | 0.869 |
| Hydrophobic moment | 0.244 |
| Microorganisms | Bacterium | TroHepc2-22 MIC (μM) | TroHepc2-22 MBC (μM) | Hepc-25 MIC (μM) | Hepc-25 MBC (μM) |
|---|---|---|---|---|---|
| Gram-positive bacteria | Streptococcus agalactiae | 16 | 32 | >256 | >256 |
| Staphylococcus aureus | 8 | 16 | >256 | >256 | |
| Gram-negative bacteria | Edwardsiella piscicida | 32 | 64 | >256 | >256 |
| Vibrio harveyi | 16 | 32 | 128 | >256 | |
| Vibrio alginolyticus | 64 | 128 | >256 | >256 | |
| Escherichia coli | 128 | 256 | >256 | >256 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Zhou, Y.; Zhang, H.; Du, X.; Cao, Z.; Wu, Y.; Liu, C.; Sun, Y. Antibacterial Activity and Mechanisms of TroHepc2-22, a Derived Peptide of Hepcidin2 from Golden Pompano (Trachinotus ovatus). Int. J. Mol. Sci. 2023, 24, 9251. https://doi.org/10.3390/ijms24119251
Zhang Z, Zhou Y, Zhang H, Du X, Cao Z, Wu Y, Liu C, Sun Y. Antibacterial Activity and Mechanisms of TroHepc2-22, a Derived Peptide of Hepcidin2 from Golden Pompano (Trachinotus ovatus). International Journal of Molecular Sciences. 2023; 24(11):9251. https://doi.org/10.3390/ijms24119251
Chicago/Turabian StyleZhang, Zhengshi, Yongcan Zhou, Han Zhang, Xiangyu Du, Zhenjie Cao, Ying Wu, Chunsheng Liu, and Yun Sun. 2023. "Antibacterial Activity and Mechanisms of TroHepc2-22, a Derived Peptide of Hepcidin2 from Golden Pompano (Trachinotus ovatus)" International Journal of Molecular Sciences 24, no. 11: 9251. https://doi.org/10.3390/ijms24119251
APA StyleZhang, Z., Zhou, Y., Zhang, H., Du, X., Cao, Z., Wu, Y., Liu, C., & Sun, Y. (2023). Antibacterial Activity and Mechanisms of TroHepc2-22, a Derived Peptide of Hepcidin2 from Golden Pompano (Trachinotus ovatus). International Journal of Molecular Sciences, 24(11), 9251. https://doi.org/10.3390/ijms24119251







