Label-Free Imaging Techniques to Evaluate Metabolic Changes Caused by Toxic Liver Injury in PCLS
Abstract
1. Introduction
2. Results and Discussion
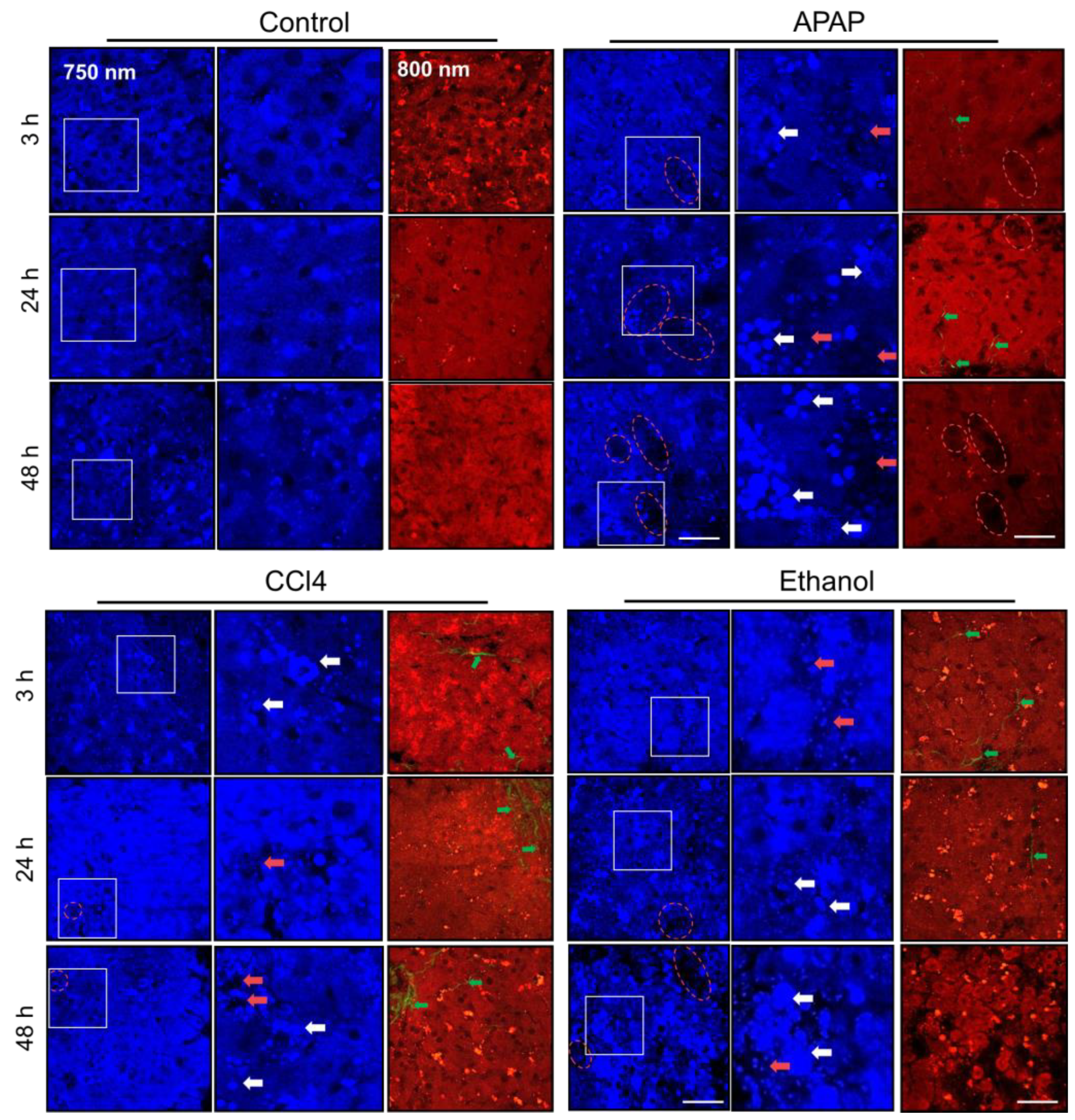
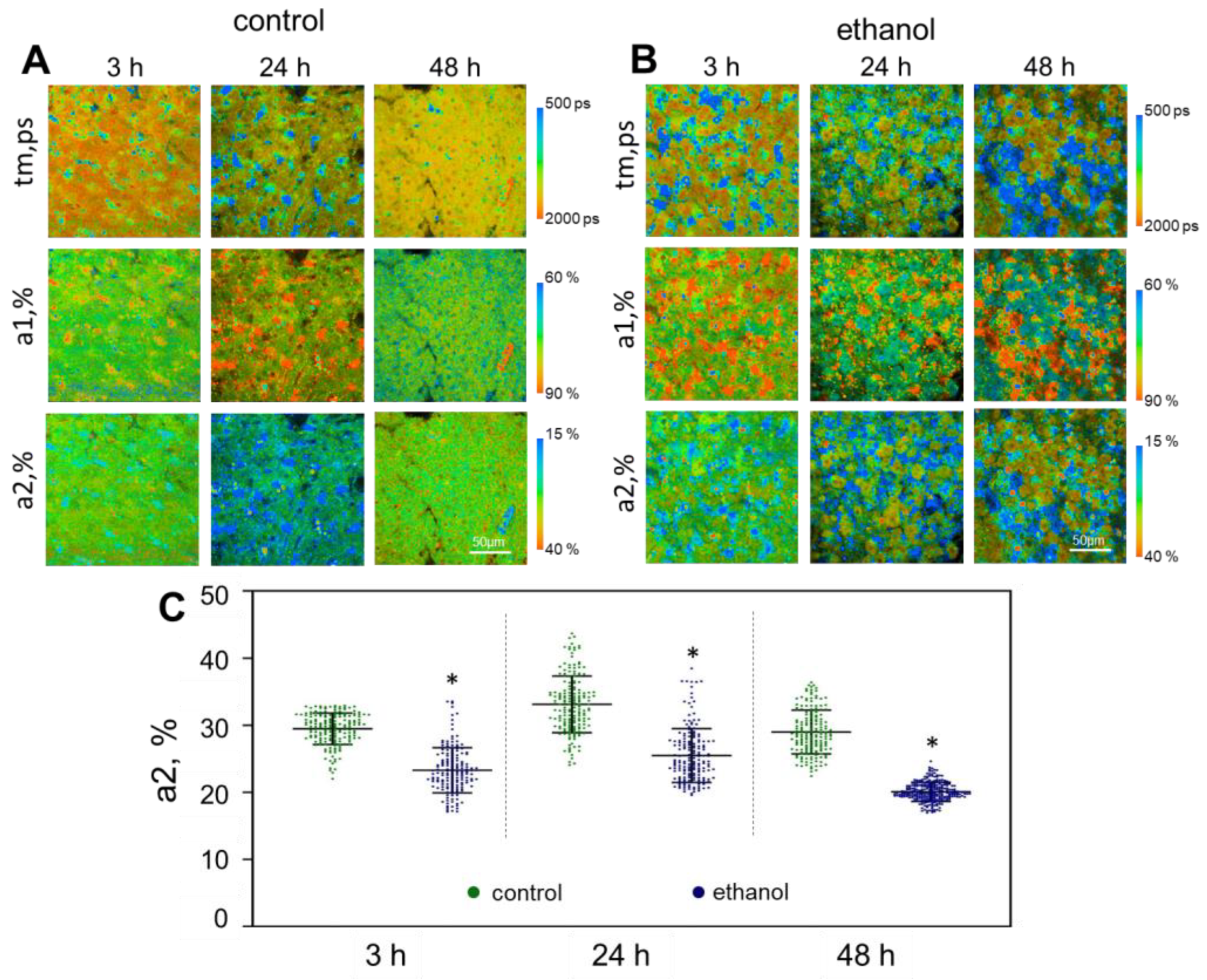
2.1. Multiphoton Microscopy and SHG
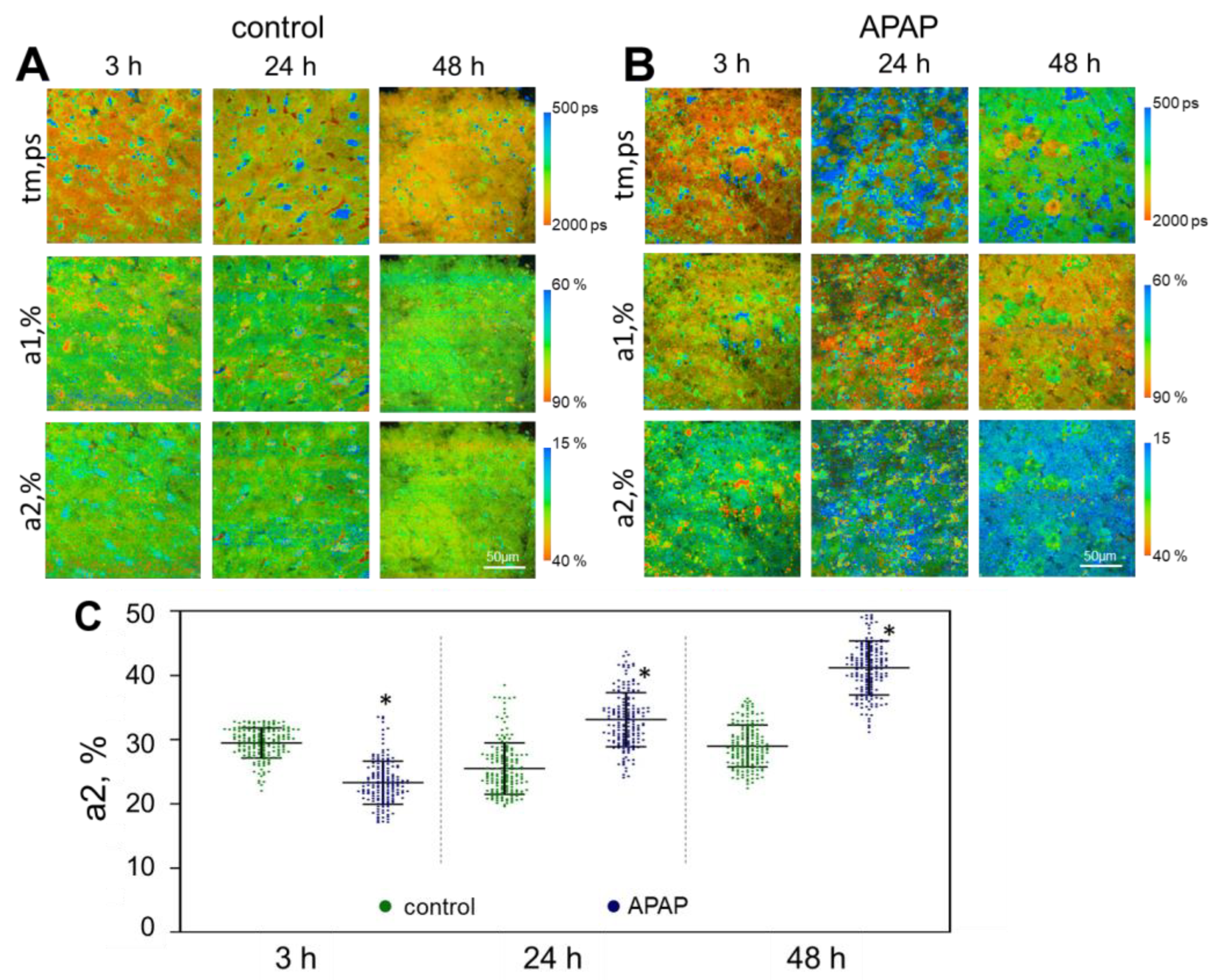
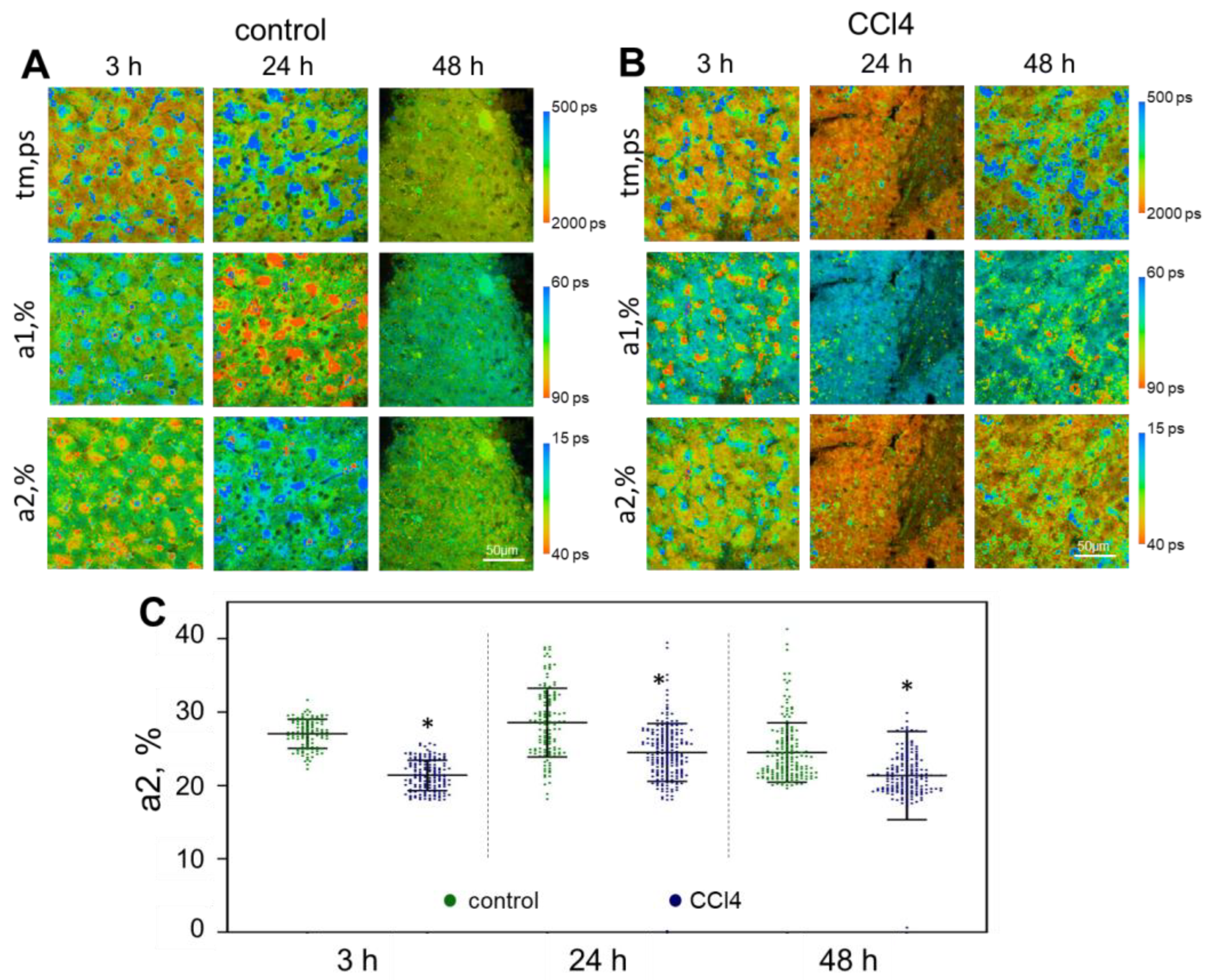
2.2. FLIM Analysis
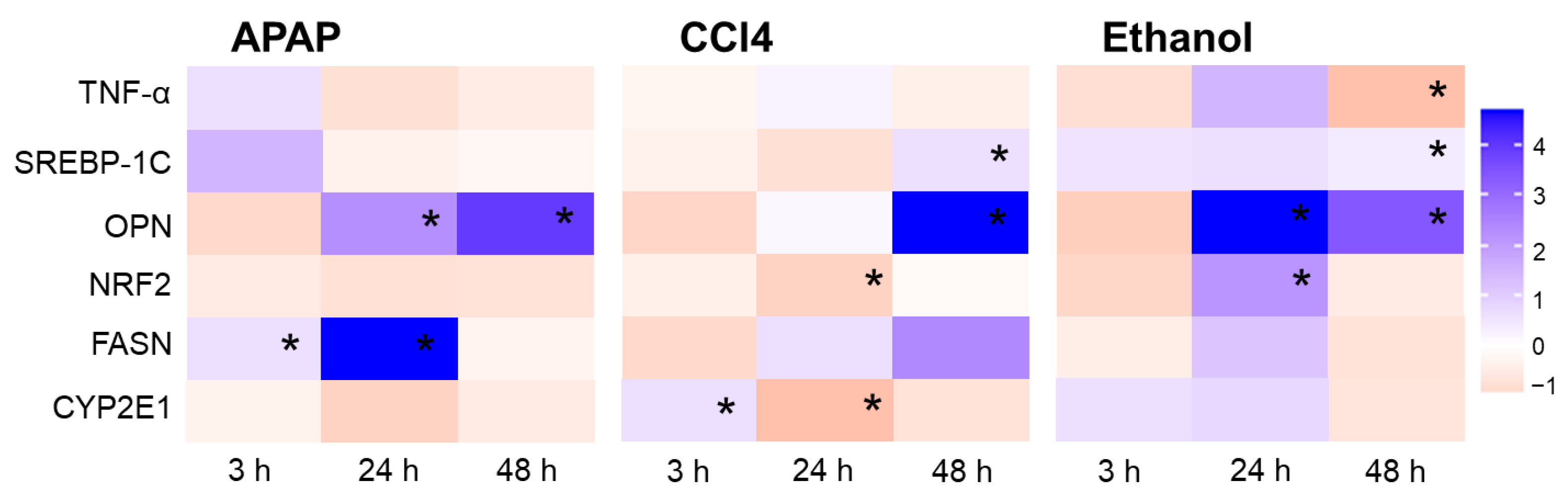
2.3. Real-Time PCR
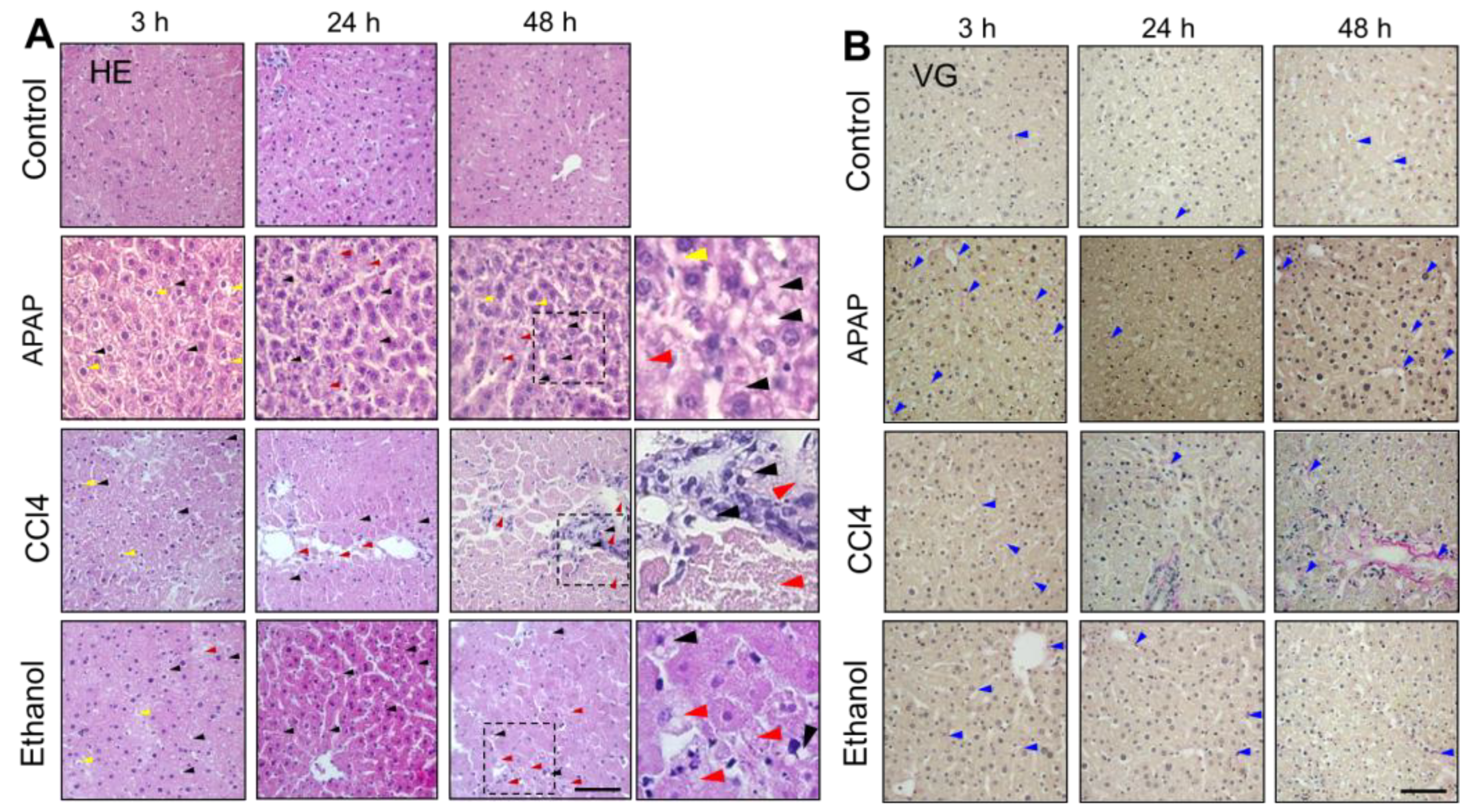
2.4. Histological Analysis
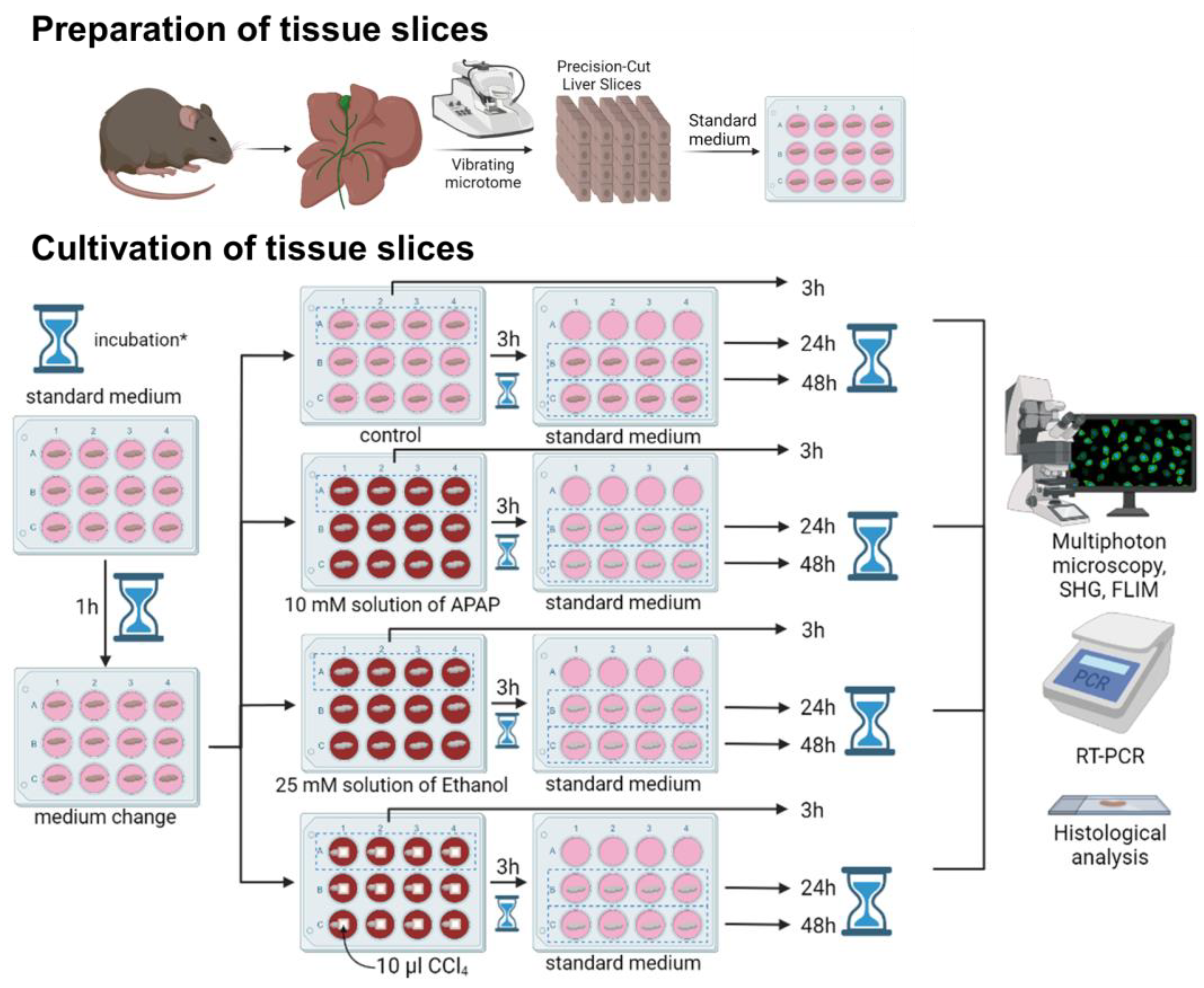
3. Materials and Methods
3.1. Precision Cut Liver Slices
3.2. Multiphoton Microscopy
3.3. Real-Time PCR
3.4. Histological Analysis
3.5. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Hoover, E.E.; Squier, J.A. Advances in multiphoton microscopy technology. Nat. Photonics 2013, 7, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Skala, M.C.; Riching, K.M.; Gendron-Fitzpatrick, A.; Eickhoff, J.; Eliceiri, K.W.; White, J.G.; Ramanujam, N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. USA 2007, 104, 19494–19499. [Google Scholar] [CrossRef] [PubMed]
- Chorvat, D.; Chorvatova, A. Multi-wavelength fluorescence lifetime spectroscopy: A new approach to the study of endogenous fluorescence in living cells and tissues. Laser Phys. Lett. 2009, 6, 175–193. [Google Scholar] [CrossRef]
- Van Manen, H.J.; Verkuijlen, P.; Wittendorp, P.; Subramaniam, V.; Van den Berg, T.K.; Roos, D.; Otto, C. Refractive index sensing of green fluorescent proteins in living cells using fluorescence lifetime imaging microscopy. Biophys. J. 2008, 94, 67–69. [Google Scholar] [CrossRef]
- Gailhouste, L.; Le Grand, Y.; Odin, C.; Guyader, D.; Turlin, B.; Ezan, F.; Désille, Y.; Guilbert, T.; Bessard, A.; Frémin, C.; et al. Fibrillar collagen scoring by second harmonic microscopy: A new tool in the assessment of liver fibrosis. J. Hepatol. 2010, 52, 398–406. [Google Scholar] [CrossRef]
- Williams, R.M.; Zipfel, W.R.; Webb, W.W. Interpreting second-harmonic generation images of collagen I fibrils. Biophys. J. 2005, 88, 1377–1386. [Google Scholar] [CrossRef]
- Becker, W. Fluorescence lifetime imaging–techniques and applications. J. Microsc. 2012, 247, 119–136. [Google Scholar] [CrossRef]
- Berezin, M.Y.; Achilefu, S. Fluorescence lifetime measurements and biological imaging. Chem. Rev. 2010, 110, 2641–2684. [Google Scholar] [CrossRef]
- Zheng, J.I.E. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation. Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef]
- Blacker, T.S.; Duchen, M.R. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic. Biol. Med. 2016, 100, 53–65. [Google Scholar] [CrossRef]
- Roberts, M.S.; Dancik, Y.; Prow, T.W.; Thorling, C.A.; Lin, L.L.; Grice, J.E.; Robertson, T.A.; König, K.; Becker, W. Non-invasive imaging of skin physiology and percutaneous penetration using fluorescence spectral and lifetime imaging with multiphoton and confocal microscopy. Eur. J. Pharm. Biopharm. 2011, 77, 469–488. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liang, X.; Mohammed, Y.H.; Thomas, J.A.; Bridle, K.R.; Thorling, C.A.; Grice, J.E.; Xu, Z.P.; Liu, X.; Crawford, D.H.; et al. Real-time histology in liver disease using multiphoton microscopy with fluorescence lifetime imaging. Biomed. Opt. Express 2015, 6, 780–792. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Thorling, C.A.; Liang, X.; Bridle, K.R.; Grice, J.E.; Zhu, Y.; Crawford, D.H.G.; Xu, Z.P.; Liu, X.; Roberts, M.S. Diagnostic imaging and therapeutic application of nanoparticles targeting the liver. J. Mater. Chem. B 2015, 3, 939–958. [Google Scholar] [CrossRef] [PubMed]
- Thorling, C.A.; Crawford, D.; Burczynski, F.J.; Liu, X.; Liau, I.; Roberts, M.S. Multiphoton microscopy in defining liver function. J. Biomed. Opt. 2014, 19, 090901. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, R.; Bridle, K.R.; Jayachandran, A.; Thomas, J.A.; Zhang, W.; Yuan, J.; Xu, Z.P.; Crawford, D.H.G.; Liang, X.; et al. Two-photon dual imaging platform for in vivo monitoring cellular oxidative stress in liver injury. Sci. Rep. 2017, 7, 45374. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, D.; Rodimova, S.; Gulin, A.; Reunov, D.; Bobrov, N.; Polozova, A.; Vasin, A.; Shcheslavskiy, V.I.; Vdovina, N.; Zagainov, V.E.; et al. Metabolic imaging and secondary ion mass spectrometry to define the structure and function of liver with acute and chronic pathology. J. Biomed. Opt. 2020, 25, 014508. [Google Scholar] [CrossRef]
- Rodimova, S.; Kuznetsova, D.; Bobrov, N.; Elagin, V.; Shcheslavskiy, V.; Zagainov, V.; Zagaynova, E. Mapping metabolism of liver tissue using two-photon FLIM. Biomed. Opt. Express 2020, 11, 4458–4470. [Google Scholar] [CrossRef]
- Prins, G.H.; Luangmonkong, T.; Oosterhuis, D.; Mutsaers, H.A.; Dekker, F.J.; Olinga, P. A pathophysiological model of non-alcoholic fatty liver disease using precision-cut liver slices. Nutrients 2019, 11, 507. [Google Scholar] [CrossRef]
- Palma, E.; Doornebal, E.J.; Chokshi, S. Precision-cut liver slices: A versatile tool to advance liver research. Hepatol. Int. 2019, 13, 51–57. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, L.; Liu, W.; Ma, X.; Cen, J.; Sun, Z.; Wang, C.; Feng, S.; Zhang, Z.; Yue, L.; et al. In vitro expansion of primary human hepatocytes with efficient liver repopulation capacity. Cell Stem Cell 2018, 23, 806–819. [Google Scholar] [CrossRef]
- Ramboer, E.; Vanhaecke, T.; Rogiers, V.; Vinken, M. Immortalized human hepatic cell lines for in vitro testing and research purposes. In Protocols in In Vitro Hepatocyte Research; Springer: Cham, Switzerland, 2015; pp. 53–76. [Google Scholar]
- LeCluyse, E.L.; Alexandre, E.; Hamilton, G.A.; Viollon-Abadie, C.; Coon, D.J.; Jolley, S.; Richert, L. Isolation and culture of primary human hepatocytes. In Basic Cell Culture Protocols; Springer: Cham, Switzerland, 2005; pp. 207–229. [Google Scholar]
- Liang, S.; Kisseleva, T.; Brenner, D.A. The role of NADPH oxidases (NOXs) in liver fibrosis and the activation of myofibroblasts. Front. Physiol. 2016, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Klassen, L.W.; Thiele, G.M.; Duryee, M.J.; Schaffert, C.S.; DeVeney, A.L.; Hunter, C.D.; Olinga, P.; Tuma, D.J. An in vitro method of alcoholic liver injury using precision-cut liver slices from rats. Biochem. Pharmacol. 2008, 76, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Van de Bovenkamp, M.; Groothuis, G.M.; Draaisma, A.L.; Merema, M.T.; Bezuijen, J.I.; van Gils, M.J.; Meijer, D.K.F.; Friedman, S.L.; Olinga, P. Precision-cut liver slices as a new model to study toxicity-induced hepatic stellate cell activation in a physiologic milieu. Toxicol. Sci. 2005, 85, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.A.; Ahmad, M.Z.; Khan, J.A.; Arshad, M.I. Crosstalk of liver immune cells and cell death mechanisms in different murine models of liver injury and its clinical relevance. Hepatob. Pancreat. Dis. 2017, 16, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Wei, Y.H.; Guo, H.W. Reduced nicotinamide adenine dinucleotide (NADH) fluorescence for the detection of cell death. Anti-Cancer Agents Med. Chem. 2009, 9, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Buschke, D.G.; Squirrell, J.M.; Fong, J.J.; Eliceiri, K.W.; Ogle, B.M. Cell death, non-invasively assessed by intrinsic fluorescence intensity of NADH, is a predictive indicator of functional differentiation of embryonic stem cells. Biol. Cell 2012, 104, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Bogaczewicz, J.; Tokarska, K.; Wozniacka, A. Changes of NADH fluorescence from the skin of patients with systemic lupus erythematosus. BioMed Res. Int. 2019, 2019, 5897487. [Google Scholar] [CrossRef]
- Ullah, H.; Khan, A.; Baig, M.W.; Ullah, N.; Ahmed, N.; Tipu, M.K.; Ali, H.; Khan, S. Poncirin attenuates CCL4-induced liver injury through inhibition of oxidative stress and inflammatory cytokines in mice. BMC Complem Med. Terap. 2020, 20, 115. [Google Scholar] [CrossRef]
- Amer, M.A.; Othman, A.I.; El-Missiry, M.A.; Farag, A.A.; Amer, M.E. Proanthocyanidins attenuated liver damage and suppressed fibrosis in CCl4-treated rats. Environ. Sci. Pollut. Res. 2022, 29, 91127–91138. [Google Scholar] [CrossRef]
- Jiang, Y.; Fan, X.; Wang, Y.; Tan, H.; Chen, P.; Zeng, H.; Huang, M.; Bi, H. Hepato-protective effects of six schisandra lignans on acetaminophen-induced liver injury are partially associated with the inhibition of CYP-mediated bioactivation. Chem. Biol. Interact. 2015, 231, 83–89. [Google Scholar] [CrossRef]
- Rodimova, S.; Bobrov, N.; Mozherov, A.; Elagin, V.; Karabut, M.; Shchechkin, I.; Kozlov, D.; Krylov, D.; Gavrina, A.; Zagainov, V.; et al. Optical Biomedical Imaging Reveals Criteria for Violated Liver Regenerative Potential. Cells 2023, 12, 479. [Google Scholar] [CrossRef] [PubMed]
- Ranjit, S.; Dvornikov, A.; Dobrinskikh, E.; Wang, X.; Luo, Y.; Levi, M.; Gratton, E. Measuring the effect of a Western diet on liver tissue architecture by FLIM autofluorescence and harmonic generation microscopy. Biomed. Opt. Express 2017, 8, 3143–3154. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Fan, X.; Wang, Y.; Chen, P.; Zeng, H.; Tan, H.; Gonzalez, F.J.; Huang, M.; Bi, H. Schisandrol B protects against acetaminophen-induced hepatotoxicity by inhibition of CYP-mediated bioactivation and regulation of liver regeneration. Toxicol. Sci. 2015, 143, 107–115. [Google Scholar] [CrossRef] [PubMed]
- McGill, M.R.; Lebofsky, M.; Norris, H.R.; Slawson, M.H.; Bajt, M.L.; Xie, Y.; Williams, C.D.; Wilkins, D.G.; Rollins, D.E.; Jaeschke, H. Plasma and liver acetaminophen-protein adduct levels in mice after acetaminophen treatment: Dose–response, mechanisms, and clinical implications. Toxicol. Appl. Pharm. 2013, 269, 240–249. [Google Scholar] [CrossRef]
- Moyer, A.M.; Fridley, B.L.; Jenkins, G.D.; Batzler, A.J.; Pelleymounter, L.L.; Kalari, K.R.; Ji, Y.; Chai, Y.; Nordgren, K.K.; Weinshilboum, R.M. Acetaminophen-NAPQI hepatotoxicity: A cell line model system genome-wide association study. Toxicol. Sci. 2011, 120, 33–41. [Google Scholar] [CrossRef]
- Barr, T.; Helms, C.; Grant, K.; Messaoudi, I. Opposing effects of alcohol on the immune system. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2016, 65, 242–251. [Google Scholar] [CrossRef]
- Zakhari, S. Overview: How is alcohol metabolized by the body? Alcohol. Res. Health 2006, 29, 245. [Google Scholar]
- Wang, S.; Shi, X.L.; Feng, M.; Wang, X.; Zhang, Z.H.; Zhao, X.; Ding, Y.T. Puerarin protects against CCl4-induced liver fibrosis in mice: Possible role of PARP-1 inhibition. Int. Immunopharmacol. 2016, 38, 238–245. [Google Scholar] [CrossRef]
- Heard, K.J. Acetylcysteine for acetaminophen poisoning. N. Engl. J. Med. 2008, 359, 285–922. [Google Scholar] [CrossRef]
- Smeitink, J.; van den Heuvel, L.; DiMauro, S. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2001, 2, 342–352. [Google Scholar] [CrossRef]
- Carvalho, N.R.; da Rosa, E.F.; Da Silva, M.H.; Tassi, C.C.; Dalla Corte, C.L.; Carbajo-Pescador, S.; Mauriz, J.L.; González-Gallego, J.; Soares, F.A. New therapeutic approach: Diphenyl diselenide reduces mitochondrial dysfunction in acetaminophen-induced acute liver failure. PLoS ONE 2013, 8, e81961. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Jaeschke, H. A mitochondrial journey through acetaminophen hepatotoxicity. Food Chem. Toxicol. 2020, 140, 111282. [Google Scholar] [CrossRef] [PubMed]
- Manibusan, M.K.; Odin, M.; Eastmond, D.A. Postulated carbon tetrachloride mode of action: A review. J. Environ. Sci. Heal. C 2007, 25, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Jaeschke, H.; McGill, M.R.; Ramachandran, A. Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: Lessons learned from acetaminophen hepatotoxicity. Drug Metab. Rev. 2012, 44, 88–106. [Google Scholar] [CrossRef] [PubMed]
- Musicco, C.; Cormio, A.; Calvaruso, M.A.; Iommarini, L.; Gasparre, G.; Porcelli, A.M.; Timperio, A.M.; Zolla, L.; Gadaleta, M.N. Analysis of the mitochondrial proteome of cybrid cells harbouring a truncative mitochondrial DNA mutation in respiratory complex I. Mol. Biosyst. 2014, 10, 1313–1319. [Google Scholar] [CrossRef]
- Hua, H.; Zhang, Z.; Qian, Y.; Yuan, H.; Ge, W.; Huang, S.; Zhang, A.; Zhang, Y.; Jia, Z.; Ding, G. Inhibition of the mitochondrial complex-1 protects against carbon tetrachloride-induced acute liver injury. Biomed. Pharmacother. 2019, 115, 108948. [Google Scholar] [CrossRef]
- Yang, S.; Tan, T.M.C.; Wee, A.; Leow, C.K. Mitochondrial respiratory function and antioxidant capacity in normal and cirrhotic livers following partial hepatectomy. Cell. Mol. Life Sci. 2004, 61, 220–229. [Google Scholar] [CrossRef]
- Goodman, R.P.; Markhard, A.L.; Shah, H.; Sharma, R.; Skinner, O.S.; Clish, C.B.; Deik, A.; Patgiri, A.; Hsu, Y.-H.H.; Masia, R.; et al. Hepatic NADH reductive stress underlies common variation in metabolic traits. Nature 2020, 583, 122–126. [Google Scholar] [CrossRef]
- Tan, H.; He, Q.; Li, R.; Lei, F.; Lei, X. Trillin reduces liver chronic inflammation and fibrosis in carbon tetrachloride (CCl4) induced liver injury in mice. Immunol. Investig. 2016, 45, 371–382. [Google Scholar] [CrossRef]
- Li, Q.; Chen, F.; Wang, F. The immunological mechanisms and therapeutic potential in drug-induced liver injury: Lessons learned from acetaminophen hepatotoxicity. Cell Biosci. 2022, 12, 187. [Google Scholar] [CrossRef]
- Dentin, R.; Girard, J.; Postic, C. Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): Two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie 2005, 87, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yao, Y.; Wei, Z.Y.; Wang, S.X.; Wu, Y.C.; Hu, Y.; Yang, C.C.; Min, J.L.; Li, L.Y.; Zhou, H. Deletion of p38γ attenuates ethanol consumption-and acetaminophen-induced liver injury in mice through promoting Dlg1. Acta Pharmacol. Sin. 2022, 43, 1733–1748. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Xue, W.; Han, B.; Yang, F.; Yin, Y.; Hu, C. Acetaminophen aggravates fat accumulation in NAFLD by inhibiting autophagy via the AMPK/mTOR pathway. Eur. J. Pharmacol. 2019, 850, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wang, J.; Xue, W.; Xin, J.; Shi, C.; Wen, J.; Feng, X.; Huang, Y.; Hu, C. Caveolin-1 attenuates acetaminophen aggravated lipid accumulation in alcoholic fatty liver by activating mitophagy via the Pink-1/Parkin pathway. Eur. J. Pharmacol. 2021, 908, 174324. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Meng, X.; Zhang, J.; Zhao, Y. Hepatoprotective effect of different combinations of 18α-and 18β-Glycyrrhizic acid against CCl4-induced liver injury in rats. Biomed. Pharmacother. 2020, 122, 109354. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Jeong, S.; Xia, Q.; Kong, X. Role of osteopontin in liver diseases. Int. J. Biol. Sci. 2016, 12, 1121. [Google Scholar] [CrossRef]
- Ramaiah, S.K.; Rittling, S. Role of osteopontin in regulating hepatic inflammatory responses and toxic liver injury. Expert Opin. Drug Met. 2007, 3, 519–526. [Google Scholar] [CrossRef]
- Ingawale, D.K.; Mandlik, S.K.; Naik, S.R. Models of hepatotoxicity and the underlying cellular, biochemical and immunological mechanism(s): A critical discussion. Environ. Toxicol. Pharmacol. 2014, 37, 118–133. [Google Scholar] [CrossRef]
- Gong, P.; Cederbaum, A.I. Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology 2006, 43, 144–153. [Google Scholar] [CrossRef]
- Jadeja, R.N.; Upadhyay, K.K.; Devkar, R.V.; Khurana, S. Naturally occurring Nrf2 activators: Potential in treatment of liver injury. Oxid. Med. Cell. Longev. 2016, 2016, 3453926. [Google Scholar] [CrossRef]
- Sun, J.; Fu, J.; Li, L.; Chen, C.; Wang, H.; Hou, Y.; Xu, Y.; Pi, J. Nrf2 in alcoholic liver disease. Toxicol. Appl. Pharm. 2018, 357, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Hamada, K.; Wang, P.; Xia, Y.; Yan, N.; Takahashi, S.; Krausz, K.W.; Hao, H.; Yan, T.; Gonzalez, F.J. Withaferin A alleviates ethanol-induced liver injury by inhibiting hepatic lipogenesis. Food Chem. Toxicol. 2022, 160, 112807. [Google Scholar] [CrossRef] [PubMed]
- Ghanim, B.Y.; Ahmad, M.; Abdallah, Q.; Khaleel, A.; Qinna, N.A. Transcriptional profiling of drug-induced liver injury biomarkers: Association of hepatic Srebf1/Pparα signaling and crosstalk of thrombin, alcohol dehydrogenase, MDR and DNA damage regulators. Mol. Cell. Biochem. 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.J. Role of cytochromes P450 in chemical toxicity and oxidative stress: Studies with CYP2E1. Mutat. Res-Fund. Mol. Mech. Mutagen. 2005, 569, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.K. Relevance of CYP2E1 to non-alcoholic fatty liver disease. In Cytochrome P450 2E1: Its Role in Disease and Drug Metabolism; Springer: Cham, Switzerland, 2013; pp. 165–175. [Google Scholar]
- Lu, Y.; Zhuge, J.; Wang, X.; Bai, J.; Cederbaum, A.I. Cytochrome P450 2E1 contributes to ethanol-induced fatty liver in mice. Hepatology 2008, 47, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Massart, J.; Begriche, K.; Fromenty, B. Cytochrome P450 2E1 should not be neglected for acetaminophen-induced liver injury in metabolic diseases with altered insulin levels or glucose homeostasis. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101470. [Google Scholar] [CrossRef]
- Chhimwal, J.; Sharma, S.; Kulurkar, P.; Patial, V. Crocin attenuates CCl4-induced liver fibrosis via PPAR-γ mediated modulation of inflammation and fibrogenesis in rats. Hum. Exp. Toxicol. 2020, 39, 1639–1649. [Google Scholar] [CrossRef]
- Zhang, X.; Kuang, G.; Wan, J.; Jiang, R.; Ma, L.; Gong, X.; Liu, X. Salidroside protects mice against CCl4-induced acute liver injury via down-regulating CYP2E1 expression and inhibiting NLRP3 inflammasome activation. Int. Immunopharmacol. 2020, 85, 106662. [Google Scholar] [CrossRef]
- Leung, T.M.; Nieto, N. CYP2E1 and oxidant stress in alcoholic and non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 395–398. [Google Scholar] [CrossRef]
- Papackova, Z.; Heczkova, M.; Dankova, H.; Sticova, E.; Lodererova, A.; Bartonova, L.; Poruba, M.; Cahova, M. Silymarin prevents acetaminophen-induced hepatotoxicity in mice. PLoS ONE 2018, 13, e0191353. [Google Scholar] [CrossRef]
- Gill, P.; Bhattacharyya, S.; McCullough, S.; Letzig, L.; Mishra, P.J.; Luo, C.; Dweep, H.; James, L. MicroRNA regulation of CYP 1A2, CYP3A4 and CYP2E1 expression in acetaminophen toxicity. Sci. Rep. 2017, 7, 12331. [Google Scholar] [CrossRef] [PubMed]
- French, S.W. The importance of CYP2E1 in the pathogenesis of alcoholic liver disease and drug toxicity and the role of the proteasome. In Cytochrome P450 2E1: Its Role in Disease and Drug Metabolism; Springer: Cham, Switzerland, 2013; pp. 145–164. [Google Scholar]
- Xie, W.; Jiang, Z.; Wang, J.; Zhang, X.; Melzig, M.F. Protective effect of hyperoside against acetaminophen (APAP) induced liver injury through enhancement of APAP clearance. Chem. Biol. Interact. 2016, 246, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.K.; Mohammed, S.A.; Khalaf, G.; Fikry, H. Role of bone marrow mesenchymal stem cells in the treatment of CCL4 induced liver fibrosis in albino rats: A histological and immunohistochemical study. Int. J. Stem Cell 2014, 7, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Cresci, G.A.; Glueck, B.; McMullen, M.R.; Xin, W.; Allende, D.; Nagy, L.E. Prophylactic tributyrin treatment mitigates chronic-binge ethanol-induced intestinal barrier and liver injury. J. Gastroenterol. Hepatol. 2017, 32, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Jaeschke, H.; Williams, C.D.; Ramachandran, A.; Bajt, M.L. Acetaminophen hepatotoxicity and repair: The role of sterile inflammation and innate immunity. Liver Int. 2012, 32, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Vatakuti, S.; Schoonen, W.G.; Elferink, M.L.; Groothuis, G.M.; Olinga, P. Acute toxicity of CCl4 but not of paracetamol induces a transcriptomic signature of fibrosis in precision-cut liver slices. Toxicol. Vitro 2015, 29, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Van Midwoud, P.M.; Merema, M.T.; Verweij, N.; Groothuis, G.M.; Verpoorte, E. Hydrogel embedding of precision-cut liver slices in a microfluidic device improves drug metabolic activity. Biotechnol. Bioeng. 2011, 108, 1404–1412. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Cardiff, R.D.; Miller, C.H.; Munn, R.J. Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harb. Prot. 2014, 2014, 411. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodimova, S.; Mozherov, A.; Elagin, V.; Karabut, M.; Shchechkin, I.; Kozlov, D.; Krylov, D.; Gavrina, A.; Bobrov, N.; Zagainov, V.; et al. Label-Free Imaging Techniques to Evaluate Metabolic Changes Caused by Toxic Liver Injury in PCLS. Int. J. Mol. Sci. 2023, 24, 9195. https://doi.org/10.3390/ijms24119195
Rodimova S, Mozherov A, Elagin V, Karabut M, Shchechkin I, Kozlov D, Krylov D, Gavrina A, Bobrov N, Zagainov V, et al. Label-Free Imaging Techniques to Evaluate Metabolic Changes Caused by Toxic Liver Injury in PCLS. International Journal of Molecular Sciences. 2023; 24(11):9195. https://doi.org/10.3390/ijms24119195
Chicago/Turabian StyleRodimova, Svetlana, Artem Mozherov, Vadim Elagin, Maria Karabut, Ilya Shchechkin, Dmitry Kozlov, Dmitry Krylov, Alena Gavrina, Nikolai Bobrov, Vladimir Zagainov, and et al. 2023. "Label-Free Imaging Techniques to Evaluate Metabolic Changes Caused by Toxic Liver Injury in PCLS" International Journal of Molecular Sciences 24, no. 11: 9195. https://doi.org/10.3390/ijms24119195
APA StyleRodimova, S., Mozherov, A., Elagin, V., Karabut, M., Shchechkin, I., Kozlov, D., Krylov, D., Gavrina, A., Bobrov, N., Zagainov, V., Zagaynova, E., & Kuznetsova, D. (2023). Label-Free Imaging Techniques to Evaluate Metabolic Changes Caused by Toxic Liver Injury in PCLS. International Journal of Molecular Sciences, 24(11), 9195. https://doi.org/10.3390/ijms24119195






