Molecular and Genetics-Based Systems for Tracing the Evolution and Exploring the Mechanisms of Human Norovirus Infections
Abstract
1. Norovirus
2. Evolution and Outbreaks
3. Technologies for the Detection and Analysis of Noroviruses
3.1. Nested Polymerase Chain Reaction (PCR) with Sanger Sequencing
3.2. Reverse-Transcription Quantitative Polymerase Chain Reaction Assays (RT-qPCR)
3.3. Digital Polymerase Chain Reaction (dPCR)
3.4. Enzyme Immunoassay (EIA)
3.5. Next-Generation Sequencing (NGS)
3.6. Aptamer-Based Detection of Specific Genotypes
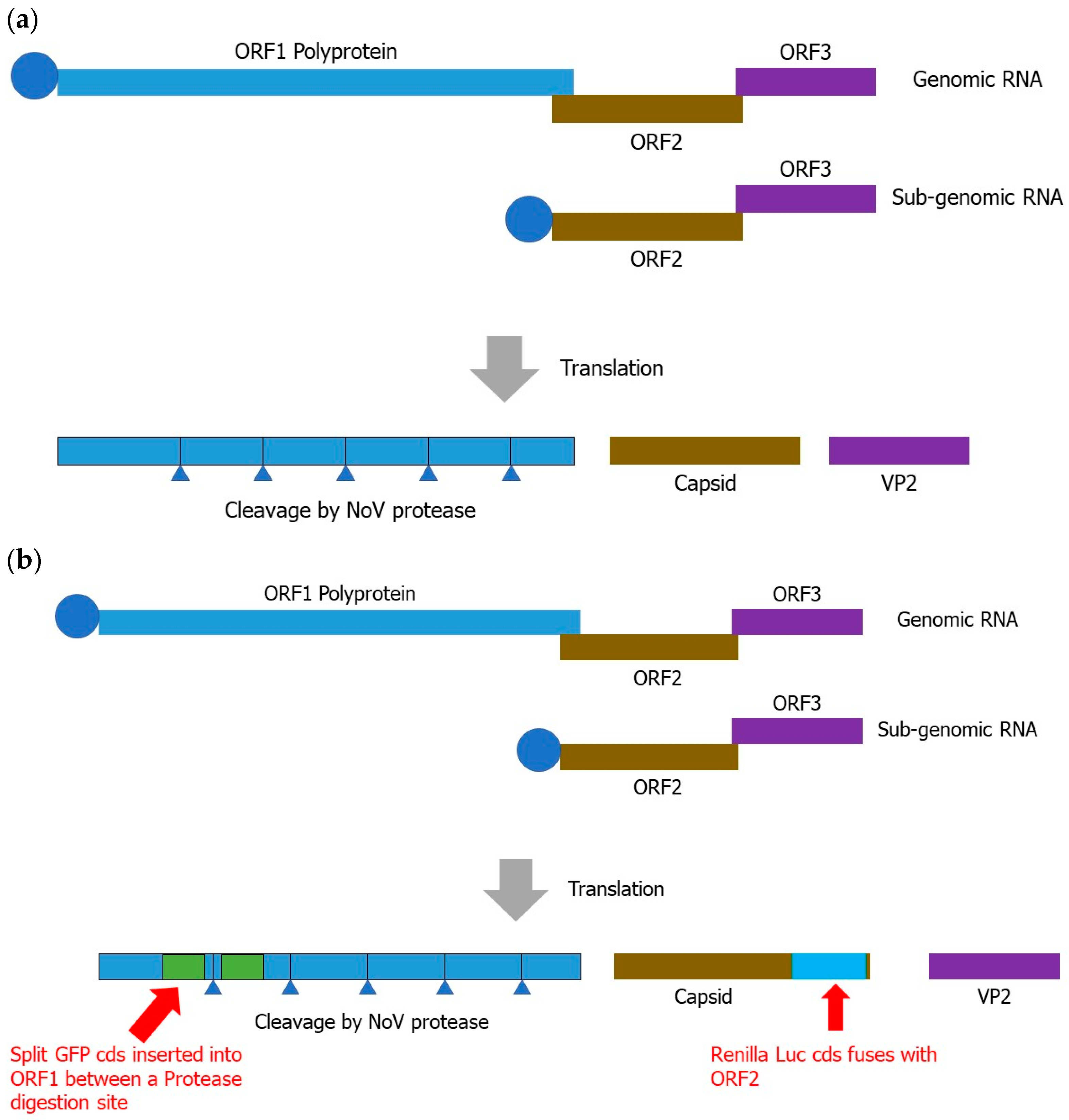
3.7. Reverse Genetics Techniques
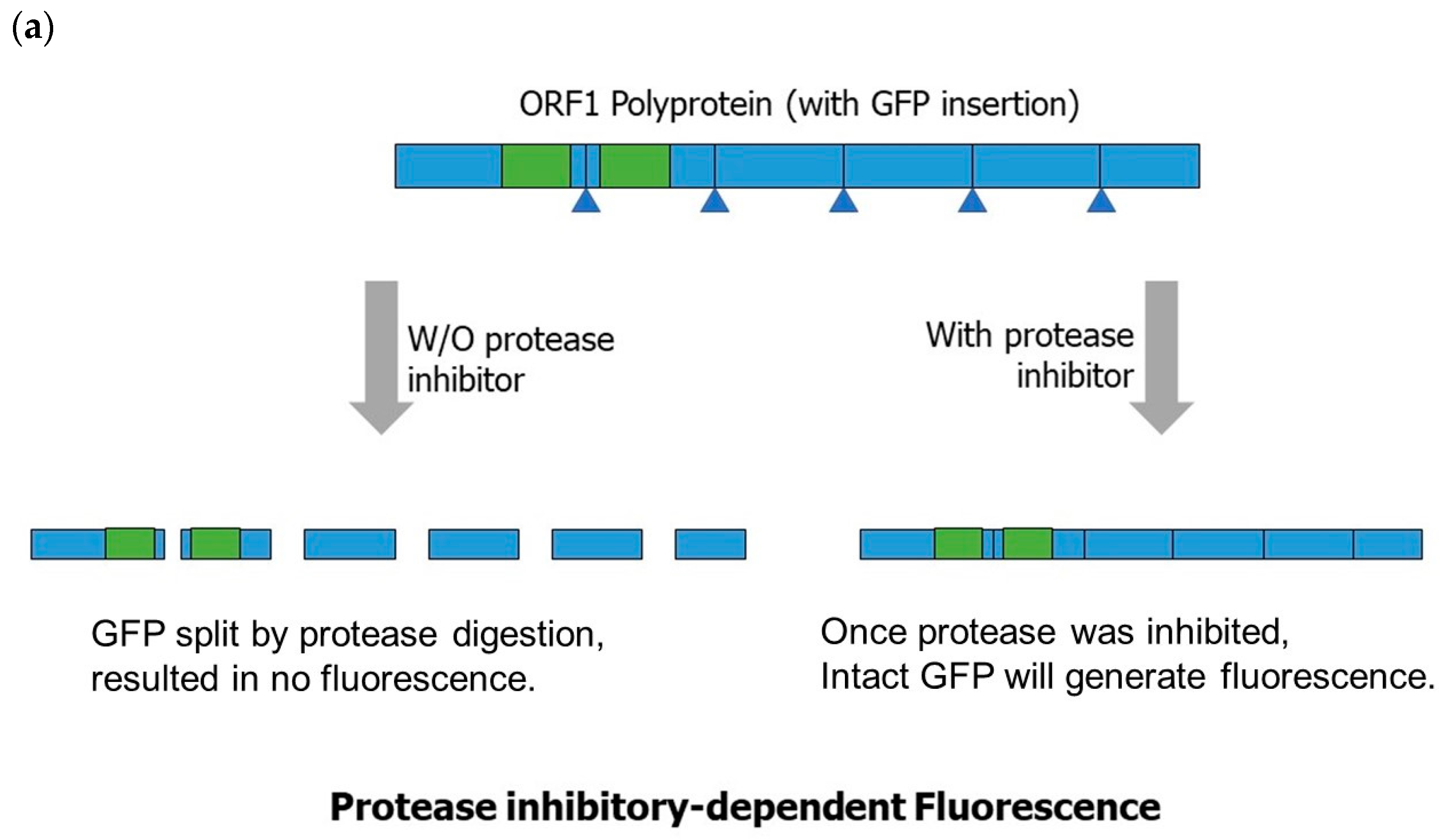
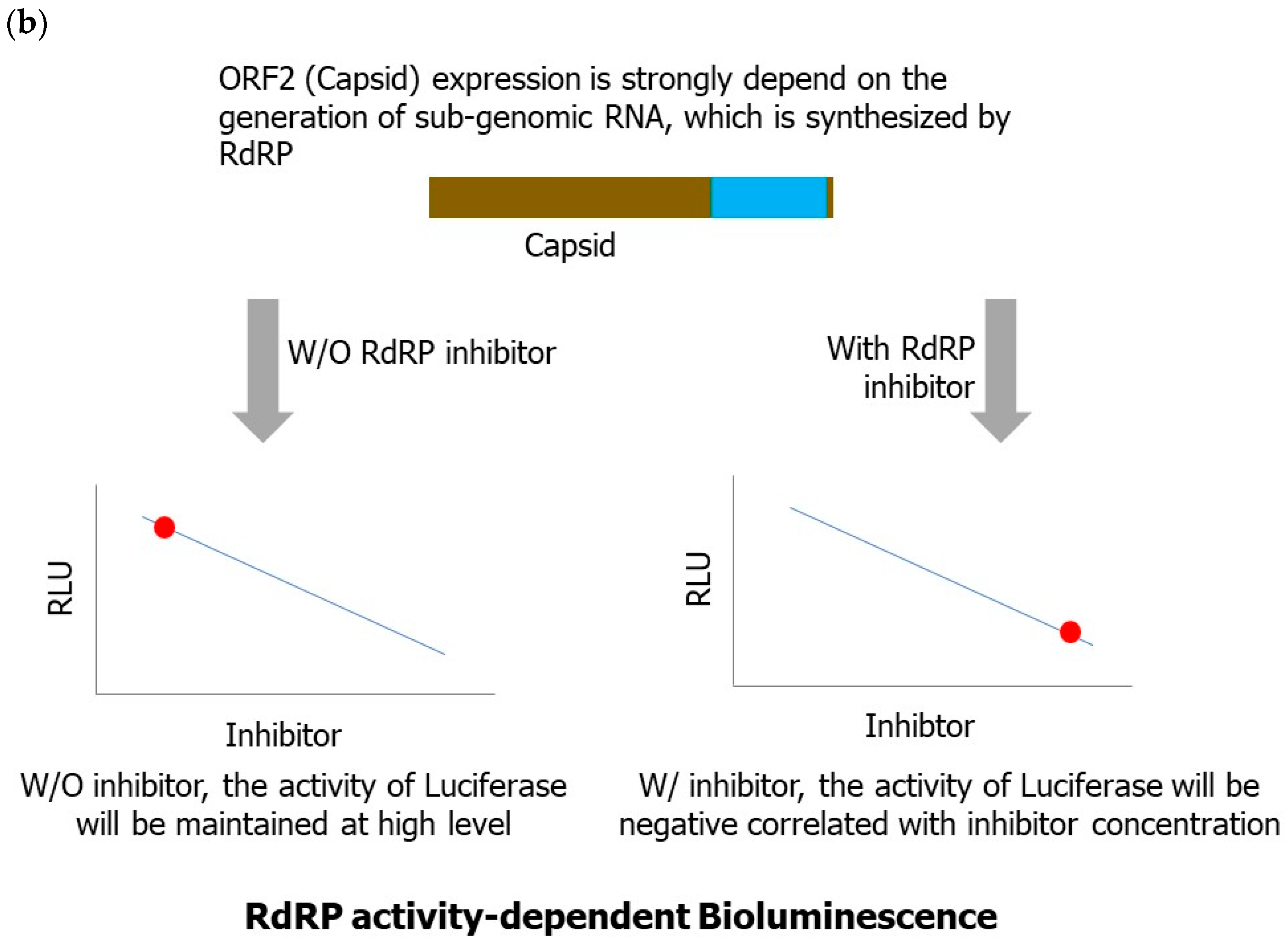
3.7.1. Reverse-Genetics-Based Norovirus Inhibitor Screening (NoVIS)
3.7.2. Current Status of Reverse Genetics for Human Norovirus
| Techniques | Detection Principles | Advantages | Disadvantages | Time | Cost | Application | Ref. |
|---|---|---|---|---|---|---|---|
| Nested PCR plus Sanger sequencing | Amplification of the VP1 region for Sanger sequencing | Highly sensitive, more high-quality and cost-effective than NGS | Time-consuming and labor-intensive | Moderately fast | Inexpensive | Provides sequence information and monitors evolution | [37] |
| RT-qPCR | PCR-based (nuclear acid amplification) | Highly sensitive; rapid results; compatible with various sample types | Lower resolution than sequencing; susceptible to contamination | Rapid | Inexpensive | Rapid detection and surveillance Known genogroup identification | [40] |
| RT-dPCR | PCR-based, microfluidic droplet platform | Improved sensitivity, precision, accuracy, and multiplexing compared to qPCR | More time-consuming and expensive than conventional PCR; technique expertise | Moderately fast | Inexpensive | Absolute quantification of norovirus | [49] |
| EIA | Antibody-based (VLP antibodies) | Rapid screening; simple and cost-effective Kits detect several different norovirus genotypes | Limited sensitivity and specificity Reliance on antibodies Escapes recognition from antigenic changes | Rapid | Inexpensive | Combined with RT-qPCR Negative samples should be confirmed a second time | [55] |
| NGS/TGS | Target and metagenomic sequencing | Highly sensitive and accurate High-throughput and comprehensive | Time-consuming, high-cost Requires high-quality samples | Lengthy | Expensive | Genetic diversity and evolution Recombination or synergism information | [57,58,59] |
| Aptamers | Synthetic oligonucleotides bind to target molecules (such as antibody-based molecules) | Faster and affordable More stable than antibodies Easy to modify and develop novel aptamers | Limited sensitivity and specificity Temperature affects binding capacity | Rapid | Inexpensive | High-throughput screening and diagnostics of emerging norovirus strains | [63,64,65] |
| Reverse genetics | Generates viral genomic RNA transcripts within a host cell | Eliminates interference from heterologous viruses Ensures the quality of RNA transcripts | Time-consuming and requires technical expertise Requires particular plasmid vectors and bacterial strains | Lengthy | Expensive | Viral genomic structure, virus–host interaction and pathogenesis, vaccine development | [75,76,77,78] |
4. Recent Advances in Norovirus Evolution and Recombination
4.1. Norovirus Evolution
4.2. Recombination of Norovirus
5. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dolin, R.; Blacklow, N.R.; DuPont, H.; Buscho, R.F.; Wyatt, R.G.; Kasel, J.A.; Hornick, R.; Chanock, R.M. Biological properties of Norwalk agent of acute infectious nonbacterial gastroenteritis. Proc. Soc. Exp. Biol. Med. 1972, 140, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Vinjé, J. Advances in laboratory methods for detection and typing of norovirus. J. Clin. Microbiol. 2015, 53, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Lambden, P.R.; Caul, E.O.; Ashley, C.R.; Clarke, I.N. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 1993, 259, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.W.; Shan, K.H.; Chan, P.K.S. Structure and Genotypes of Noroviruses. In The Norovirus; Academic Press: Cambridge, MA, USA, 2017; pp. 51–63. [Google Scholar] [CrossRef]
- Hardy, M.E. Norovirus protein structure and function. FEMS Microbiol. Lett. 2005, 253, 1–8. [Google Scholar] [CrossRef]
- Ettayebi, K.; Hardy, M.E. Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J. Virol. 2003, 77, 11790–11797. [Google Scholar] [CrossRef]
- Yen, J.B.; Chen, L.W.; Wei, L.H.; Hung, C.H.; Wang, S.S.; Lin, C.L.; Chang, P.J. Identification and Characterization of Human Norovirus NTPase Regions Required for Lipid Droplet Localization, Cellular Apoptosis, and Interaction with the Viral P22 Protein. Microbiol. Spectr. 2021, 9, e0042221. [Google Scholar] [CrossRef]
- Daughenbaugh, K.F.; Wobus, C.E.; Hardy, M.E. VPg of murine norovirus binds translation initiation factors in infected cells. Virol. J. 2006, 3, 33. [Google Scholar] [CrossRef]
- Chang, K.O.; Takahashi, D.; Prakash, O.; Kim, Y. Characterization and inhibition of norovirus proteases of genogroups I and II using a fluorescence resonance energy transfer assay. Virology 2012, 423, 125–133. [Google Scholar] [CrossRef]
- Eden, J.S.; Sharpe, L.J.; White, P.A.; Brown, A.J. Norovirus RNA-dependent RNA polymerase is phosphorylated by an important survival kinase, Akt. J. Virol. 2011, 85, 10894–10898. [Google Scholar] [CrossRef]
- Prasad, B.V.; Hardy, M.E.; Dokland, T.; Bella, J.; Rossmann, M.G.; Estes, M.K. X-ray crystallographic structure of the Norwalk virus capsid. Science 1999, 286, 287–290. [Google Scholar] [CrossRef]
- Marionneau, S.; Ruvoën, N.; Le Moullac-Vaidye, B.; Clement, M.; Cailleau-Thomas, A.; Ruiz-Palacois, G.; Huang, P.; Jiang, X.; Le Pendu, J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 2002, 122, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Hutson, A.M.; Atmar, R.L.; Marcus, D.M.; Estes, M.K. Norwalk virus-like particle hemagglutination by binding to h histo-blood group antigens. J. Virol. 2003, 77, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Hutson, A.M.; Estes, M.K.; Prasad, B.V. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. USA 2008, 105, 9175–9180. [Google Scholar] [CrossRef] [PubMed]
- Kroneman, A.; Vega, E.; Vennema, H.; Vinjé, J.; White, P.A.; Hansman, G.; Green, K.; Martella, V.; Katayama, K.; Koopmans, M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013, 158, 2059–2068. [Google Scholar] [CrossRef]
- La Rosa, G.; Fontana, S.; Di Grazia, A.; Iaconelli, M.; Pourshaban, M.; Muscillo, M. Molecular identification and genetic analysis of Norovirus genogroups I and II in water environments: Comparative analysis of different reverse transcription-PCR assays. Appl. Environ. Microbiol. 2007, 73, 4152–4161. [Google Scholar] [CrossRef]
- Patel, M.M.; Widdowson, M.A.; Glass, R.I.; Akazawa, K.; Vinjé, J.; Parashar, U.D. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 2008, 14, 1224–1231. [Google Scholar] [CrossRef]
- Siebenga, J.J.; Vennema, H.; Zheng, D.P.; Vinjé, J.; Lee, B.E.; Pang, X.L.; Ho, E.C.; Lim, W.; Choudekar, A.; Broor, S.; et al. Norovirus illness is a global problem: Emergence and spread of norovirus GII.4 variants, 2001–2007. J. Infect. Dis. 2009, 200, 802–812. [Google Scholar] [CrossRef]
- Lam, T.T.; Zhu, H.; Smith, D.K.; Guan, Y.; Holmes, E.C.; Pybus, O.G. The recombinant origin of emerging human norovirus GII.4/2008: Intra-genotypic exchange of the capsid P2 domain. J. Gen. Virol. 2012, 93, 817–822. [Google Scholar] [CrossRef]
- Phan, T.G.; Kuroiwa, T.; Kaneshi, K.; Ueda, Y.; Nakaya, S.; Nishimura, S.; Yamamoto, A.; Sugita, K.; Nishimura, T.; Yagyu, F.; et al. Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J. Med. Virol. 2006, 78, 971–978. [Google Scholar] [CrossRef]
- Lindesmith, L.C.; Donaldson, E.F.; Lobue, A.D.; Cannon, J.L.; Zheng, D.P.; Vinje, J.; Baric, R.S. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008, 5, e31. [Google Scholar] [CrossRef]
- Debbink, K.; Lindesmith, L.C.; Donaldson, E.F.; Costantini, V.; Beltramello, M.; Corti, D.; Swanstrom, J.; Lanzavecchia, A.; Vinjé, J.; Baric, R.S. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J. Infect. Dis. 2013, 208, 1877–1887. [Google Scholar] [CrossRef]
- Hoffmann, D.; Hutzenthaler, M.; Seebach, J.; Panning, M.; Umgelter, A.; Menzel, H.; Protzer, U.; Metzler, D. Norovirus GII.4 and GII.7 capsid sequences undergo positive selection in chronically infected patients. Infect. Genet. Evol. 2012, 12, 461–466. [Google Scholar] [CrossRef]
- Bull, R.A.; Eden, J.S.; Rawlinson, W.D.; White, P.A. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 2010, 6, e1000831. [Google Scholar] [CrossRef]
- Noda, M.; Fukuda, S.; Nishio, O. Statistical analysis of attack rate in norovirus foodborne outbreaks. Int. J. Food Microbiol. 2008, 122, 216–220. [Google Scholar] [CrossRef]
- Kendra, J.A.; Tohma, K.; Ford-Siltz, L.A.; Lepore, C.J.; Parra, G.I. Antigenic cartography reveals complexities of genetic determinants that lead to antigenic differences among pandemic GII.4 noroviruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2015874118. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.J.; Gray, J.J.; Gallimore, C.I.; Xerry, J.; Iturriza-Gómara, M. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS ONE 2008, 3, e1485. [Google Scholar] [CrossRef]
- Motoya, T.; Nagasawa, K.; Matsushima, Y.; Nagata, N.; Ryo, A.; Sekizuka, T.; Yamashita, A.; Kuroda, M.; Morita, Y.; Suzuki, Y.; et al. Molecular Evolution of the VP1 Gene in Human Norovirus GII.4 Variants in 1974–2015. Front. Microbiol. 2017, 8, 2399. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; White, P.A. Mechanisms of GII.4 norovirus evolution. Trends Microbiol. 2011, 19, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Tohma, K.; Lepore, C.J.; Gao, Y.; Ford-Siltz, L.A.; Parra, G.I. Population Genomics of GII.4 Noroviruses Reveal Complex Diversification and New Antigenic Sites Involved in the Emergence of Pandemic Strains. mBio 2019, 10, e02202-19. [Google Scholar] [CrossRef]
- Bok, K.; Abente, E.J.; Realpe-Quintero, M.; Mitra, T.; Sosnovtsev, S.V.; Kapikian, A.Z.; Green, K.Y. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 2009, 83, 11890–11901. [Google Scholar] [CrossRef]
- Mathijs, E.; Muylkens, B.; Mauroy, A.; Ziant, D.; Delwiche, T.; Thiry, E. Experimental evidence of recombination in murine noroviruses. J. Gen. Virol. 2010, 91, 2723–2733. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Tanaka, M.M.; White, P.A. Norovirus recombination. J. Gen. Virol. 2007, 88, 3347–3359. [Google Scholar] [CrossRef] [PubMed]
- Mahar, J.E.; Bok, K.; Green, K.Y.; Kirkwood, C.D. The importance of intergenic recombination in norovirus GII.3 evolution. J. Virol. 2013, 87, 3687–3698. [Google Scholar] [CrossRef]
- Bull, R.A.; Hansman, G.S.; Clancy, L.E.; Tanaka, M.M.; Rawlinson, W.D.; White, P.A. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 2005, 11, 1079–1085. [Google Scholar] [CrossRef]
- Saito, M.; Tsukagoshi, H.; Ishigaki, H.; Aso, J.; Ishii, H.; Okayama, K.; Ryo, A.; Ishioka, T.; Kuroda, M.; Saruki, N.; et al. Molecular evolution of the capsid (VP1) region in human norovirus genogroup II genotype 3. Heliyon 2020, 6, e03835. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Kageyama, T.; Fukushi, S.; Hoshino, F.B.; Shinohara, M.; Uchida, K.; Natori, K.; Takeda, N.; Katayama, K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 2002, 100, 107–114. [Google Scholar] [CrossRef]
- Schmid, M.; Oehme, R.; Schalasta, G.; Brockmann, S.; Kimmig, P.; Enders, G. Fast detection of Noroviruses using a real-time PCR assay and automated sample preparation. BMC Infect. Dis. 2004, 4, 15. [Google Scholar] [CrossRef]
- Hoehne, M.; Schreier, E. Detection of Norovirus genogroup I and II by multiplex real-time RT- PCR using a 3′-minor groove binder-DNA probe. BMC Infect. Dis. 2006, 6, 69. [Google Scholar] [CrossRef]
- Ramanan, P.; Espy, M.J.; Khare, R.; Binnicker, M.J. Detection and differentiation of norovirus genogroups I and II from clinical stool specimens using real-time PCR. Diagn. Microbiol. Infect. Dis. 2017, 87, 325–327. [Google Scholar] [CrossRef]
- Rutjes, S.A.; van den Berg, H.H.; Lodder, W.J.; de Roda Husman, A.M. Real-time detection of noroviruses in surface water by use of a broadly reactive nucleic acid sequence-based amplification assay. Appl. Environ. Microbiol. 2006, 72, 5349–5358. [Google Scholar] [CrossRef]
- Park, Y.; Cho, Y.H.; Jee, Y.; Ko, G. Immunomagnetic separation combined with real-time reverse transcriptase PCR assays for detection of norovirus in contaminated food. Appl. Environ. Microbiol. 2008, 74, 4226–4230. [Google Scholar] [CrossRef]
- Rodríguez, R.A.; Thie, L.; Gibbons, C.D.; Sobsey, M.D. Reducing the effects of environmental inhibition in quantitative real-time PCR detection of adenovirus and norovirus in recreational seawaters. J. Virol. Methods 2012, 181, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Glass, R.I.; Parashar, U.D.; Estes, M.K. Norovirus gastroenteritis. N. Engl. J. Med. 2009, 361, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Jothikumar, N.; Lowther, J.A.; Henshilwood, K.; Lees, D.N.; Hill, V.R.; Vinjé, J. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl. Environ. Microbiol. 2005, 71, 1870–1875. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, J.; Zhang, S.; Yang, S.; Wang, X.; Han, Y.; Shen, Z.; Xu, X. Simultaneous quantification of hepatitis A virus and norovirus genogroup I and II by triplex droplet digital PCR. Food Microbiol. 2022, 103, 103933. [Google Scholar] [CrossRef]
- Gyawali, P.; Croucher, D.; Hewitt, J. Preliminary evaluation of BioFire FilmArray(®) Gastrointestinal Panel for the detection of noroviruses and other enteric viruses from wastewater and shellfish. Environ. Sci. Pollut. Res. Int. 2018, 25, 27657–27661. [Google Scholar] [CrossRef]
- Buss, S.N.; Leber, A.; Chapin, K.; Fey, P.D.; Bankowski, M.J.; Jones, M.K.; Rogatcheva, M.; Kanack, K.J.; Bourzac, K.M. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J. Clin. Microbiol. 2015, 53, 915–925. [Google Scholar] [CrossRef]
- Monteiro, S.; Santos, R. Nanofluidic digital PCR for the quantification of Norovirus for water quality assessment. PLoS ONE 2017, 12, e0179985. [Google Scholar] [CrossRef]
- Persson, S.; Eriksson, R.; Lowther, J.; Ellström, P.; Simonsson, M. Comparison between RT droplet digital PCR and RT real-time PCR for quantification of noroviruses in oysters. Int. J. Food Microbiol. 2018, 284, 73–83. [Google Scholar] [CrossRef]
- Polo, D.; Schaeffer, J.; Fournet, N.; Le Saux, J.C.; Parnaudeau, S.; McLeod, C.; Le Guyader, F.S. Digital PCR for Quantifying Norovirus in Oysters Implicated in Outbreaks, France. Emerg. Infect. Dis. 2016, 22, 2189–2191. [Google Scholar] [CrossRef]
- Tan, D.M.; Lyu, S.L.; Liu, W.; Zeng, X.Y.; Lan, L.; Qu, C.; Zhuge, S.Y.; Zhong, Y.X.; Xie, Y.H.; Li, X.G. Utility of Droplet Digital PCR Assay for Quantitative Detection of Norovirus in Shellfish, from Production to Consumption in Guangxi, China. Biomed. Environ. Sci. 2018, 31, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Xiang, X.; Xue, L.; Cai, W.; Gao, J.; Yang, J.; Liang, Y.; Wang, L.; Chen, M.; Pang, R.; et al. Development of a novel RAA-based microfluidic chip for absolute quantitative detection of human norovirus. Microchem. J. 2021, 164, 106050. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, W.; Li, H.; Li, N.; Lin, J.-M. Advances in droplet digital polymerase chain reaction on microfluidic chips. Lab Chip 2023, 23, 1258–1278. [Google Scholar] [CrossRef]
- Costantini, V.; Grenz, L.; Fritzinger, A.; Lewis, D.; Biggs, C.; Hale, A.; Vinjé, J. Diagnostic Accuracy and Analytical Sensitivity of IDEIA Norovirus Assay for Routine Screening of Human Norovirus. J. Clin. Microbiol. 2010, 48, 2770–2778. [Google Scholar] [CrossRef]
- Chan, M.C.W.; Sung, J.J.Y.; Lam, R.K.Y.; Chan, P.K.S.; Lee, N.L.S.; Lai, R.W.M.; Leung, W.K. Fecal Viral Load and Norovirus-associated Gastroenteritis. Emerg. Infect. Dis. 2006, 12, 1278–1280. [Google Scholar] [CrossRef]
- Cotten, M.; Petrova, V.; Phan, M.V.; Rabaa, M.A.; Watson, S.J.; Ong, S.H.; Kellam, P.; Baker, S. Deep sequencing of norovirus genomes defines evolutionary patterns in an urban tropical setting. J. Virol. 2014, 88, 11056–11069. [Google Scholar] [CrossRef]
- Brown, J.R.; Roy, S.; Ruis, C.; Yara Romero, E.; Shah, D.; Williams, R.; Breuer, J. Norovirus Whole-Genome Sequencing by SureSelect Target Enrichment: A Robust and Sensitive Method. J. Clin. Microbiol. 2016, 54, 2530–2537. [Google Scholar] [CrossRef] [PubMed]
- Fonager, J.; Stegger, M.; Rasmussen, L.D.; Poulsen, M.W.; Rønn, J.; Andersen, P.S.; Fischer, T.K. A universal primer-independent next-generation sequencing approach for investigations of norovirus outbreaks and novel variants. Sci. Rep. 2017, 7, 813. [Google Scholar] [CrossRef] [PubMed]
- Flint, A.; Reaume, S.; Harlow, J.; Hoover, E.; Weedmark, K.; Nasheri, N. Genomic analysis of human noroviruses using combined Illumina-Nanopore data. Virus Evol. 2021, 7, veab079. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of Aptamers in Virus Detection and Antiviral Therapy. Front. Microbiol. 2019, 10, 1462. [Google Scholar] [CrossRef] [PubMed]
- Schilling-Loeffler, K.; Rodriguez, R.; Williams-Woods, J. Target Affinity and Structural Analysis for a Selection of Norovirus Aptamers. Int. J. Mol. Sci. 2021, 22, 8868. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Escudero-Abarca, B.I.; Suh, S.H.; Jaykus, L.A. Generation and characterization of nucleic acid aptamers targeting the capsid P domain of a human norovirus GII.4 strain. J. Biotechnol. 2015, 209, 41–49. [Google Scholar] [CrossRef]
- Beier, R.; Pahlke, C.; Quenzel, P.; Henseleit, A.; Boschke, E.; Cuniberti, G.; Labudde, D. Selection of a DNA aptamer against norovirus capsid protein VP1. FEMS Microbiol. Lett. 2014, 351, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Abarca, B.I.; Suh, S.H.; Moore, M.D.; Dwivedi, H.P.; Jaykus, L.-A. Selection, Characterization and Application of Nucleic Acid Aptamers for the Capture and Detection of Human Norovirus Strains. PLoS ONE 2014, 9, e106805. [Google Scholar] [CrossRef]
- Conrad, R.C.; Giver, L.; Tian, Y.; Ellington, A.D. In vitro selection of nucleic acid aptamers that bind proteins. Methods Enzymol. 1996, 267, 336–367. [Google Scholar] [CrossRef] [PubMed]
- Kulbachinskiy, A.V. Methods for selection of aptamers to protein targets. Biochemistry 2007, 72, 1505–1518. [Google Scholar] [CrossRef]
- Li, H.-Y.; Jia, W.-N.; Li, X.-Y.; Zhang, L.; Liu, C.; Wu, J. Advances in detection of infectious agents by aptamer-based technologies. Emerg. Microbes Infect. 2020, 9, 1671–1681. [Google Scholar] [CrossRef]
- Weng, X.; Neethirajan, S. Aptamer-based fluorometric determination of norovirus using a paper-based microfluidic device. Microchim. Acta 2017, 184, 4545–4552. [Google Scholar] [CrossRef]
- Asanaka, M.; Atmar, R.L.; Ruvolo, V.; Crawford, S.E.; Neill, F.H.; Estes, M.K. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. USA 2005, 102, 10327–10332. [Google Scholar] [CrossRef]
- Katayama, K.; Hansman, G.S.; Oka, T.; Ogawa, S.; Takeda, N. Investigation of norovirus replication in a human cell line. Arch. Virol. 2006, 151, 1291–1308. [Google Scholar] [CrossRef] [PubMed]
- Robilotti, E.; Deresinski, S.; Pinsky, B.A. Norovirus. Clin. Microbiol. Rev. 2015, 28, 134–164. [Google Scholar] [CrossRef]
- Chaudhry, Y.; Nayak, A.; Bordeleau, M.E.; Tanaka, J.; Pelletier, J.; Belsham, G.J.; Roberts, L.O.; Goodfellow, I.G. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 2006, 281, 25315–25325. [Google Scholar] [CrossRef] [PubMed]
- Arias, A.; Ureña, L.; Thorne, L.; Yunus, M.A.; Goodfellow, I. Reverse genetics mediated recovery of infectious murine norovirus. J. Vis. Exp. 2012, 64, e4145. [Google Scholar] [CrossRef]
- Hulo, C.; de Castro, E.; Masson, P.; Bougueleret, L.; Bairoch, A.; Xenarios, I.; Le Mercier, P. ViralZone: A knowledge resource to understand virus diversity. Nucleic Acids Res. 2011, 39, D576–D582. [Google Scholar] [CrossRef] [PubMed]
- Almazán, F.; Sola, I.; Zuñiga, S.; Marquez-Jurado, S.; Morales, L.; Becares, M.; Enjuanes, L. Coronavirus reverse genetic systems: Infectious clones and replicons. Virus Res. 2014, 189, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Aubry, F.; Nougairède, A.; Gould, E.A.; de Lamballerie, X. Flavivirus reverse genetic systems, construction techniques and applications: A historical perspective. Antivir. Res. 2015, 114, 67–85. [Google Scholar] [CrossRef]
- Hall, R.A.; Nisbet, D.J.; Pham, K.B.; Pyke, A.T.; Smith, G.A.; Khromykh, A.A. DNA vaccine coding for the full-length infectious Kunjin virus RNA protects mice against the New York strain of West Nile virus. Proc. Natl. Acad. Sci. USA 2003, 100, 10460–10464. [Google Scholar] [CrossRef]
- Cockrell, A.S.; Beall, A.; Yount, B.; Baric, R. Efficient Reverse Genetic Systems for Rapid Genetic Manipulation of Emergent and Preemergent Infectious Coronaviruses. Methods Mol. Biol. 2017, 1602, 59–81. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, S.; Zhang, X.; Su, L.; Ni, J.; Zhang, Y.; Fang, L. Recent insights into reverse genetics of norovirus. Virus Res. 2023, 325, 199046. [Google Scholar] [CrossRef]
- Katayama, K.; Murakami, K.; Sharp, T.M.; Guix, S.; Oka, T.; Takai-Todaka, R.; Nakanishi, A.; Crawford, S.E.; Atmar, R.L.; Estes, M.K. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc. Natl. Acad. Sci. USA 2014, 111, E4043–E4052. [Google Scholar] [CrossRef]
- Oliveira, L.M.; Blawid, R.; Orílio, A.F.; Andrade, B.Y.G.; Souza, A.C.A.; Nagata, T. Development of an infectious clone and replicon system of norovirus GII.4. J. Virol. Methods 2018, 258, 49–53. [Google Scholar] [CrossRef] [PubMed]
- White, P.A. Evolution of norovirus. Clin. Microbiol. Infect. 2014, 20, 741–745. [Google Scholar] [CrossRef]
- Yu, J.M.; Liang, Z.Y.; Guo, K.; Sun, X.M.; Zhang, Q.; Dong, Y.J.; Duan, Z.J. Intra-Host Evolution of Norovirus GII.4 in a Chronic Infected Patient with Hematopoietic Stem Cell Transplantation. Front. Microbiol. 2020, 11, 375. [Google Scholar] [CrossRef] [PubMed]
- van Beek, J.; de Graaf, M.; Smits, S.; Schapendonk, C.M.E.; Verjans, G.; Vennema, H.; van der Eijk, A.A.; Phan, M.V.T.; Cotten, M.; Koopmans, M. Whole-Genome Next-Generation Sequencing to Study Within-Host Evolution of Norovirus (NoV) Among Immunocompromised Patients with Chronic NoV Infection. J. Infect. Dis. 2017, 216, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
- Tohma, K.; Saito, M.; Pajuelo, M.J.; Mayta, H.; Zimic, M.; Lepore, C.J.; Ford-Siltz, L.A.; Gilman, R.H.; Parra, G.I. Viral intra-host evolution in immunocompetent children contributes to human norovirus diversification at the global scale. Emerg. Microbes Infect. 2021, 10, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Siebenga, J.J.; Beersma, M.F.; Vennema, H.; van Biezen, P.; Hartwig, N.J.; Koopmans, M. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: A model for in vivo molecular evolution. J. Infect. Dis. 2008, 198, 994–1001. [Google Scholar] [CrossRef]
- Kobayashi, M.; Matsushima, Y.; Motoya, T.; Sakon, N.; Shigemoto, N.; Okamoto-Nakagawa, R.; Nishimura, K.; Yamashita, Y.; Kuroda, M.; Saruki, N.; et al. Molecular evolution of the capsid gene in human norovirus genogroup II. Sci. Rep. 2016, 6, 29400. [Google Scholar] [CrossRef]
- Carlsson, B.; Lindberg, A.M.; Rodriguez-Díaz, J.; Hedlund, K.O.; Persson, B.; Svensson, L. Quasispecies dynamics and molecular evolution of human norovirus capsid P region during chronic infection. J. Gen. Virol. 2009, 90, 432–441. [Google Scholar] [CrossRef]
- Etherington, G.J.; Dicks, J.; Roberts, I.N. High throughput sequence analysis reveals hitherto unreported recombination in the genus Norovirus. Virology 2006, 345, 88–95. [Google Scholar] [CrossRef]
- Rohayem, J.; Münch, J.; Rethwilm, A. Evidence of recombination in the norovirus capsid gene. J. Virol. 2005, 79, 4977–4990. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, T.; Wang, H.; Shen, H. Intergenotype Recombination among New Norovirus GII.4 Variants in the Complete Genome. Intervirology 2017, 60, 138–143. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.-C.; Bai, G.-H.; Lin, P.-C.; Chen, C.-Y.; Hsu, Y.-H.; Lee, Y.-C.; Chen, S.-Y. Molecular and Genetics-Based Systems for Tracing the Evolution and Exploring the Mechanisms of Human Norovirus Infections. Int. J. Mol. Sci. 2023, 24, 9093. https://doi.org/10.3390/ijms24109093
Lin S-C, Bai G-H, Lin P-C, Chen C-Y, Hsu Y-H, Lee Y-C, Chen S-Y. Molecular and Genetics-Based Systems for Tracing the Evolution and Exploring the Mechanisms of Human Norovirus Infections. International Journal of Molecular Sciences. 2023; 24(10):9093. https://doi.org/10.3390/ijms24109093
Chicago/Turabian StyleLin, Sheng-Chieh, Geng-Hao Bai, Pei-Chun Lin, Chung-Yung Chen, Yi-Hsiang Hsu, Yuan-Chang Lee, and Shih-Yen Chen. 2023. "Molecular and Genetics-Based Systems for Tracing the Evolution and Exploring the Mechanisms of Human Norovirus Infections" International Journal of Molecular Sciences 24, no. 10: 9093. https://doi.org/10.3390/ijms24109093
APA StyleLin, S.-C., Bai, G.-H., Lin, P.-C., Chen, C.-Y., Hsu, Y.-H., Lee, Y.-C., & Chen, S.-Y. (2023). Molecular and Genetics-Based Systems for Tracing the Evolution and Exploring the Mechanisms of Human Norovirus Infections. International Journal of Molecular Sciences, 24(10), 9093. https://doi.org/10.3390/ijms24109093







