The Mechanism of Tigecycline Resistance in Acinetobacter baumannii Revealed by Proteomic and Genomic Analysis
Abstract
1. Introduction
2. Results
2.1. Selection of Tigecycline-Resistant and Tigecycline-Susceptible Strains from Tigecycline-Susceptible and Tigecycline-Resistant Strains, Respectively
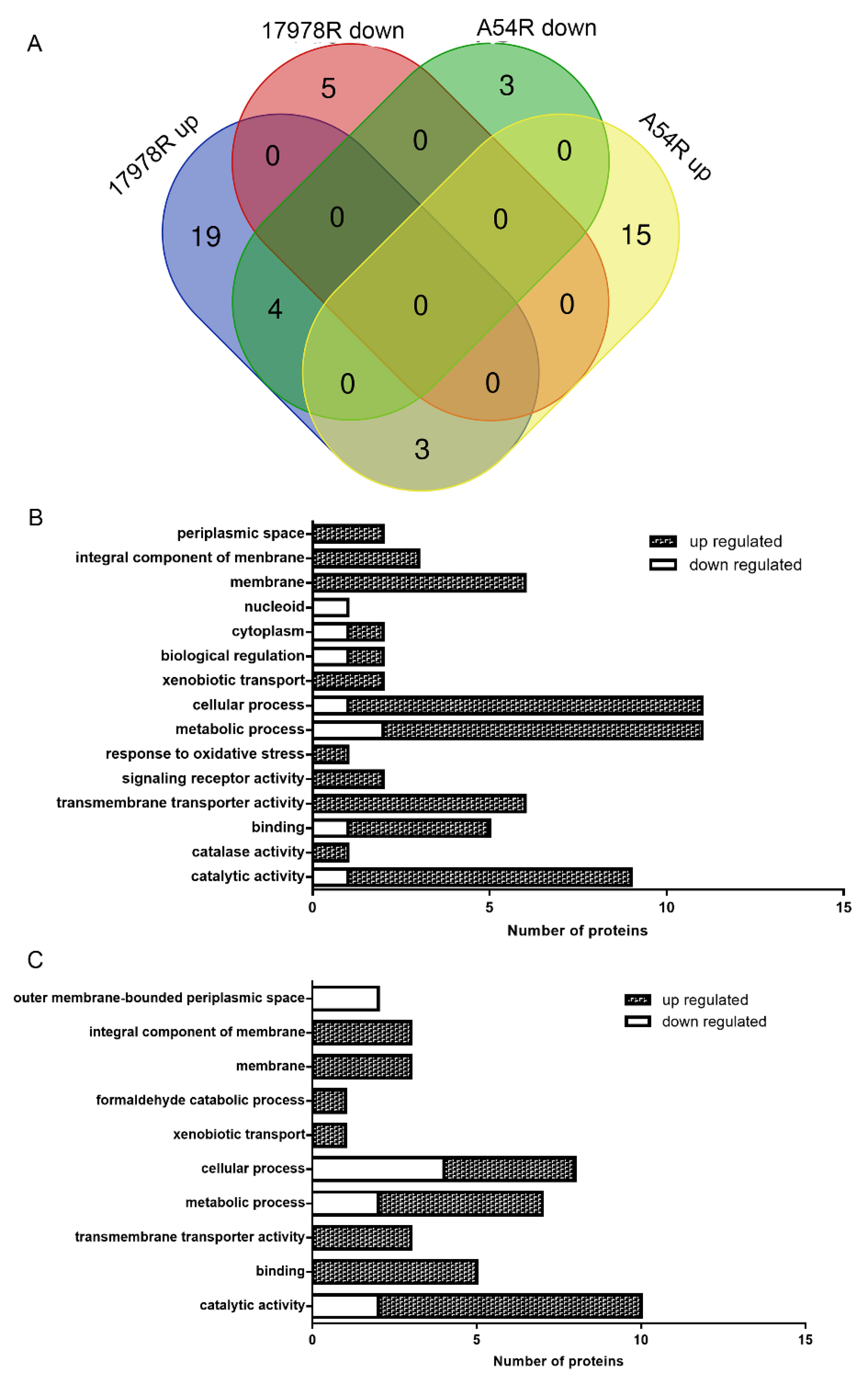
2.2. Proteomic Variations Associated with Tigecycline Resistance
2.3. Efflux Pump Plays Important Roles in Tigecycline Resistance
2.4. Genomic Variation of Tigecycline-Susceptible and Tigecycline-Resistant Strains before and after Selection
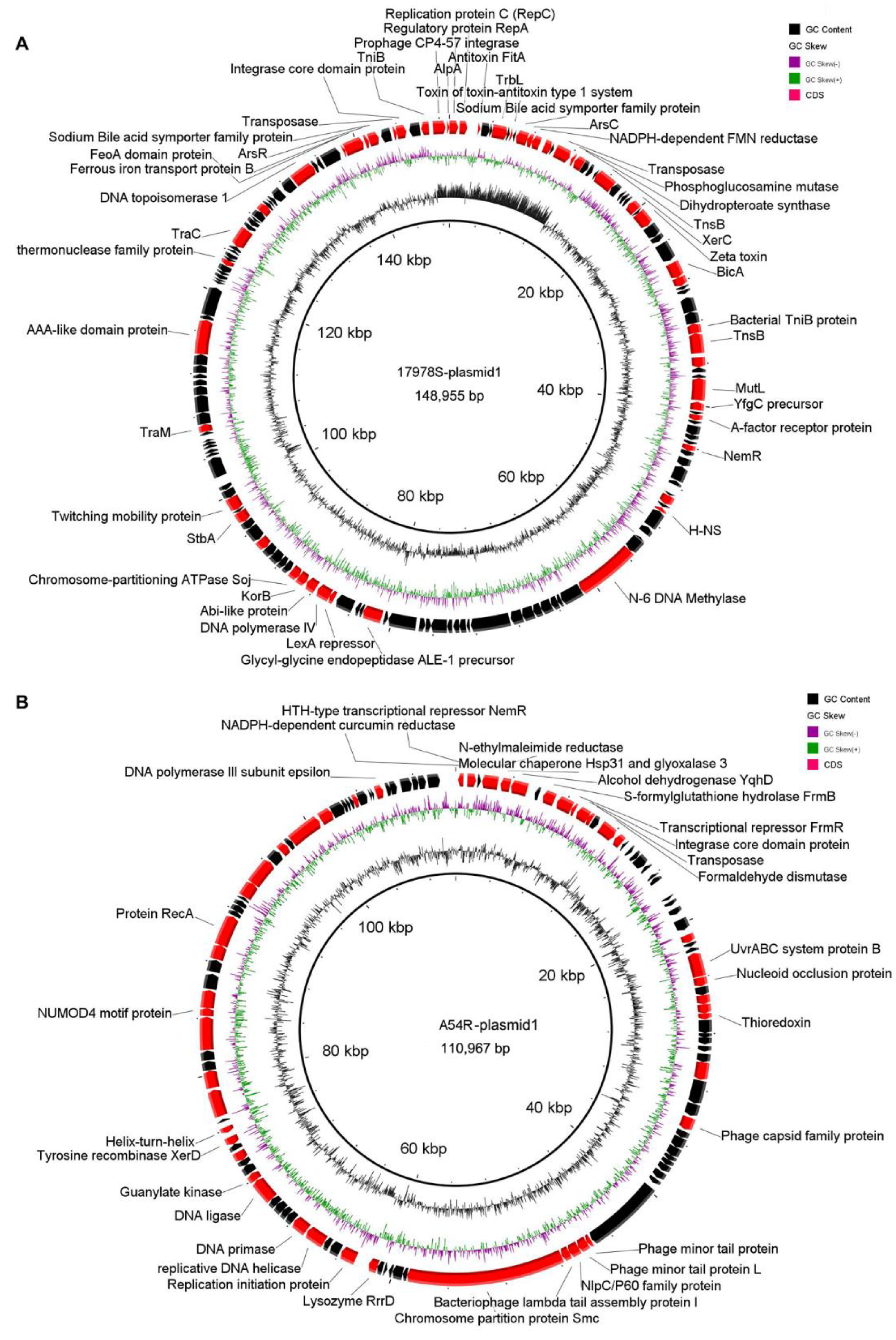
2.4.1. Plasmid Variation before and after Selection
2.4.2. Chromosome Variation before and after Selection
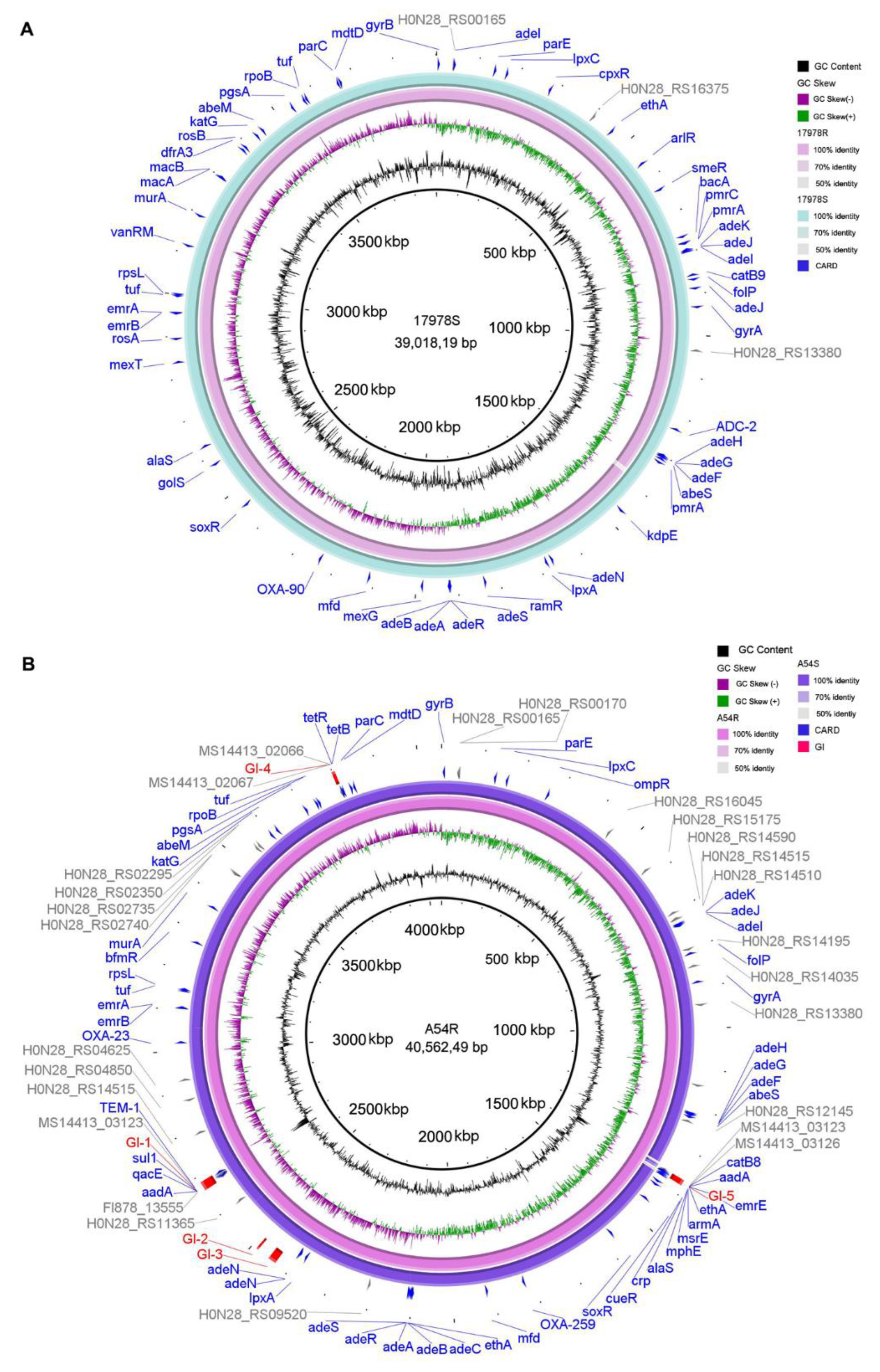
2.5. Variation of Antibiotic Resistance Genes, Virulence Factors, and Genomic Islands
2.6. Variation of IS Elements before and after Selection
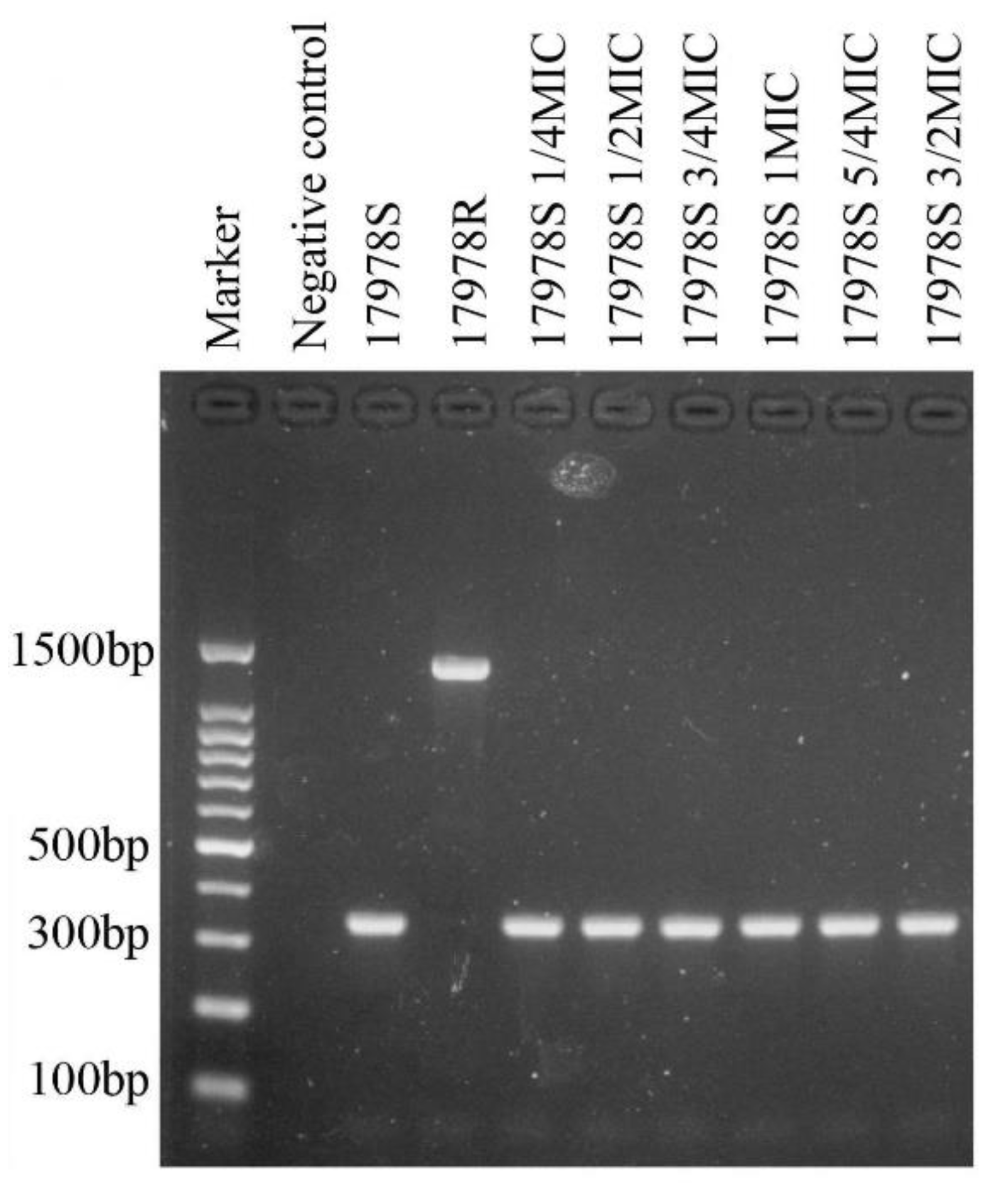
2.7. Deletion of Hns Decreased Tigecycline Sensitivity in 17978S
2.8. Proteomic Analysis of 17978S and Hns-Deleted Strains
2.9. Complement of AcrR to 17978R Reduces Tigecycline Resistance
3. Discussion
4. Materials and Methods
4.1. Bacteria Strains and Selection of Tigecycline Resistance Varied Strains
4.2. Stability of the Antibiotic Resistance of the Selected Strains
4.3. Protein Sample Preparation
4.4. Protein Digestion, iTRAQ Labeling and LC-MS/MS
4.5. Downstream Processing and Analysis of Proteomics Data
4.6. Efflux Pump Inhibition Experiment
4.7. Whole Genome Sequencing
4.8. Gene Knockout and Complement
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vázquez-López, R.; Solano-Gálvez, S.G.; Juárez Vignon-Whaley, J.J.; Jiménez-Rojas, V.; Torres-González, P.; Garza-Ramos, U.; Silva-Sánchez, J. Acinetobacter baumannii Resistance: A Real Challenge for Clinicians. Antibiotics 2020, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Tankovic, J.; Legrand, P.; De Gatines, G.; Chemineau, V.; Brun-Buisson, C.; Duval, J.; Courvalin, P. Characterization of a Hospital Outbreak of Imipenem-Resistant Acinetobacter baumannii by Phenotypic and Genotypic Typing Methods. J. Clin. Microbiol. 1994, 32, 2677–2681. [Google Scholar] [CrossRef] [PubMed]
- Volkers, G.; Palm, G.J.; Weiss, M.S.; Wright, G.D.; Hinrichs, W. Structural Basis for a New Tetracycline Resistance Mechanism Relying on the TetX Monooxygenase. FEBS Lett. 2011, 585, 1061–1066. [Google Scholar] [CrossRef]
- Ruzin, A.; Keeney, D.; Bradford, P.A. AdeABC Multidrug Efflux Pump Is Associated with Decreased Susceptibility to Tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii Complex. J. Antimicrob. Chemother. 2007, 59, 1001–1004. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, X.; Chen, L.; Han, L.; Guo, T.; Chen, Y.; Lou, T.; Wu, N.; Yu, F. In Vitro Activity of Various Antibiotics in Combination with Tigecycline Against Acinetobacter baumannii: A Systematic Review and Meta-Analysis. Microb. Drug Resist. 2017, 23, 982–993. [Google Scholar] [CrossRef]
- Pournaras, S.; Koumaki, V.; Gennimata, V.; Kouskouni, E.; Dimitroulia, E.; Tsakris, A. In Vitro Activity of Tigecycline Against Acinetobacter baumannii: Global Epidemiology and Resistance Mechanisms. Adv. Exp. Med. Biol. 2016, 897, 1–14. [Google Scholar]
- Ruzin, A.; Immermann, F.W.; Bradford, P.A. RT-PCR and Statistical Analyses of adeABC Expression in Clinical Isolates of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex. Microb. Drug Resist. 2010, 16, 87–89. [Google Scholar] [CrossRef]
- Rumbo, C.; Gato, E.; López, M.; Ruiz de Alegría, C.; Fernández-Cuenca, F.; Martínez-Martínez, L.; Vila, J.; Pachón, J.; Cisneros, J.M.; Rodríguez-Baño, J.; et al. Contribution of Efflux Pumps, Porins, and β-Lactamases to Multidrug Resistance in Clinical Isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013, 57, 5247–5257. [Google Scholar] [CrossRef]
- Yoon, E.J.; Courvalin, P.; Grillot-Courvalin, C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 2013, 57, 2989–2995. [Google Scholar] [CrossRef]
- Deng, M.; Zhu, M.H.; Li, J.J.; Bi, S.; Sheng, Z.K.; Hu, F.S.; Zhang, J.P.; Chen, W.; Zhang, X.F.; Wang, L.X. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob. Agents Chemother. 2014, 58, 297–303. [Google Scholar] [CrossRef]
- Coyne, S.; Guigon, G.; Courvalin, P.; Perichon, B. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob. Agents Chemother. 2010, 54, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, D.; Lv, Y.; Cao, X.; Jia, J.; Chen, Y.; Chen, Y.; Wang, C.; Liu, X.; Liu, H.; et al. Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 2019, 64, e01393-19. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Li, X.; Zhou, H.; Jiang, Y.; Chen, Y.; Hua, X.; Li, M.; Li, J.; Yu, Y. Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J. Antimicrob. Chemother. 2014, 69, 72–76. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Ji, J.; Chen, Y.; Jiang, Y.; Jiang, X.; Chen, Y.; Li, J.; Yu, Y. Tigecycline resistance in Acinetobacter baumannii mediated by frameshift mutation in plsC, encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 625–631. [Google Scholar] [CrossRef]
- Li, X.; Quan, J.; Yang, Y.; Ji, J.; Liu, L.; Fu, Y.; Hua, X.; Chen, Y.; Jiang, Y.; Yu, Y. Abrp, a new gene, confers reduced susceptibility to tetracycline, glycylcine, chloramphenicol and fosfomycin classes in Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1371–1375. [Google Scholar] [CrossRef]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Quan, J.; Hua, X.; Zhang, X.; Zhang, H.; Cui, L.; Zhou, Y. Complete Genome Sequence of Acinetobacter baumannii XH386 (ST208), a Multi-Drug Resistant Bacteria Isolated from Pediatric Hospital in China. Genom. Data 2016, 7, 269–274. [Google Scholar] [CrossRef]
- Osman, D.; Piergentili, C.; Chen, J.; Sayer, L.N.; Usón, I.; Huggins, T.G.; Robinson, N.J.; Pohl, E.; Daldal, F. The Effectors and Sensory Sites of Formaldehyde-Responsive Regulator FrmR and Metal-Sensing Variant. J. Biol. Chem. 2016, 291, 19502–19516. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Ambrose, S.J.; Hall, R.M. A Large Conjugative Acinetobacter baumannii Plasmid Carrying the sul2 Sul-phonamide and strAB Streptomycin Resistance Genes. Plasmid 2016, 87–88, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Sadler, M.; Mormile, M.R.; Frank, R.L. Characterization of the IS200/IS605 Insertion Sequence Family in Halanaerobium Hydrogeniformans. Genes 2020, 11, 529. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Tang, H. ISEScan: Automated Identification of Insertion Sequence Elements in Prokaryotic Genomes. Bioinformatics 2017, 33, 3340–3347. [Google Scholar] [CrossRef] [PubMed]
- Odenholt, I. Pharmacodynamic Effects of Subinhibitory Antibiotic Concentrations. Int. J. Antimicrob. Agents 2001, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, D.; Le, C.; Pimentel, C.; Lee, S.; Lee, J.; Song, W.; Skoglund, E.; Unemo, M.; Ahn, S. Histone-Like Nucleoid-Structuring Protein (H-NS) Regulatory Role in Antibiotic Resistance in Acinetobacter baumannii. Sci. Rep. 2021, 11, 18414. [Google Scholar] [CrossRef]
- Le, C.; Pimentel, C.; Tuttobene, M.R.; Kim, S.; Unemo, M.; Ahn, S. Involvement of the Histone-Like Nucleoid Structuring Protein (H-NS) in Acinetobacter baumannii’s Natural Transformation. Pathogens 2021, 10, 1048. [Google Scholar] [CrossRef]
- Lippa, A.M.; Gebhardt, M.J.; Dove, S.L. H-NS-like proteins in Pseudomonas aeruginosa coordinately silence intragenic transcription. Mol. Microbiol. 2021, 115, 1138–1151. [Google Scholar] [CrossRef] [PubMed]
- Samantarrai, D.; Lakshman Sagar, A.; Gudla, R.; Panda, A.K.; Mohapatra, S.; Nayak, B.; Patra, J.K. TonB-Dependent Transporters in Sphingomonads: Unraveling Their Distribution and Function in Environmental Adaptation. Microorganisms 2020, 8, 3. [Google Scholar] [CrossRef]
- Hatefi Oskuei, R.; Darvish Alipour Astaneh, S.; Rasooli, I. A conserved region of Acinetobacter trimeric autotransporter adhesion, Ata, provokes suppression of Acinetobacter baumannii virulence. Arch. Microbiol. 2021, 203, 3483–3493. [Google Scholar] [CrossRef]
- Deng, W.; Li, C.; Xie, J. The underlying mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell Signal. 2013, 25, 1608–1613. [Google Scholar] [CrossRef]
- Subhadra, B.; Kim, J.; Kim, D.H.; Ko, K.S. Local Repressor AcrR Regulates AcrAB Efflux Pump Required for Biofilm Formation and Virulence in Acinetobacter nosocomialis. Front. Cell. Infect. Microbiol. 2018, 8, 270. [Google Scholar] [CrossRef]
- Yuhan, Y.; Ziyun, Y.; Yongbo, Z.; Qian, Z.; Jinyu, H.; Changqing, B. Over expression of AdeABC and AcrAB-TolC efflux systems confers tigecycline resistance in clinical isolates of Acinetobacter baumannii and Klebsiella pneumoniae. Rev. Soc. Bras. Med. Trop. 2016, 49, 165–171. [Google Scholar] [CrossRef]
- Geisinger, E.; Huo, W.; Hernandez-Bird, J.; Isberg, R.R. Acinetobacter baumannii: Envelope Determinants That Control Drug Resistance, Virulence, and Surface Variability. Annu. Rev. Microbiol. 2019, 73, 481–506. [Google Scholar] [CrossRef]
- Roca, I.; Espinal, P.; Vila-Farrés, X.; Vila, J. The Acinetobacter baumannii Oxymoron: Commensal Hospital Dweller Turned Pan-Drug-Resistant Menace. Front. Microbiol. 2012, 3, 148. [Google Scholar] [CrossRef] [PubMed]
- Tomaras, A.P.; Dorsey, C.W.; Edelmann, R.E.; Actis, L.A. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: Involvement of a novel chaperone-usher pili assembly system. Microbiology 2003, 149, 3473–3484. [Google Scholar] [CrossRef] [PubMed]
- Abirami, G.; Durgadevi, R.; Velmurugan, P.; Ravi, A.V. Gene Expressing Analysis Indicates the Role of Pyrogallol as a Novel Antibiofilm and Antivirulence Agent against Acinetobacter baumannii. Arch. Microbiol. 2021, 203, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.-L.; Xu, Y.-H.; She, T.-T. LC-MS/MS: A Rapid and Simple New Method for the Determination of Carbapenem β-Lactamases. Genet. Mol. Res. 2015, 14, 14457–14468. [Google Scholar] [CrossRef]
- Liu, D.; Liu, Z.S.; Hu, P.; Cai, L.; Fu, B.Q.; Li, Y.S.; Lu, S.Y.; Liu, N.N.; Ma, X.L.; Chi, D.; et al. Characterization of Surface Antigen Protein 1 (SurA1) from Acinetobacter baumannii and Its Role in Virulence and Fitness. Vet. Microbiol. 2016, 186, 126–138. [Google Scholar] [CrossRef]
- Wang, P.; Wang, H.; Liu, C.; Feng, C.; Lu, Q.; Zou, Q. Fitness Costs of Tigecycline Resistance in Acinetobacter baumannii and the Resistance Mechanism Revealed by a Transposon Mutation Library. Antibiotics 2022, 11, 10. [Google Scholar] [CrossRef]
- Weber, B.S.; Ly, P.M.; Irwin, J.N.; Pukatzki, S.; Feldman, M.F. A Multidrug Resistance Plasmid Contains the Molecular Switch for Type VI Secretion in Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA 2015, 112, 9442–9447. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial Insertion Sequences: Their Genomic Impact and Diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Huang, F.; Fitchett, N.; Razo-Gutierrez, C.; Le, C.; Martinez, J.; Ra, G.; Lopez, C.; Gonzalez, L.J.; Sieira, R.; Vila, A.J.; et al. The H-NS Regulator Plays a Role in the Stress Induced by Carbapenemase Expression in Acinetobacter baumannii. mSphere 2020, 5, e00724-20. [Google Scholar] [CrossRef]
- Corvec, S.; Caroff, N.; Espaze, E.; Giraudeau, C.; Drugeon, H.; Reynaud, A. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 2003, 52, 629–635. [Google Scholar] [CrossRef]
- Héritier, C.; Poirel, L.; Nordmann, P. Cephalosporinase Over-Expression Resulting from Insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 2006, 12, 123–130. [Google Scholar] [CrossRef]
- Lopes, B.S.; Amyes, S.G.B. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J. Med. Microbiol. 2012, 61, 1103–1108. [Google Scholar] [CrossRef]
- Nishino, K.; Hayashi-Nishino, M.; Yamaguchi, A. H-NS modulates multidrug resistance of Salmonella enterica serovar Typhimurium by repressing multidrug efflux genes acrEF. Antimicrob. Agents Chemother. 2009, 53, 3541–3543. [Google Scholar] [CrossRef]
- Nishino, K.; Yamaguchi, A. Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J. Bacteriol. 2004, 186, 1423–1429. [Google Scholar] [CrossRef]
- Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 126. [Google Scholar] [CrossRef] [PubMed]
- Eijkelkamp, B.A.; Hassan, K.A.; Paulsen, I.T.; Brown, M.H. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genom. 2011, 12, 126. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.X.; Li, X.Z.; Mistry, A.; Srikumar, R.; Zhang, L.; Lomovskaya, O.; Poole, K. Influence of the tonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1998, 42, 2225–2231. [Google Scholar] [CrossRef]
- Subhadra, B.; Surendran, S.; Lim, B.R.; Balamurugan, K. Regulation of the AcrAB Efflux System by the Quorum-Sensing Regulator AnoR in Acinetobacter nosocomialis. J. Microbiol. 2020, 58, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Baugh, S.; Ekanayaka, A.S.; Piddock, L.J.V.; Webber, M.A. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother. 2012, 67, 2409–2417. [Google Scholar] [CrossRef]
- Yang, N.; Liu, Y.; He, P.; Liu, Q.; Zhao, P.; Xu, T.; Wang, L.; Feng, Y. iTRAQ-Based Differential Proteomic Analysis Reveals the Pathways Associated with Tigecycline Resistance in Acinetobacter baumannii. Cell. Physiol. Biochem. 2018, 51, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef] [PubMed]
- Massouras, A.; Hens, K.; Gubelmann, C.; Uplekar, S.; Decouttere, F.; Rougemont, J.; Deplancke, B. Primer-initiated sequence synthesis to detect and assemble structural variants. Nat. Methods 2010, 7, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Amin, I.M.; Richmond, G.E.; Sen, P.; Koh, T.H.; Piddock, L.J.; Chua, K.L. A method for generating marker-less gene deletions in multidrug-resistant Acinetobacter baumannii. BMC Microbiol. 2013, 13, 158. [Google Scholar] [CrossRef]


| Stains | Without Inhibitor | +PAβN | +CCCP |
|---|---|---|---|
| 17978S | 0.5 | 0.5 | 0.5 |
| 17978R | 128 | 64 | 16 |
| A54S | 8 | 8 | 8 |
| A54R | 1 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Wang, L.; Wang, P.; Xiao, D.; Zou, Q. The Mechanism of Tigecycline Resistance in Acinetobacter baumannii Revealed by Proteomic and Genomic Analysis. Int. J. Mol. Sci. 2023, 24, 8652. https://doi.org/10.3390/ijms24108652
Liu C, Wang L, Wang P, Xiao D, Zou Q. The Mechanism of Tigecycline Resistance in Acinetobacter baumannii Revealed by Proteomic and Genomic Analysis. International Journal of Molecular Sciences. 2023; 24(10):8652. https://doi.org/10.3390/ijms24108652
Chicago/Turabian StyleLiu, Cunwei, Lei Wang, Ping Wang, Di Xiao, and Qinghua Zou. 2023. "The Mechanism of Tigecycline Resistance in Acinetobacter baumannii Revealed by Proteomic and Genomic Analysis" International Journal of Molecular Sciences 24, no. 10: 8652. https://doi.org/10.3390/ijms24108652
APA StyleLiu, C., Wang, L., Wang, P., Xiao, D., & Zou, Q. (2023). The Mechanism of Tigecycline Resistance in Acinetobacter baumannii Revealed by Proteomic and Genomic Analysis. International Journal of Molecular Sciences, 24(10), 8652. https://doi.org/10.3390/ijms24108652





