Clinical Characterization of Targetable Mutations (BRAF V600E and KRAS G12C) in Advanced Colorectal Cancer—A Nation-Wide Study
Abstract
1. Introduction
2. Results
2.1. Population Characteristics
2.2. BRAF V600E Mutation
2.3. KRAS G12C Mutation
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Didkowska, J.; Wojciechowska, U.; Michałek, I.; Caetano dos Santos, F.; Olasek, P. NOWOTWORY ZŁOŚLIWE W POLSCE W 2019 ROKU (CANCER IN POLAND IN 2019); Polish National Cancer Registry: Warsaw, Poland, 2021; ISBN 0867–8251. [Google Scholar]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS Mutation: From Undruggable to Druggable in Cancer. Signal Transduct. Target. Ther. 2021, 6, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020, 80, 2669–2974. [Google Scholar] [CrossRef] [PubMed]
- Bylsma, L.C.; Gillezeau, C.; Garawin, T.A.; Kelsh, M.A.; Fryzek, J.P.; Sangaré, L.; Lowe, K.A.; Kimberly Lowe, C.A. Prevalence of RAS and BRAF Mutations in Metastatic Colorectal Cancer Patients by Tumor Sidedness: A Systematic Review and Meta-Analysis. Cancer Med. 2019, 9, 1044–1057. [Google Scholar] [CrossRef]
- Park, H.S.; Chun, Y.J.; Kim, H.S.; Kim, J.H.; Lee, C.K.; Beom, S.H.; Shin, S.J.; Ahn, J.B. Clinical Features and KRAS Mutation in Colorectal Cancer with Bone Metastasis. Sci. Rep. 2020, 10, 21180. [Google Scholar] [CrossRef]
- Kamphues, C.; Kadowaki, S.; Amini, N.; van den Berg, I.; Wang, J.; Andreatos, N.; Sakamoto, Y.; Ogura, T.; Kakuta, M.; Theochari, M.; et al. The Interplay of KRAS Mutational Status with Tumor Laterality in Non-Metastatic Colorectal Cancer: An International, Multi-Institutional Study in Patients with Known KRAS, BRAF, and MSI Status. J. Surg. Oncol. 2020, 123, 1005–1014. [Google Scholar] [CrossRef]
- Tran, C.G.; Goffredo, P.; Mott, S.L.; Hart, A.; You, Y.N.; Vauthey, J.N.; Weigel, R.J.; Hassan, I. The Impact of KRAS Mutation, Microsatellite Instability, and Tumor Laterality on the Prognosis of Nonmetastatic Colon Cancer. Surgery 2022, 171, 657–665. [Google Scholar] [CrossRef]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the Undruggable RAS: Mission Possible? Nat. Rev. Drug Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF Gene in Human Cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Potocki, P.M.; Wysocki, P.J. BRAF—A New Therapeutic Target in Colorectal Cancer. Oncol. Clin. Pract. 2018, 14, 86–95. [Google Scholar]
- Yao, Z.; Torres, N.M.; Tao, A.; Gao, Y.; Luo, L.; Li, Q.; De, E.; Abdel-wahab, O.; Solit, D.B.; Poulikakos, P. BRAF Mutants Evade ERK Dependent Feedback by Different Mechanisms That Determine Their Sensitivity to Pharmacologic Inhibition Zhan. Cancer Cell 2015, 28, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E–Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Klute, K.; Garrett-Mayer, E.; Halabi, S.; Mangat, P.K.; Nazemzadeh, R.; Yost, K.J.; Butler, N.L.; Perla, V.; Schilsky, R.L. Cobimetinib plus Vemurafenib (C+V) in Patients (Pts) with Colorectal Cancer (CRC) with BRAF V600E Mutations: Results from the TAPUR Study. J. Clin. Oncol. 2020, 38, 122. [Google Scholar] [CrossRef]
- Wang, J.; Shen, J.; Huang, C.; Cao, M.; Shen, L. Clinicopathological Significance of BRAFV600E Mutation in Colorectal Cancer: An Updated Meta-Analysis. J. Cancer 2019, 10, 2332–2341. [Google Scholar] [CrossRef]
- Wójcik, P.; Okoń, K.; Osuch, C.; Klimkowska, A.; Tomaszewska, R. BRAF Mutations in Sporadic Colorectal Carcinoma from Polish Patients. Polish J. Pathol. 2010, 61, 23–26. [Google Scholar]
- Wojas-Krawczyk, K.; Kalinka-Warzocha, E.; Reszka, K.; Nicoś, M.; Szumiło, J.; Mańdziuk, S.; Szczepaniak, K.; Kupnicka, D.; Lewandowski, R.; Milanowski, J.; et al. Analysis of KRAS, NRAS, BRAF, and PIK3CA Mutations Could Predict Metastases in Colorectal Cancer: A Preliminary Study. Adv. Clin. Exp. Med. 2019, 28, 67–73. [Google Scholar] [CrossRef]
- Bożyk, A.; Krawczyk, P.; Reszka, K.; Krukowska, K.; Kolak, A.; Mańdziuk, S.; Wojas-Krawczyk, K.; Ramlau, R.; Milanowski, J. Correlation between KRAS, NRAS and BRAF Mutations and Tumor Localizations in Patients with Primary and Metastatic Colorectal Cancer. Arch. Med. Sci. 2022, 18, 1221. [Google Scholar] [CrossRef]
- Martianov, A.S.; Mitiushkina, N.V.; Ershova, A.N.; Martynenko, D.E.; Bubnov, M.G.; Amankwah, P.; Yanus, G.A.; Aleksakhina, S.N.; Tiurin, V.I.; Venina, A.R.; et al. KRAS, NRAS, BRAF, HER2 and MSI Status in a Large Consecutive Series of Colorectal Carcinomas. Int. J. Mol. Sci. 2023, 24, 4868. [Google Scholar] [CrossRef]
- Clancy, C.; Burke, J.P.; Kalady, M.F.; Coffey, J.C. BRAF Mutation Is Associated with Distinct Clinicopathological Characteristics in Colorectal Cancer: A Systematic Review and Meta-Analysis. Color. Dis. 2013, 15, e711–e718. [Google Scholar] [CrossRef]
- Cremolini, C.; Di Bartolomeo, M.; Amatu, A.; Antoniotti, C.; Moretto, R.; Berenato, R.; Perrone, F.; Tamborini, E.; Aprile, G.; Lonardi, S.; et al. BRAF Codons 594 and 596 Mutations Identify a New Molecular Subtype of Metastatic Colorectal Cancer at Favorable Prognosis. Ann. Oncol. 2015, 26, 2092–2097. [Google Scholar] [CrossRef]
- Jones, J.C.; Renfro, L.A.; Al-Shamsi, H.O.; Schrock, A.B.; Rankin, A.; Zhang, B.Y.; Kasi, P.M.; Voss, J.S.; Leal, A.D.; Sun, J.; et al. Non-V600 BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J. Clin. Oncol. 2017, 35, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.D.; Tejpar, S.; Delorenzi, M.; Yan, P.; Fiocca, R.; Klingbiel, D.; Dietrich, D.; Biesmans, B.; Bodoky, G.; Barone, C.; et al. Prognostic Role of KRAS and BRAF in Stage II and III Resected Colon Cancer: Results of the Translational Study on the PETACC-3, EORTC 40993, SAKK 60-00 Trial. J. Clin. Oncol. 2010, 28, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Fariña-Sarasqueta, A.; van Lijnschoten, G.; Moerland, E.; Creemers, G.; Lemmens, V.E.P.P.; Rutten, H.J.T.; van den Brule, A.J.C. The BRAF V600E Mutation Is an Independent Prognostic Factor for Survival in Stage II and Stage III Colon Cancer Patients. Ann. Oncol. 2010, 21, 2396–2402. [Google Scholar] [CrossRef] [PubMed]
- Venderbosch, S.; Nagtegaal, I.D.; Maughan, T.S.; Smith, C.G.; Cheadle, J.P.; Fisher, D.; Kaplan, R.; Quirke, P.; Seymour, M.T.; Richman, S.D.; et al. Mismatch Repair Status and BRAF Mutation Status in Metastatic Colorectal Cancer Patients: A Pooled Analysis of the CAIRO, CAIRO2, COIN, and FOCUS Studies. Clin. Cancer Res. 2014, 20, 5322–5330. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, S.; Ooki, A.; Yamaguchi, K.; Kakuta, M.; Takahashi, S.; Takahashi, A.; Arai, Y.; Akagi, K.; Shigenori Kadowaki, J.; Muro, K.; et al. Prognostic Value of KRAS and BRAF Mutations in Curatively Resected Colorectal Cancer Observational Study. World J. Gastroenterol. 2015, 21, 1275–1283. [Google Scholar] [CrossRef]
- Seligmann, J.F.; Fisher, D.; Smith, C.G.; Richman, S.D.; Elliott, F.; Brown, S.; Adams, R.; Maughan, T.; Quirke, P.; Cheadle, J.; et al. Investigating the Poor Outcomes of BRAF -Mutant Advanced Colorectal Cancer: Analysis from 2530 Patients in Randomised Clinical Trials. Ann. Oncol. 2017, 28, 562–568. [Google Scholar] [CrossRef]
- Lee, S.M.; Sung, C.O. Comprehensive Analysis of Mutational and Clinicopathologic Characteristics of Poorly Differentiated Colorectal Neuroendocrine Carcinomas. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Klempner, S.J.; Gershenhorn, B.; Tran, P.; Lee, T.K.; Erlander, M.G.; Gowen, K.; Schrock, A.B.; Morosini, D.; Ross, J.S.; Stephens, P.J.; et al. BRAFV600E Mutations in High-Grade Colorectal Neuroendocrine Tumors May Predict Responsiveness to BRAF-MEK Combination Therapy. Cancer Discov. 2016, 6, 594–600. [Google Scholar] [CrossRef]
- Nakano, M.; Shimada, Y.; Matsumoto, Y.; Saiki, T.; Zhou, Q.; Sasaki, K.; Moriyama, M.; Yoshihara, K.; Natsumeda, M.; Kuriyama, Y.; et al. Efficacy of BRAF Inhibitor and Anti-EGFR Antibody in Colorectal Neuroendocrine Carcinoma. Clin. J. Gastroenterol. 2022, 15, 413–418. [Google Scholar] [CrossRef]
- U.S. Food and Drug Afministration FDA Grants Accelerated Approval to Dabrafenib in Combination with Trametinib for Unresectable or Metastatic Solid Tumors with BRAF V600E Mutation. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-dabrafenib-combination-trametinib-unresectable-or-metastatic-solid (accessed on 7 May 2023).
- Scartozzi, M.; Giampieri, R.; Aprile, G.; Iacono, D.; Santini, D.; Dell’Aquila, E.; Silvestris, N.; Gnoni, A.; Bonotto, M.; Puzzoni, M.; et al. The Distinctive Molecular, Pathological and Clinical Characteristics of BRAF-Mutant Colorectal Tumors. Expert Rev. Mol. Diagn. 2015, 15, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Chida, K.; Kotani, D.; Masuishi, T.; Kawakami, T.; Kawamoto, Y.; Kato, K.; Fushiki, K.; Sawada, K.; Kumanishi, R.; Shirasu, H.; et al. The Prognostic Impact of KRAS G12C Mutation in Patients with Metastatic Colorectal Cancer: A Multicenter Retrospective Observational Study. Oncologist 2021, 26, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.T.; Coker, O.; Chowdhury, S.; Shen, J.P.; Morris, V.K.; Dasari, A.; Raghav, K.; Nusrat, M.; Kee, B.; Parseghian, C.; et al. Comprehensive Clinical and Molecular Characterization of KRAS G12C -Mutant Colorectal Cancer. JCO Precis. Oncol. 2021, 5, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Osterlund, E.; Ristimäki, A.; Kytölä, S.; Kuopio, T.; Heervä, E.; Muhonen, T.; Halonen, P.; Kallio, R.; Soveri, L.-M.; Sundström, J.; et al. KRAS-G12C Mutation in One Real-Life and Three Population-Based Nordic Cohorts of Metastatic Colorectal Cancer. Front. Oncol. 2022, 12, 826073. [Google Scholar] [CrossRef] [PubMed]
- Schirripa, M.; Nappo, F.; Cremolini, C.; Salvatore, L.; Rossini, D.; Bensi, M.; Businello, G.; Pietrantonio, F.; Randon, G.; Fucà, G.; et al. KRAS G12C Metastatic Colorectal Cancer: Specific Features of a New Emerging Target Population. Clin. Colorectal Cancer 2020, 19, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Giampieri, R.; Lupi, A.; Ziranu, P.; Bittoni, A.; Pretta, A.; Pecci, F.; Persano, M.; Giglio, E.; Copparoni, C.; Crocetti, S.; et al. Retrospective Comparative Analysis of KRAS G12C vs. Other KRAS Mutations in MCRC Patients Treated with First-Line Chemotherapy Doublet + Bevacizumab. Front. Oncol. 2021, 11, 736104. [Google Scholar] [CrossRef]
- Roussille, P.; Tachon, G.; Villalva, C.; Milin, S.; Frouin, E.; Godet, J.; Berger, A.; Emambux, S.; Petropoulos, C.; Wager, M.; et al. Pathological and Molecular Characteristics of Colorectal Cancer with Brain Metastases. Cancers 2018, 10, 504. [Google Scholar] [CrossRef]
- Cui, W.; Franchini, F.; Alexander, M.; Officer, A.; Wong, H.L.; IJzerman, M.; Desai, J.; Solomon, B.J. Real World Outcomes in KRAS G12C Mutation Positive Non-Small Cell Lung Cancer. Lung Cancer 2020, 146, 310–317. [Google Scholar] [CrossRef]
- Vassella, E.; Kashani, E.; Zens, P.; Kündig, A.; Fung, C.; Scherz, A.; Herrmann, E.; Ermis, E.; Schmid, R.A.; Berezowska, S. Mutational Profiles of Primary Pulmonary Adenocarcinoma and Paired Brain Metastases Disclose the Importance of KRAS Mutations. Eur. J. Cancer 2021, 159, 227–236. [Google Scholar] [CrossRef]
- Skoulidis, F.; Li, B.T.; Dy, G.K.; Price, T.J.; Falchook, G.S.; Wolf, J.; Italiano, A.; Schuler, M.; Borghaei, H.; Barlesi, F.; et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 2021, 384, 2371–2381. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2017. [Google Scholar]
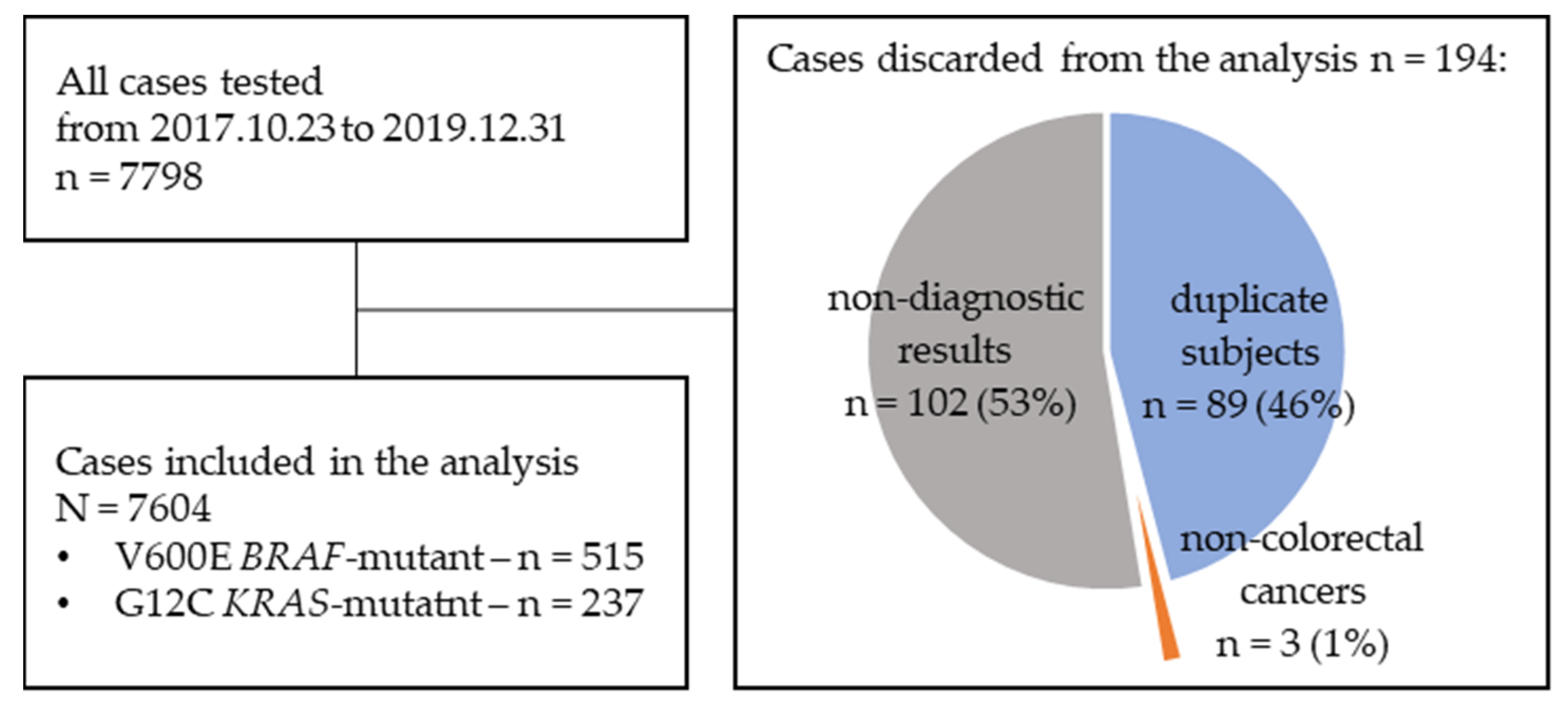

| Parameter | Populations Defined by Mutation Types | ||||
|---|---|---|---|---|---|
| Overall | BRAF V600E | KRAS G12C | Other KRAS | ||
| (N = 7604) | (N = 515) | (N = 237) | (N = 3591) | ||
| Sex | Female | 3132 (41.19%) | 295 (57.28%) | 101 (42.62%) | 1527 (42.52%) |
| Male | 4471 (58.81%) | 220 (42.72%) | 136 (57.38%) | 2064 (57.48%) | |
| Age (years) | <30 | 21 (0.28%) | 0 (0.00%) | 0 (0.00%) | 10 (0.28%) |
| 31–40 | 115 (1.51%) | 6 (1.17%) | 6 (2.53%) | 49 (1.36%) | |
| 41–50 | 432 (5.68%) | 22 (4.27%) | 13 (5.49%) | 190 (5.29%) | |
| 51–60 | 1298 (17.07%) | 74 (14.37%) | 36 (15.19%) | 588 (16.37%) | |
| 61–70 | 3192 (41.98%) | 222 (43.11%) | 111 (46.84%) | 1486 (41.38%) | |
| 71–80 | 2173 (28.58%) | 156 (30.29%) | 64 (27.00%) | 1070 (29.80%) | |
| over 80 | 373 (4.91%) | 35 (6.80%) | 7 (2.95%) | 198 (5.51%) | |
| Tumour stage | T1 | 39 (0.98%) | 0 (0.00%) | 1 (0.83%) | 25 (1.29%) |
| T2 | 291 (7.29%) | 10 (3.33%) | 11 (9.09%) | 144 (7.44%) | |
| T3 | 2611 (65.37%) | 175 (58.33%) | 81 (66.94%) | 1243 (64.20%) | |
| T4 | 1039 (26.01%) | 115 (38.33%) | 28 (23.14%) | 516 (26.65%) | |
| Tis | 14 (0.35%) | 0 (0.00%) | 0 (0.00%) | 8 (0.41%) | |
| Nodal stage (0 vs. 1 vs. 2) | N0 | 1006 (26.18%) | 57 (19.79%) | 37 (31.09%) | 486 (26.00%) |
| N1 | 1460 (37.99%) | 94 (32.64%) | 43 (36.13%) | 740 (39.59%) | |
| N2 | 1377 (35.83%) | 137 (47.57%) | 39 (32.77%) | 643 (34.40%) | |
| Histology | Adenocarcinoma NOS | 6364 (89.93%) | 362 (78.02%) | 196 (88.69%) | 3108 (88.52%) |
| Partially mucinous | 632 (8.93%) | 89 (58.17%) | 23 (56.10%) | 382 (56.26%) | |
| Mucinous | 35 (0.49%) | 6 (3.92%) | 0 (0.00%) | 10 (1.47%) | |
| Cribriform or comedo | 32 (0.45%) | 2 (1.31%) | 2 (4.88%) | 9 (1.33%) | |
| MANEC | 14 (0.2%) | 5 (3.27%) | 0 (0.00%) | 2 (0.29%) | |
| Grade | G1 | 590 (12.14%) | 26 (7.60%) | 17 (10.90%) | 299 (12.88%) |
| G2 | 3509 (72.22%) | 181 (52.92%) | 122 (78.21%) | 1708 (73.59%) | |
| G3 | 756 (15.56%) | 134 (39.18%) | 17 (10.90%) | 313 (13.49%) | |
| G4 | 4 (0.08%) | 1 (0.29%) | 0 (0.00%) | 1 (0.04%) | |
| Primary tumour sidedness | Left colon or rectum | 4827 (75.46%) | 129 (30.57%) | 163 (82.32%) | 2255 (74.20%) |
| Right colon | 1570 (24.54%) | 293 (69.43%) | 35 (17.68%) | 784 (25.80%) | |
| Sample type | Endoscopic biopsy | 1388 (22.91%) | 73 (17.76%) | 45 (22.84%) | 654 (22.58%) |
| Needle biopsy | 218 (3.6%) | 7 (1.70%) | 12 (6.09%) | 90 (3.11%) | |
| Surgery | 4452 (73.49%) | 331 (80.54%) | 140 (71.07%) | 2152 (74.31%) | |
| Sample source | Primary | 4968 (87.88%) | 346 (90.58%) | 153 (83.61%) | 2376 (88.43%) |
| Metastatic | 685 (12.12%) | 36 (9.42%) | 30 (16.39%) | 311 (11.57%) | |
| Location of metastatic samples | Local recurrence | 81 (11.82%) | 3 (0.79%) | 5 (2.73%) | 39 (1.45%) |
| Liver | 266 (38.83%) | 11 (2.88%) | 11 (6.01%) | 108 (4.02%) | |
| Lung | 73 (10.66%) | 3 (0.79%) | 1 (0.55%) | 42 (1.56%) | |
| Peritoneal | 148 (21.61%) | 12 (3.14%) | 5 (2.73%) | 76 (2.83%) | |
| Nodal | 31 (4.53%) | 3 (0.79%) | 2 (1.09%) | 10 (0.37%) | |
| Ovarian | 33 (4.82%) | 2 (0.52%) | 1 (0.55%) | 10 (0.37%) | |
| Small intestine | 26 (3.8%) | 2 (0.52%) | 2 (1.09%) | 12 (0.45%) | |
| CNS | 5 (0.73%) | 0 (0%) | 2 (1.09%) | 2 (0.07%) | |
| Skin/subcutaneous | 22 (3.21%) | 0 (0%) | 1 (0.55%) | 12 (0.45%) | |
| Parameter | Group | p | ||
|---|---|---|---|---|
| BRAF V600E Mutation (N = 515) | No BRAF V600E Mutation (N = 7089) | |||
| Tumour stage | T1 | 0 (0.00%) | 39 (1.06%) | p < 0.001 * |
| T2 | 10 (3.33%) | 281 (7.61%) | ||
| T3 | 175 (58.33%) | 2436 (65.94%) | ||
| T4 | 115 (38.33%) | 924 (25.01%) | ||
| Tis | 0 (0.00%) | 14 (0.38%) | ||
| Nodal stage | N0 | 57 (19.32%) | 949 (25.91%) | p < 0.001 * |
| N1a | 40 (13.56%) | 476 (12.99%) | ||
| N1b | 43 (14.58%) | 692 (18.89%) | ||
| N1c | 11 (3.73%) | 198 (5.41%) | ||
| N2a | 55 (18.64%) | 594 (16.22%) | ||
| N2b | 82 (27.80%) | 646 (17.64%) | ||
| Nx | 7 (2.37%) | 108 (2.95%) | ||
| Nodal stage (0 vs. 1 vs. 2) | N0 | 57 (19.79%) | 949 (26.69%) | p < 0.001 * |
| N1 | 94 (32.64%) | 1366 (38.42%) | ||
| N2 | 137 (47.57%) | 1240 (34.88%) | ||
| Nodal stage (positive vs. negative) | Negative | 57 (19.79%) | 949 (26.69%) | p = 0.013 * |
| Positive | 231 (80.21%) | 2606 (73.31%) | ||
| Angioinvasion | No | 41 (21.69%) | 775 (34.63%) | p < 0.001 * |
| Yes | 148 (78.31%) | 1463 (65.37%) | ||
| Perineural invasion | No | 44 (38.94%) | 696 (51.98%) | p = 0.01 * |
| Yes | 69 (61.06%) | 643 (48.02%) | ||
| Biopsy vs. surgery for primary samples | Biopsy | 80 (19.46%) | 1526 (27.02%) | p = 0.001 * |
| Surgery | 331 (80.54%) | 4121 (72.98%) | ||
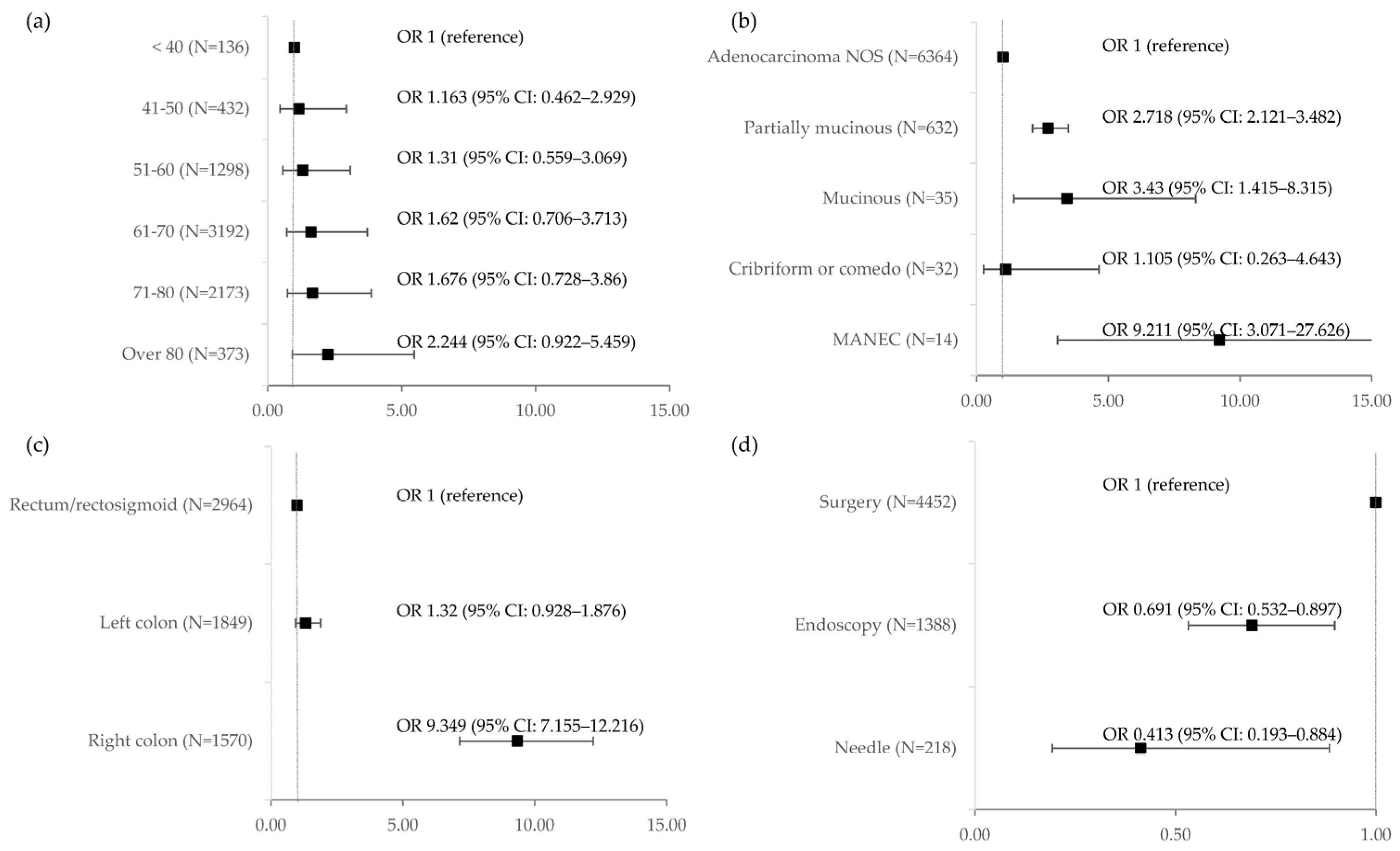
| Trait | Group | BRAF V600E | OR | 95% CI | p | |
|---|---|---|---|---|---|---|
| Sex | Male (N = 4471) | 220 (4.92%) | 1 | ref. | ||
| Female (N = 3132) | 295 (9.42%) | 2.009 | 1.677 | 2.408 | <0.001 | |
| Age (years) | <40 (N = 136) | 6 (4.41%) | 1 | ref. | ||
| 41–50 (N = 432) | 22 (5.09%) | 1.163 | 0.462 | 2.929 | 0.749 | |
| 51–60 (N = 1298) | 74 (5.70%) | 1.31 | 0.559 | 3.069 | 0.534 | |
| 61–70 (N = 3192) | 222 (6.95%) | 1.62 | 0.706 | 3.713 | 0.255 | |
| 71–80 (N = 2173) | 156 (7.18%) | 1.676 | 0.728 | 3.86 | 0.225 | |
| Over 80 (N = 373) | 35 (9.38%) | 2.244 | 0.922 | 5.459 | 0.075 | |
| Histology | Adenocarcinoma NOS (N = 6364) | 362 (5.69%) | 1 | ref. | ||
| Partially mucinous (N = 632) | 89 (14.08%) | 2.718 | 2.121 | 3.482 | <0.001 | |
| Mucinous (N = 35) | 6 (17.14%) | 3.43 | 1.415 | 8.315 | 0.006 | |
| Cribriform or comedo (N = 32) | 2 (6.25%) | 1.105 | 0.263 | 4.643 | 0.891 | |
| MANEC (N = 14) | 5 (35.71%) | 9.211 | 3.071 | 27.626 | <0.001 | |
| Mucous component | No (N = 6555) | 385 (5.87%) | 1 | ref. | ||
| Yes (N = 1049) | 130 (12.39%) | 2.267 | 1.837 | 2.798 | <0.001 | |
| Signet cells presence | No (N = 7187) | 474 (6.60%) | 1 | ref. | ||
| Yes (N = 417) | 41 (9.83%) | 1.544 | 1.104 | 2.16 | 0.011 | |
| Grade | G1 (N = 590) | 26 (4.41%) | 1 | ref. | ||
| G2 (N = 3509) | 181 (5.16%) | 1.18 | 0.775 | 1.797 | 0.441 | |
| G3 (N = 756) | 134 (17.72%) | 4.673 | 3.024 | 7.221 | <0.001 | |
| G4 (N = 4) | 1 (25.00%) | 7.231 | 0.727 | 71.909 | 0.091 | |
| Primary tumour localization | Rectum/rectosigmoid (N = 2964) | 71 (2.40%) | 1 | ref. | ||
| Left colon (N = 1849) | 58 (3.14%) | 1.32 | 0.928 | 1.876 | 0.122 | |
| Right colon (N = 1570) | 293 (18.66%) | 9.349 | 7.155 | 12.216 | <0.001 | |
| Sample type | Surgery (N = 4452) | 331 (7.43%) | 1 | ref. | ||
| Endoscopy (N = 1388) | 73 (5.26%) | 0.691 | 0.532 | 0.897 | 0.006 | |
| Needle (N = 218) | 7 (3.21%) | 0.413 | 0.193 | 0.884 | 0.023 | |
| Sample origin | Primary (N = 4968) | 346 (6.96%) | 1 | ref. | ||
| Metastatic (N = 685) | 36 (5.26%) | 0.741 | 0.521 | 1.055 | 0.096 | |
| Biopsy vs. surgery for metastatic tissues | Surgery (N = 854) | 59 (6.91%) | 1 | ref. | ||
| Biopsy (N = 259) | 8 (3.09%) | 0.429 | 0.202 | 0.911 | 0.028 | |
| Parameter | Group | |||||
|---|---|---|---|---|---|---|
| KRAS G12C (N = 237) | No KRAS G12C (N = 7367) | p | Other KRAS (N = 3591) | p | ||
| Tumour stage | T1 | 1 (0.83%) | 38 (0.98%) | p = 0.835 | p = 0.859 | |
| T2 | 11 (9.09%) | 280 (7.23%) | 144 (7.44%) | |||
| T3 | 81 (66.94%) | 2530 (65.32%) | 1243 (64.20%) | |||
| T4 | 28 (23.14%) | 1011 (26.10%) | 516 (26.65%) | |||
| Tis | 0 (0.00%) | 14 (0.36%) | 8 (0.41%) | |||
| Nodal stage | N0 | 37 (30.33%) | 969 (25.26%) | p = 0.051 | 486 (25.34%) | p = 0.062 |
| N1a | 8 (6.56%) | 508 (13.24%) | 272 (14.18%) | |||
| N1b | 27 (22.13%) | 708 (18.46%) | 354 (18.46%) | |||
| N1c | 8 (6.56%) | 201 (5.24%) | 114 (5.94%) | |||
| N2a | 26 (21.31%) | 623 (16.24%) | 307 (16.01%) | |||
| N2b | 13 (10.66%) | 715 (18.64%) | 336 (17.52%) | |||
| Nx | 3 (2.46%) | 112 (2.92%) | 49 (2.55%) | |||
| Nodal stage (0 vs. 1 vs. 2) | N0 | 37 (31.09%) | 969 (26.02%) | p = 0.457 | 486 (26.00%) | p = 0.466 |
| N1 | 43 (36.13%) | 1417 (38.05%) | 740 (39.59%) | |||
| N2 | 39 (32.77%) | 1338 (35.93%) | 643 (34.40%) | |||
| Nodal stage (positive vs. negative) | Negative | 37 (31.09%) | 969 (26.02%) | p = 0.257 | 486 (26.00%) | p = 0.265 |
| Positive | 82 (68.91%) | 2755 (73.98%) | 1383 (74.00%) | |||
| Angioinvasion | No | 26 (30.95%) | 790 (33.72%) | p = 0.682 | 434 (36.94%) | p = 0.326 |
| Yes | 58 (69.05%) | 1553 (66.28%) | 741 (63.06%) | |||
| Perineural invasion | No | 25 (46.30%) | 715 (51.14%) | p = 0.575 | 375 (53.19%) | p = 0.403 |
| Yes | 29 (53.70%) | 683 (48.86%) | 330 (46.81%) | |||
| Biopsy vs. surgery for primary samples | Biopsy | 57 (28.93%) | 1549 (26.43%) | p = 0.483 | 744 (25.69%) | p = 0.357 |
| Surgery | 140 (71.07%) | 4312 (73.57%) | 2152 (74.31%) | |||
| Trait | Group | KRAS G12C | OR | 95% CI | p | |
|---|---|---|---|---|---|---|
| Sex | Male (N = 4471) | 136 (3.04%) | 1 | ref. | ||
| Female (N = 3132) | 101 (3.22%) | 1.062 | 0.818 | 1.38 | 0.651 | |
| Age (years) | <40 (N = 136) | 6 (4.41%) | 1 | ref. | ||
| 41–50 (N = 432) | 13 (3.01%) | 0.672 | 0.251 | 1.804 | 0.43 | |
| 51–60 (N = 1298) | 36 (2.77%) | 0.618 | 0.256 | 1.494 | 0.285 | |
| 61–70 (N = 3192) | 111 (3.48%) | 0.781 | 0.337 | 1.808 | 0.563 | |
| 71–80 (N = 2173) | 64 (2.95%) | 0.657 | 0.28 | 1.547 | 0.337 | |
| Over 80 (N = 373) | 7 (1.88%) | 0.414 | 0.137 | 1.256 | 0.119 | |
| Histology | Partially mucinous (N = 632) | 23 (3.64%) | 1 | ref. | ||
| Mucinous (N = 35) | 0 (0.00%) | -- | -- | -- | -- | |
| Cribriform or comedo (N = 32) | 2 (6.25%) | 1.765 | 0.398 | 7.838 | 0.455 | |
| MANEC (N = 14) | 0 (0.00%) | -- | -- | -- | -- | |
| Other (N = 527) | 16 (3.04%) | 0.829 | 0.433 | 1.586 | 0.571 | |
| Primary tumour localization | Rectum (N = 2964) | 111 (3.74%) | 1 | ref. | ||
| Left colon (N = 1849) | 51 (2.76%) | 0.729 | 0.521 | 1.021 | 0.066 | |
| Right colon (N = 1570) | 35 (2.23%) | 0.586 | 0.399 | 0.861 | 0.007 | |
| Primary sidedness | Left colon or rectum (N = 4827) | 163 (3.38%) | 1 | ref. | ||
| Right colon (N = 1570) | 35 (2.23%) | 0.652 | 0.451 | 0.944 | 0.024 | |
| Primary localization (colon vs. rectum) | Colon (N = 4149) | 107 (2.58%) | 1 | ref. | ||
| Rectum (N = 2964) | 111 (3.74%) | 1.47 | 1.122 | 1.925 | 0.005 | |
| Sample type | Surgery (N = 4452) | 140 (3.14%) | 1 | ref. | ||
| Endoscopy (N = 1388) | 45 (3.24%) | 1.032 | 0.734 | 1.452 | 0.856 | |
| Needle (N = 218) | 12 (5.50%) | 1.794 | 0.979 | 3.289 | 0.059 | |
| Sample origin (primary vs. metastatic) | Primary (N = 4968) | 153 (3.08%) | 1 | ref. | ||
| Metastatic (N = 685) | 30 (4.38%) | 1.441 | 0.966 | 2.15 | 0.073 | |
| Sample origin (detailed) | Primary (N = 4968) | 153 (3.08%) | 1 | ref. | ||
| Metastatic—other (N = 598) | 23 (3.85%) | 1.259 | 0.805 | 1.968 | 0.313 | |
| Metastatic—local (N = 82) | 5 (6.10%) | 2.044 | 0.815 | 5.121 | 0.127 | |
| Metastatic—CNS (N = 5) | 2 (40.00%) | 20.98 | 3.48 | 126.47 | 0.001 | |
| Biopsy vs. surgery for metastatic samples | Surgery (N = 854) | 32 (3.75%) | 1 | ref. | ||
| Biopsy (N = 259) | 14 (5.41%) | 1.468 | 0.771 | 2.795 | 0.243 | |
| Trait | Group | KRAS G12C | OR | 95% CI | p | |
|---|---|---|---|---|---|---|
| Sex | Male (N = 2200) | 136 (6.18%) | 1 | ref. | ||
| Female (N = 1628) | 101 (6.20%) | 1.004 | 0.77 | 1.309 | 0.978 | |
| Age (years) | <40 (N = 65) | 6 (9.23%) | 1 | ref. | ||
| 41–50 (N = 203) | 13 (6.40%) | 0.673 | 0.245 | 1.848 | 0.442 | |
| 51–60 (N = 624) | 36 (5.77%) | 0.602 | 0.244 | 1.488 | 0.272 | |
| 61–70 (N = 1597) | 111 (6.95%) | 0.735 | 0.31 | 1.739 | 0.483 | |
| 71–80 (N = 1134) | 64 (5.64%) | 0.588 | 0.245 | 1.414 | 0.236 | |
| Over 80 (N = 205) | 7 (3.41%) | 0.348 | 0.113 | 1.074 | 0.066 | |
| Histology | Partially mucinous (N = 405) | 23 (5.68%) | 1 | ref. | ||
| Mucinous (N = 10) | 0 (0.00%) | -- | -- | -- | -- | |
| Cribriform or comedo (N = 11) | 2 (18.18%) | 3.691 | 0.753 | 18.079 | 0.107 | |
| MANEC (N = 2) | 0 (0.00%) | -- | -- | -- | -- | |
| Other (N = 292) | 16 (5.48%) | 0.963 | 0.499 | 1.856 | 0.91 | |
| Primary tumour localization | Rectum (N = 1533) | 111 (7.24%) | 1 | ref. | ||
| Left colon (N = 876) | 51 (5.82%) | 0.792 | 0.562 | 1.115 | 0.182 | |
| Right colon (N = 819) | 35 (4.27%) | 0.572 | 0.387 | 0.845 | 0.005 | |
| Primary sidedness | Left colon or rectum (N = 2418) | 163 (6.74%) | 1 | ref. | ||
| Right colon (N = 819) | 35 (4.27%) | 0.618 | 0.425 | 0.898 | 0.012 | |
| Primary localization (colon vs. rectum) | Colon (N = 2060) | 107 (5.19%) | 1 | ref. | ||
| Rectum (N = 1533) | 111 (7.24%) | 1.425 | 1.083 | 1.874 | 0.011 | |
| Sample type | Surgery (N = 2292) | 140 (6.11%) | 1 | ref. | ||
| Endoscopy (N = 699) | 45 (6.44%) | 1.058 | 0.748 | 1.497 | 0.752 | |
| Needle (N = 102) | 12 (11.76%) | 2.05 | 1.096 | 3.833 | 0.025 | |
| Sample origin (primary vs. metastatic) | Primary (N = 2529) | 153 (6.05%) | 1 | ref. | ||
| Metastatic (N = 341) | 30 (8.80%) | 1.498 | 0.995 | 2.255 | 0.053 | |
| Sample origin (detailed) | Primary (N = 2529) | 153 (6.05%) | 1 | ref. | ||
| Metastatic—other (N = 293) | 23 (7.85%) | 1.323 | 0.838 | 2.087 | 0.229 | |
| Metastatic—locoregional (N = 44) | 5 (11.36%) | 1.991 | 0.774 | 5.124 | 0.153 | |
| Metastatic—CNS (N = 4) | 2 (50.00%) | 15.529 | 2.173 | 110.997 | 0.006 | |
| Biopsy vs. surgery for metastatic samples | Surgery (N = 450) | 32 (7.11%) | 1 | ref. | ||
| Biopsy (N = 124) | 14 (11.29%) | 1.662 | 0.857 | 3.224 | 0.132 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potocki, P.M.; Wójcik, P.; Chmura, Ł.; Goc, B.; Fedewicz, M.; Bielańska, Z.; Swadźba, J.; Konopka, K.; Kwinta, Ł.; Wysocki, P.J. Clinical Characterization of Targetable Mutations (BRAF V600E and KRAS G12C) in Advanced Colorectal Cancer—A Nation-Wide Study. Int. J. Mol. Sci. 2023, 24, 9073. https://doi.org/10.3390/ijms24109073
Potocki PM, Wójcik P, Chmura Ł, Goc B, Fedewicz M, Bielańska Z, Swadźba J, Konopka K, Kwinta Ł, Wysocki PJ. Clinical Characterization of Targetable Mutations (BRAF V600E and KRAS G12C) in Advanced Colorectal Cancer—A Nation-Wide Study. International Journal of Molecular Sciences. 2023; 24(10):9073. https://doi.org/10.3390/ijms24109073
Chicago/Turabian StylePotocki, Paweł M., Piotr Wójcik, Łukasz Chmura, Bartłomiej Goc, Marcin Fedewicz, Zofia Bielańska, Jakub Swadźba, Kamil Konopka, Łukasz Kwinta, and Piotr J. Wysocki. 2023. "Clinical Characterization of Targetable Mutations (BRAF V600E and KRAS G12C) in Advanced Colorectal Cancer—A Nation-Wide Study" International Journal of Molecular Sciences 24, no. 10: 9073. https://doi.org/10.3390/ijms24109073
APA StylePotocki, P. M., Wójcik, P., Chmura, Ł., Goc, B., Fedewicz, M., Bielańska, Z., Swadźba, J., Konopka, K., Kwinta, Ł., & Wysocki, P. J. (2023). Clinical Characterization of Targetable Mutations (BRAF V600E and KRAS G12C) in Advanced Colorectal Cancer—A Nation-Wide Study. International Journal of Molecular Sciences, 24(10), 9073. https://doi.org/10.3390/ijms24109073







