Characterization of Disease Resistance Induced by a Pyrazolecarboxylic Acid Derivative in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
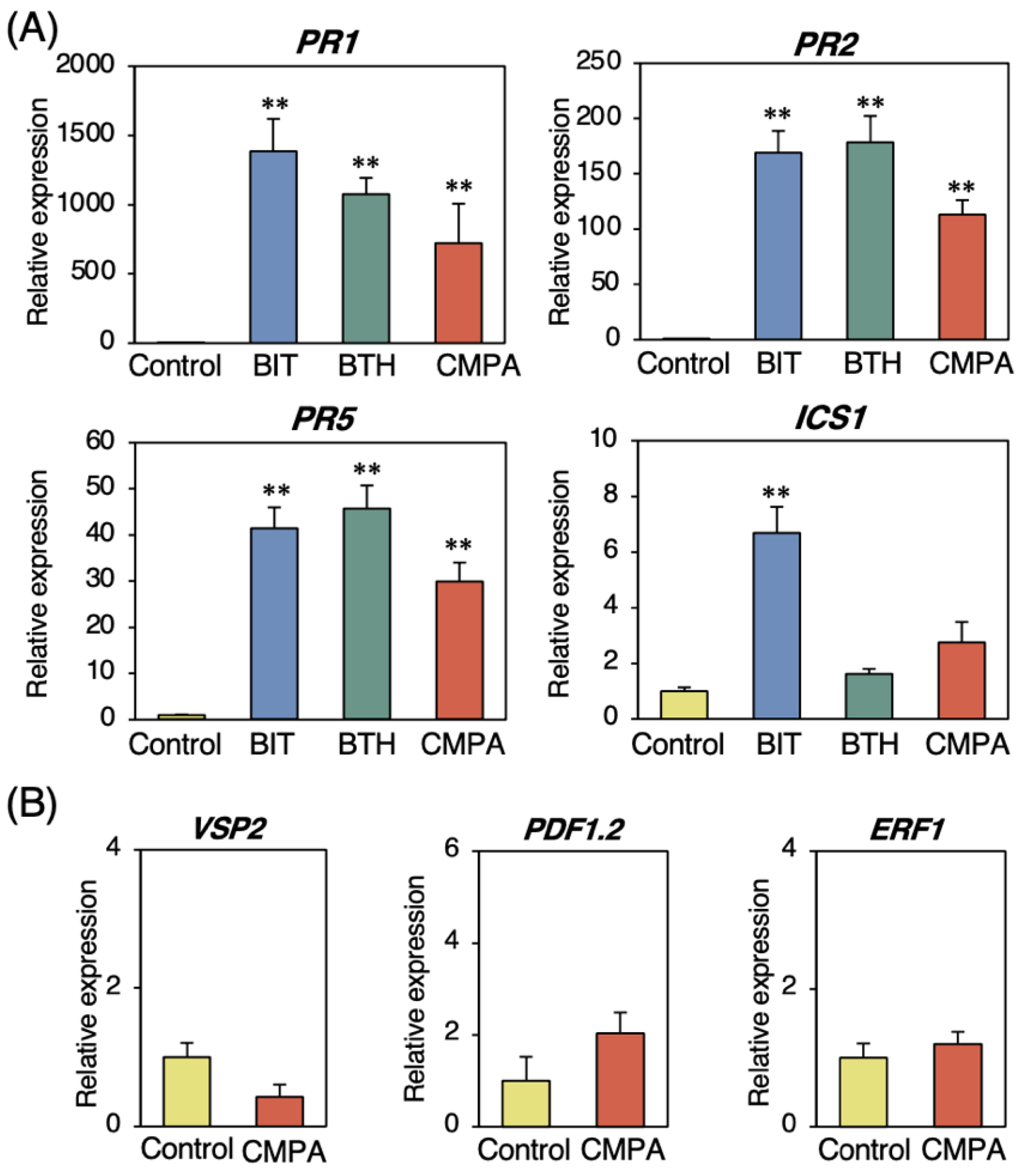
2.1. Induction of a Broad Range of Disease Resistance in Arabidopsis by CMPA
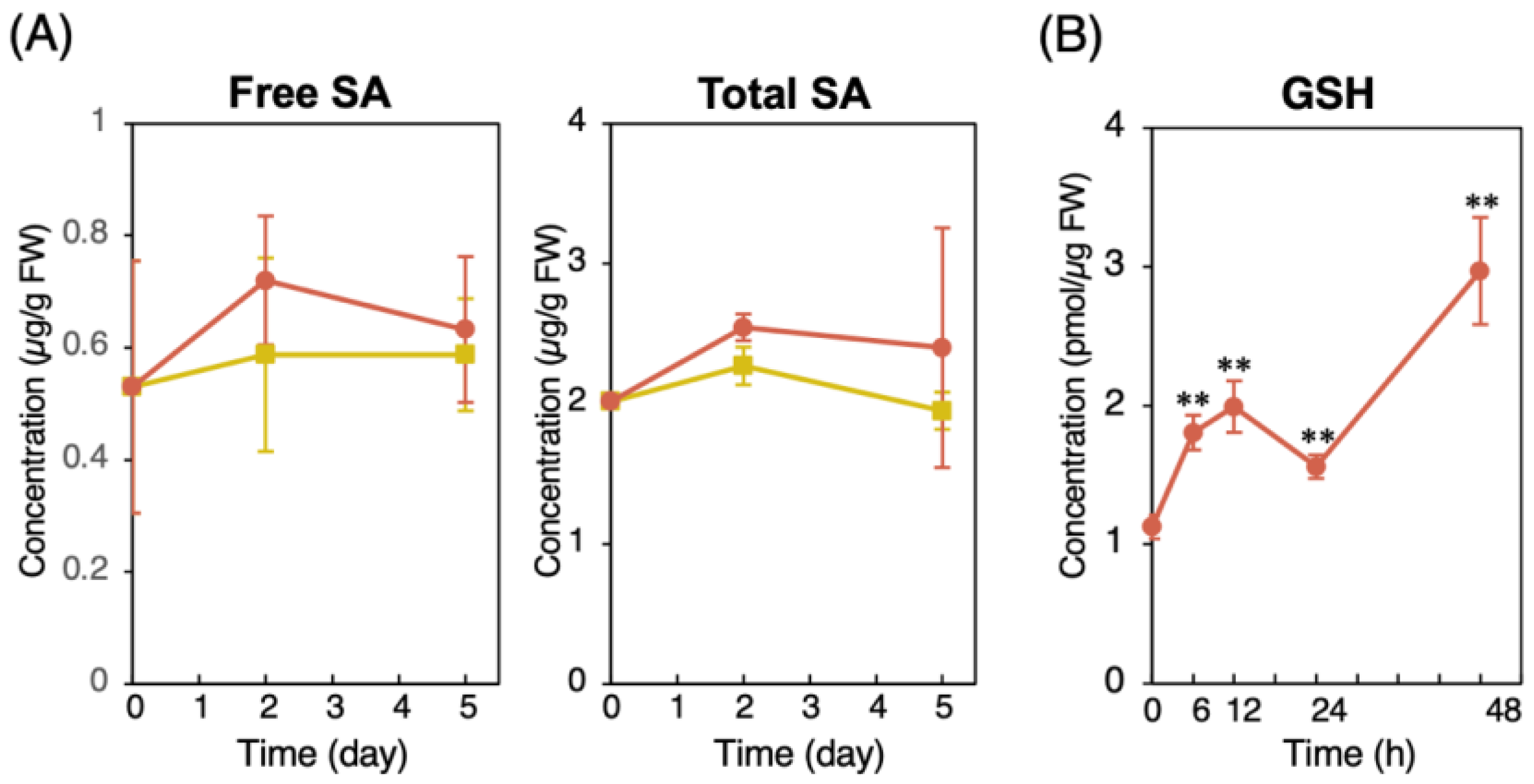
2.2. Physiological Changes Associated with CMPA-Induced Resistance
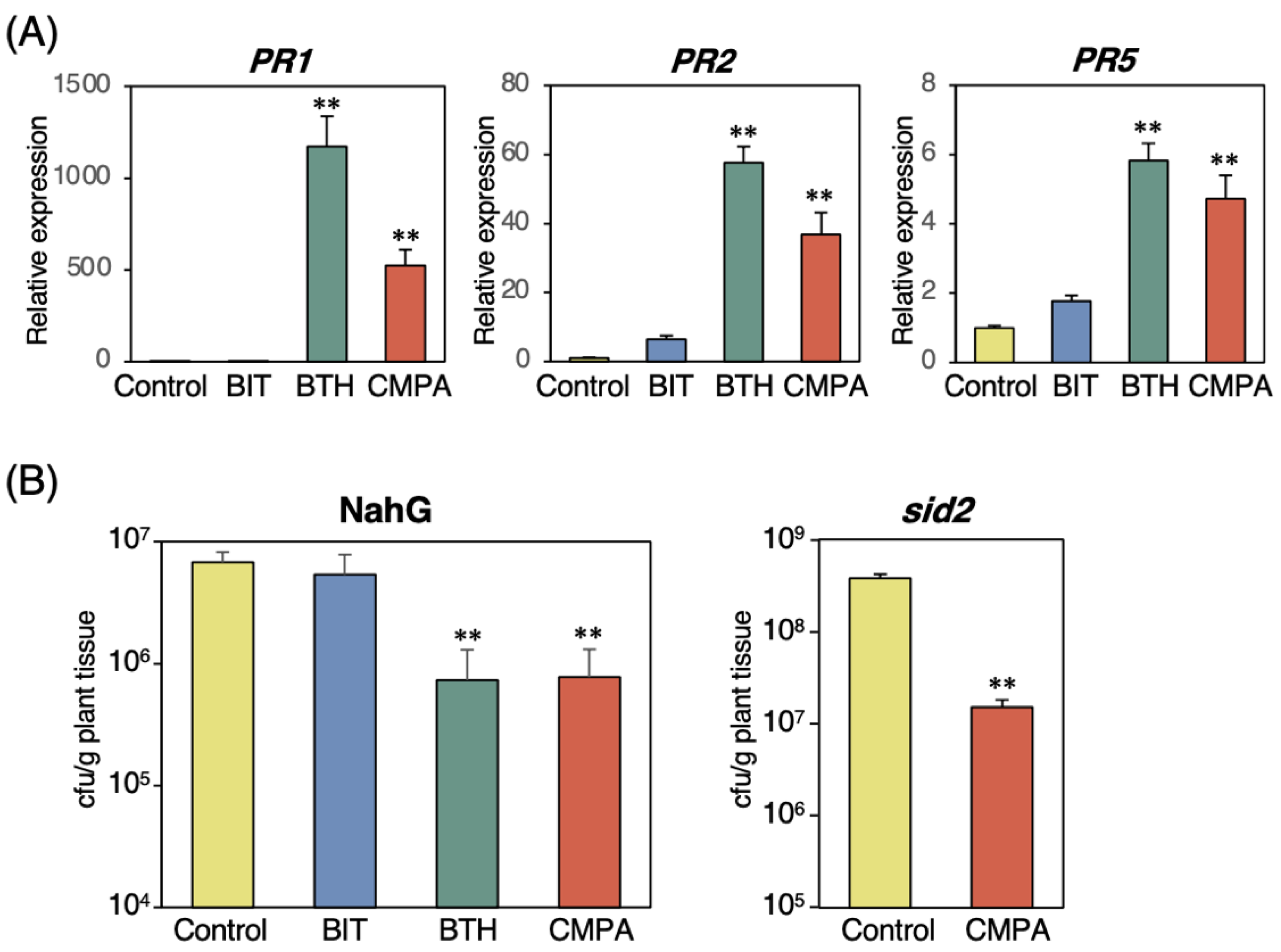
2.3. Effects of CMPA on SA-Deficient Mutant Plants
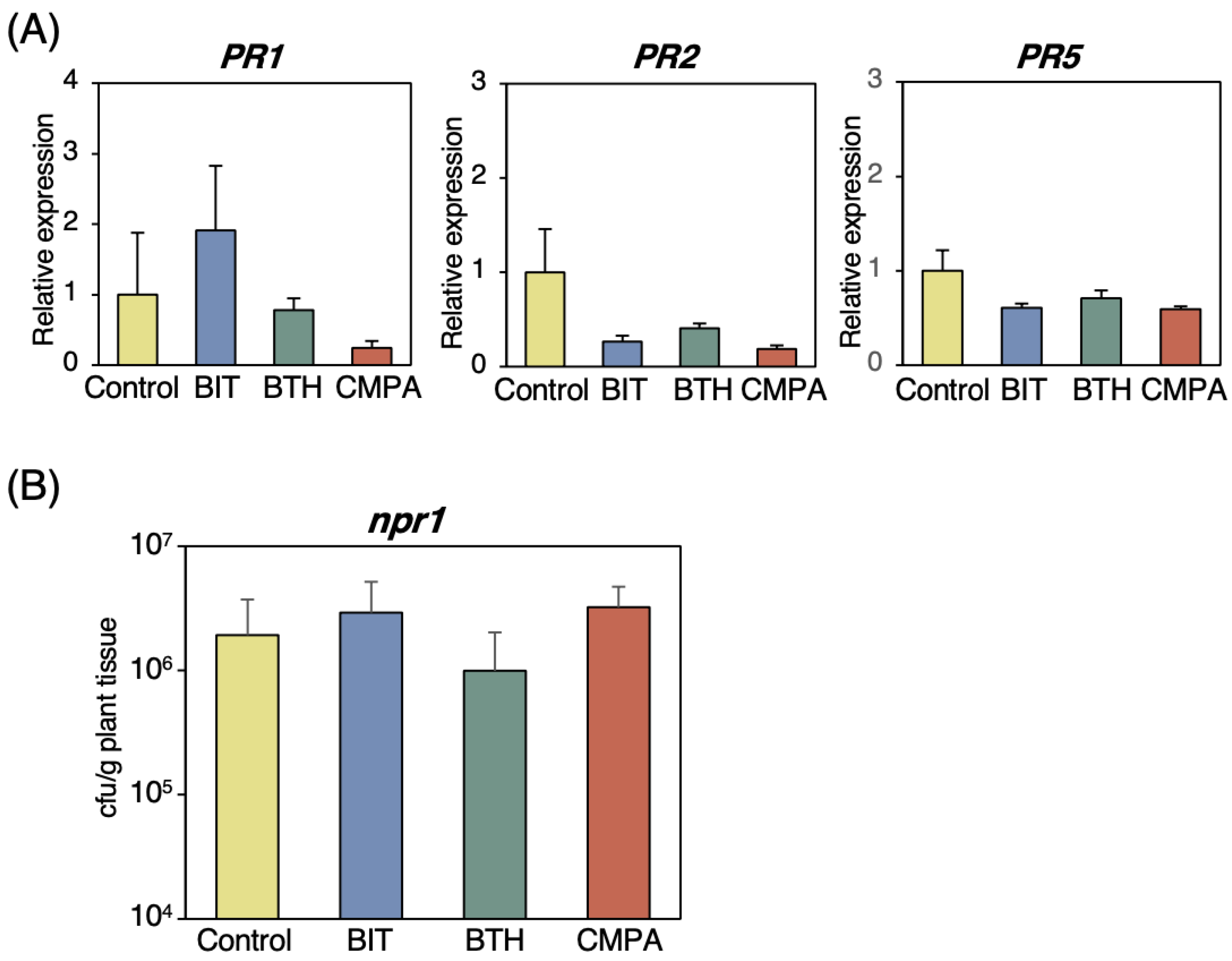
2.4. Effects of CMPA on SA Receptor-Deficient Mutant Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. Arabidopsis Pathogen Infection Assay
4.3. Analysis of Gene Expression by RT–qPCR Analysis
4.4. Extraction and Analysis of SA
4.5. Extraction and Analysis of GSH
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chester, K.S. The problem of acquired physiological immunity in plants. Q. Rev. Biol. 1933, 8, 275–324. [Google Scholar] [CrossRef]
- Durner, J.; Shah, J.; Klessig, D.F. Salicylic acid and disease resistance in plants. Trends Plant Sci. 1997, 2, 266–274. [Google Scholar] [CrossRef]
- Spoel, S.H. Signal Transduction in Systemic Immunity. Plant Cell 2019, 31, 1412–1413. [Google Scholar] [CrossRef] [PubMed]
- Métraux, J.P.; Signer, H.; Ryals, J.; Ward, E.; Wyss-Benz, M.; Gaudin, J.; Raschdorf, K.; Schmid, E.; Blum, W.; Inverardi, B. Increase in salicylic Acid at the onset of systemic acquired resistance in cucumber. Science 1990, 250, 1004–1006. [Google Scholar] [CrossRef]
- Ward, E.R.; Uknes, S.J.; Williams, S.C.; Dincher, S.S.; Wiederhold, D.L.; Alexander, D.C.; Ahl-Goy, P.; Metraux, J.P.; Ryals, J.A. Coordinate Gene Activity in Response to Agents That Induce Systemic Acquired Resistance. Plant Cell 1991, 3, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic Acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef]
- Delaney, T.P.; Uknes, S.; Vernooij, B.; Friedrich, L.; Weymann, K.; Negrotto, D.; Gaffney, T.; Gut-Rella, M.; Kessmann, H.; Ward, E.; et al. A central role of salicylic Acid in plant disease resistance. Science 1994, 266, 1247–1250. [Google Scholar] [CrossRef]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate Synthase is Required to Synthesize Salicylic Acid for Plant Defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.-K. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic Acid Biosynthesis in Plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef]
- Wang, W.; Withers, J.; Li, H.; Zwack, P.J.; Rusnac, D.V.; Shi, H.; Liu, L.; Yan, S.; Hinds, T.R.; Guttman, M.; et al. Structural basis of salicylic acid perception by Arabidopsis NPR proteins. Nature 2020, 586, 311–316. [Google Scholar] [CrossRef]
- Kumar, S.; Zavaliev, R.; Wu, Q.; Zhou, Y.; Cheng, J.; Dillard, L.; Powers, J.; Withers, J.; Zhao, J.; Guan, Z.; et al. Structural basis of NPR1 in activating plant immunity. Nature 2022, 605, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Kusajima, M.; Fujita, M.; Yamakawa, H.; Ushiwatari, T.; Mori, T.; Tsukamoto, K.; Hayashi, H.; Maruyama-Nakashita, A.; Che, F.S.; Nakashita, H. Characterization of plant immunity-activating mechanism by a pyrazole derivative. Biosci. Biotechnol. Biochem. 2020, 84, 1427–1435. [Google Scholar] [CrossRef]
- Watanabe, T.; Igarashi, H.; Matsumoto, K.; Seki, S.; Mase, S.; Sekizawa, Y. The characteristics of probenazole (oryzemate®) for the control of rice blast. J. Pestic. Sci. 1977, 2, 291–296. [Google Scholar] [CrossRef]
- Friedrich, L.; Lawton, K.; Ruessz, W.; Masner, P.; Specker, N.; Rella, M.G.; Meier, B.; Dincher, S.; Staub, T.; Uknes, S.; et al. A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 1996, 10, 61–70. [Google Scholar] [CrossRef]
- Yasuda, M.; Nakashita, H.; Yoshida, S. Tiadinil, a Novel Class of Activator of Systemic Acquired Resistance, Induces Defense Gene Expression and Disease Resistance in Tobacco. J. Pestic. Sci. 2004, 29, 46–49. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, W. Recent advances in synthetic chemical inducers of plant immunity. Front. Plant Sci. 2018, 9, 1613. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yang, S.; Zheng, W.; Wang, Y. Plant immunity inducers: From discovery to agricultural application. Stress Biol. 2022, 2, 5. [Google Scholar] [CrossRef]
- Lawton, K.A.; Friedrich, L.; Hunt, M.; Weymann, K.; Delaney, T.; Kessmann, H.; Staub, T.; Ryals, J. Benzothiadiazole Induces Disease Resistance in Arabidopsis by Activation of the Systemic Acquired Resistance Signal Transduction Pathway. Plant J. 1996, 10, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Ishikawa, A.; Jikumaru, Y.; Seki, M.; Umezawa, T.; Asami, T.; Maruyama-Nakashita, A.; Kudo, T.; Shinozaki, K.; Yoshida, S.; et al. Antagonistic Interaction between Systemic Acquired Resistance and the Abscisic Acid–mediated Abiotic Stress Response in Arabidopsis. Plant Cell 2008, 20, 1678–1692. [Google Scholar] [CrossRef]
- Yoshioka, K.; Nakashita, H.; Klessig, D.F.; Yamaguchi, I. Probenazole Induces Systemic Acquired Resistance in Arabidopsis with a Novel Type of Action. Plant J. 2001, 25, 149–157. [Google Scholar] [CrossRef]
- Nakashita, H. Studies on regulation of plant physiology by pesticides. J. Pestic. Sci. 2021, 46, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Nakashita, H.; Kusajima, M.; Kato, H.; Fujita, M. Regulation of SA-Mediated Signal Transduction in Plant Immune System. In Salicylic Acid—A Versatile Plant Growth Regulator; Hayat, S., Siddiqui, H., Damalas, C.A., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 235–247. [Google Scholar] [CrossRef]
- Nakashita, H.; Yoshioka, K.; Yasuda, M.; Nitta, T.; Arai, Y.; Yoshida, S.; Yamaguchi, I. Probenazole Induces Systemic Acquired Resistance in Tobacco through Salicylic Acid Accumulation. Physiol. Mol. Plant Pathol. 2002, 61, 197–203. [Google Scholar] [CrossRef]
- Vernooij, B.; Friedrich, L.; Goy, P.A.; Staub, T.; Kessmann, H.; Ryals, J. 2,6-dichloroisonicotinic acid-induced resistance to pathogens without the accumulation of salicylic acid. Mol. Plant Microbe Interact. 1995, 8, 228–234. [Google Scholar] [CrossRef]
- Yoshida, H.; Konishi, K.; Koike, K.; Nakagawa, T.; Sekido, S.; Yamaguchi, I. Effect of N-cyanomethyl-2-chloroisonicotinamide for control of rice blast. J. Pestic. Sci. 1990, 15, 413–417. [Google Scholar] [CrossRef]
- Nakashita, H.; Yasuda, M.; Nishioka, M.; Hasegawa, S.; Arai, Y.; Uramoto, M.; Yoshida, S.; Yamaguchi, I. Chloroisonicotinamide Derivative Induces a Broad Range of Disease Resistance in Rice and Tobacco. Plant Cell Physiol. 2002, 43, 823–831. [Google Scholar] [CrossRef]
- Yasuda, M.; Nakashita, H.; Hasegawa, S.; Nishioka, M.; Arai, Y.; Uramoto, M.; Yamaguchi, I.; Yoshida, S. N-cyanomethyl-2-chloroisonicotinamide induces systemic acquired resistance in Arabidopsis without salicylic acid accumulation. Biosci. Biotechnol. Biochem. 2003, 67, 322–328. [Google Scholar] [CrossRef]
- Nishioka, M.; Nakashita, H.; Suzuki, H.; Akiyama, S.; Yoshida, S.; Yamaguchi, I. Induction of Resistance against Rice Blast Disease by a Novel Class of Plant Activator, Pyrazolecarboxylic Acid Derivatives. J. Pestic. Sci. 2003, 28, 416–421. [Google Scholar] [CrossRef]
- Nishioka, M.; Nakashita, H.; Yasuda, M.; Yoshida, S.; Yamaguchi, I. Induction of Resistance against Rice Bacterial Leaf Blight by 3-Chloro-1-methyl-1H-pyrazole-5-carboxylic Acid. J. Pestic. Sci. 2005, 30, 47–49. [Google Scholar] [CrossRef]
- Yasuda, M.; Nishioka, M.; Nakashita, H.; Yamaguchi, I.; Yoshida, S. Pyrazolecarboxylic Acid Derivative Induces Systemic Acquired Resistance in Tobacco. Biosci. Biotechnol. Biochem. 2003, 67, 2614–2620. [Google Scholar] [CrossRef]
- Govrin, E.; Levine, A. Infection of Arabidopsis with a necrotrophic pathogen, Botrytis cinerea, elicits various defense responses but does not induce systemic acquired resistance (SAR). Plant Mol. Biol. 2002, 48, 267–276. [Google Scholar] [CrossRef]
- Ferrari, S.; Plotnikova, J.M.; De Lorenzo, G.; Ausubel, F.M. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 2003, 35, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Audenaert, K.; De Meyer, G.B.; Höfte, M.M. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002, 128, 491–501. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, G.; Capieau, K.; Audenaert, K.; Buchala, A.; Métraux, J.P.; Höfte, M. Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol. Plant Microbe Interact. 1999, 12, 450–458. [Google Scholar] [CrossRef]
- Zheng, L.; Campbell, M.; Murphy, J.; Lam, S.; Xu, J.R. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 2000, 13, 724–732. [Google Scholar] [CrossRef]
- Food Safety Commission of Japan. Dichlobentiazox (Pesticides). Food Saf. (Tokyo) 2020, 8, 6–7. [Google Scholar]
- Ando, S.; Jaskiewicz, M.; Mochizuki, S.; Koseki, S.; Miyashita, S.; Takahashi, H.; Conrath, U. Priming for enhanced ARGONAUTE2 activation accompanies induced resistance to cucumber mosaic virus in Arabidopsis thaliana. Mol. Plant Pathol. 2021, 22, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Chanda, B.; Venugopal, S.C.; Kulshrestha, S.; Navarre, D.A.; Downie, B.; Vaillancourt, L.; Kachroo, A.; Kachroo, P. Glycerol-3-phosphate levels are associated with basal resistance to the hemibiotrophic fungus Colletotrichum higginsianum in Arabidopsis. Plant Physiol. 2008, 147, 2017–2029. [Google Scholar] [CrossRef] [PubMed]
- Hevia, M.A.; Canessa, P.; Müller-Esparza, H.; Larrondo, L.F. A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 8744–8749. [Google Scholar] [CrossRef] [PubMed]
- Kusajima, M.; Fujita, M.; Soudthedlath, K.; Nakamura, H.; Yoneyama, K.; Nomura, T.; Akiyama, K.; Maruyama-Nakashita, A.; Asami, T.; Nakashita, H. Strigolactones Modulate Salicylic Acid-Mediated Disease Resistance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 5246. [Google Scholar] [CrossRef]
- Morikawa-Ichinose, T.; Kim, S.J.; Allahham, A.; Kawaguchi, R.; Maruyama-Nakashita, A. Glucosinolate Distribution in the Aerial Parts of sel1-10, a Disruption Mutant of the Sulfate Transporter SULTR1;2, in Mature Arabidopsis thaliana Plants. Plants 2019, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Ushiwatari, T.; Suyama, A.; Tominaga-Wada, R.; Wada, T.; Maruyama-Nakashita, A. Contribution of Root Hair Development to Sulfate Uptake in Arabidopsis. Plants 2019, 8, 106. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasuda, M.; Fujita, M.; Soudthedlath, K.; Kusajima, M.; Takahashi, H.; Tanaka, T.; Narita, F.; Asami, T.; Maruyama-Nakashita, A.; Nakashita, H. Characterization of Disease Resistance Induced by a Pyrazolecarboxylic Acid Derivative in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 9037. https://doi.org/10.3390/ijms24109037
Yasuda M, Fujita M, Soudthedlath K, Kusajima M, Takahashi H, Tanaka T, Narita F, Asami T, Maruyama-Nakashita A, Nakashita H. Characterization of Disease Resistance Induced by a Pyrazolecarboxylic Acid Derivative in Arabidopsis thaliana. International Journal of Molecular Sciences. 2023; 24(10):9037. https://doi.org/10.3390/ijms24109037
Chicago/Turabian StyleYasuda, Michiko, Moeka Fujita, Khamsalath Soudthedlath, Miyuki Kusajima, Hideki Takahashi, Tomoya Tanaka, Futo Narita, Tadao Asami, Akiko Maruyama-Nakashita, and Hideo Nakashita. 2023. "Characterization of Disease Resistance Induced by a Pyrazolecarboxylic Acid Derivative in Arabidopsis thaliana" International Journal of Molecular Sciences 24, no. 10: 9037. https://doi.org/10.3390/ijms24109037
APA StyleYasuda, M., Fujita, M., Soudthedlath, K., Kusajima, M., Takahashi, H., Tanaka, T., Narita, F., Asami, T., Maruyama-Nakashita, A., & Nakashita, H. (2023). Characterization of Disease Resistance Induced by a Pyrazolecarboxylic Acid Derivative in Arabidopsis thaliana. International Journal of Molecular Sciences, 24(10), 9037. https://doi.org/10.3390/ijms24109037






