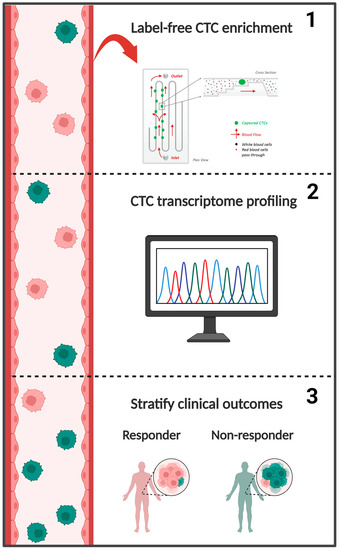
Transcriptome Profiling of Circulating Tumor Cells to Predict Clinical Outcomes in Metastatic Castration-Resistant Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. CTC Transcriptome Gene Panel
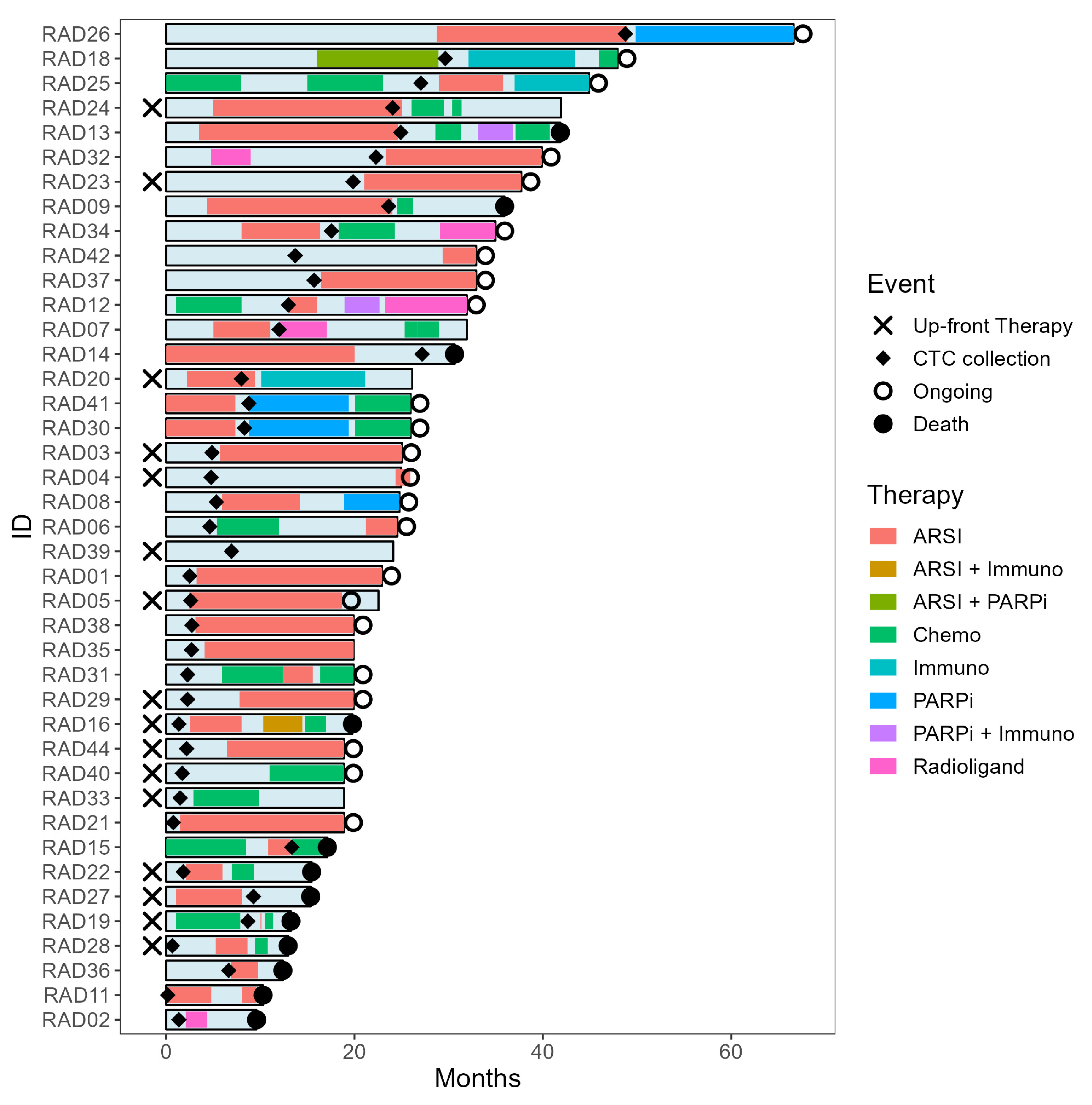
2.2. Clinical Follow-Up
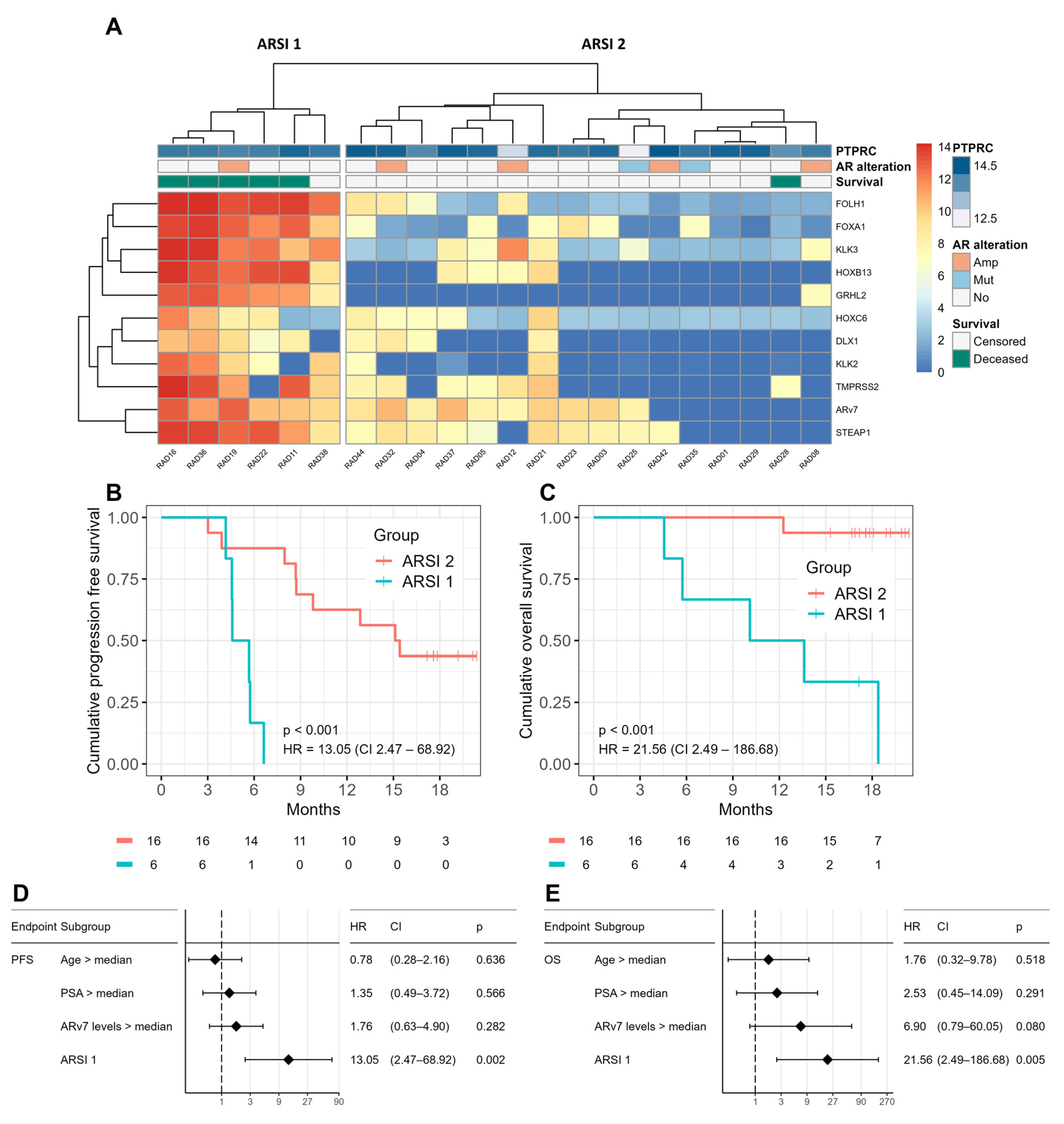
2.3. Genomic Alterations and CTC Transcriptomes
2.4. Prognostic Value of CTC Profiling
2.5. CTC Profiles Predict Therapy Response
2.6. Longitudinal CTC Profiling—A Case Discussion
3. Discussion
4. Materials and Methods
4.1. Gene Panel Development
4.2. Patient Recruitment
4.3. Healthy Donor Recruitment
4.4. Next-Generation DNA Sequencing
4.5. Presentation of Genomic Alterations
4.6. Sample Processing
4.7. Transcriptome Profiling
4.8. Clinical Follow-Up and Data Collection
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bister, K. Discovery of Oncogenes: The Advent of Molecular Cancer Research. Proc. Natl. Acad. Sci. USA 2015, 112, 15259–15260. [Google Scholar] [CrossRef] [PubMed]
- Peyraud, F.; Italiano, A. Combined Parp Inhibition and Immune Checkpoint Therapy in Solid Tumors. Cancers 2020, 12, 1502. [Google Scholar] [CrossRef] [PubMed]
- Teyssonneau, D.; Margot, H.; Cabart, M.; Anonnay, M.; Sargos, P.; Vuong, N.S.; Soubeyran, I.; Sevenet, N.; Roubaud, G. Prostate Cancer and PARP Inhibitors: Progress and Challenges. J. Hematol. Oncol. 2021, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Gilson, P.; Merlin, J.L.; Harlé, A. Deciphering Tumour Heterogeneity: From Tissue to Liquid Biopsy. Cancers 2022, 14, 1384. [Google Scholar] [CrossRef]
- Frangioni, J.V. New Technologies for Human Cancer Imaging. J. Clin. Oncol. 2008, 26, 4012–4021. [Google Scholar] [CrossRef]
- Braun, S.; Vogl, F.D.; Naume, B.; Janni, W.; Osborne, M.P.; Coombes, R.C.; Schlimok, G.; Diel, I.J.; Gerber, B.; Gebauer, G.; et al. A Pooled Analysis of Bone Marrow Micrometastasis in Breast Cancer. N. Engl. J. Med. 2005, 353, 793–802. [Google Scholar] [CrossRef]
- Riethdorf, S.; O’Flaherty, L.; Hille, C.; Pantel, K. Clinical Applications of the CellSearch Platform in Cancer Patients. Adv. Drug Deliv. Rev. 2018, 125, 102–121. [Google Scholar] [CrossRef]
- Lambros, M.B.; Seed, G.; Sumanasuriya, S.; Gil, V.; Crespo, M.; Fontes, M.; Chandler, R.; Mehra, N.; Fowler, G.; Ebbs, B.; et al. Single-Cell Analyses of Prostate Cancer Liquid Biopsies Acquired by Apheresis. Clin. Cancer Res. 2018, 24, 5635–5644. [Google Scholar] [CrossRef]
- Rushton, A.J.; Nteliopoulos, G.; Shaw, J.A.; Coombes, R.C. A Review of Circulating Tumour Cell Enrichment Technologies. Cancers 2021, 13, 970. [Google Scholar] [CrossRef]
- Casanova-Salas, I.; Athie, A.; Boutros, P.C.; Del Re, M.; Miyamoto, D.T.; Pienta, K.J.; Posadas, E.M.; Sowalsky, A.G.; Stenzl, A.; Wyatt, A.W.; et al. Quantitative and Qualitative Analysis of Blood-Based Liquid Biopsies to Inform Clinical Decision-Making in Prostate Cancer. Eur. Urol. 2021, 79, 762–771. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Li, D.; Ma, X.; Xu, K.; Ding, B.; Li, H.; Wang, Z.; Ouyang, W.; Long, G.; et al. Androgen Receptor Splice Variant 7 Predicts Shorter Response in Patients with Metastatic Hormone-Sensitive Prostate Cancer Receiving Androgen Deprivation Therapy. Eur. Urol. 2021, 79, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Veiga, T.; González-Conde, M.; León-Mateos, L.; Piñeiro-Cid, R.; Abuín, C.; Muinelo-Romay, L.; Martínez-Fernández, M.; Brea Iglesias, J.; García González, J.; Anido, U.; et al. Longitudinal CTCs Gene Expression Analysis on Metastatic Castration-Resistant Prostate Cancer Patients Treated with Docetaxel Reveals New Potential Prognosis Markers. Clin. Exp. Metastasis 2021, 38, 239–251. [Google Scholar] [CrossRef]
- Miller, M.C.; Robinson, P.S.; Wagner, C.; O’Shannessy, D.J. The ParsortixTM Cell Separation System—A Versatile Liquid Biopsy Platform. Cytom. Part A 2018, 93, 1234–1239. [Google Scholar] [CrossRef]
- Englert, D.F.; Seto, K.K.Y. Solid Phase Nucleic Acid Target Capture and Replication Using Strand Displacing Polymerases. U.S. Patent 11,060,131, 13 July 2021. [Google Scholar]
- Englert, D.; Kolesnikova, M.; Zaman, N.; Biosciences, A.; Hustler, A.; Wishart, G.; O’shannessy, D.J. Multiplex Gene Expression Using the HyCEAD Assay in CTCs Isolated with the ParsortixTM System. Cancer Res. 2019, 79, 1375. [Google Scholar] [CrossRef]
- Moore, R.G.; Khazan, N.; Coulter, M.A.; Singh, R.; Miller, M.C.; Sivagnanalingam, U.; Dubeshter, B.; Angel, C.; Liu, C.; Seto, K.; et al. Malignancy Assessment Using Gene Identification in Captured Cells Algorithm for the Prediction of Malignancy in Women with a Pelvic Mass. Obstet. Gynecol. 2022, 140, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Podolak, J.; Eilers, K.; Newby, T.; Slottke, R.; Tucker, E.; Olson, S.B.; Lue, H.-W.; Youngren, J.; Aggarwal, R.; Small, E.J.; et al. Androgen Receptor Amplification Is Concordant between Circulating Tumor Cells and Biopsies from Men Undergoing Treatment for Metastatic Castration Resistant Prostate Cancer. Oncotarget 2017, 8, 71447. [Google Scholar] [CrossRef] [PubMed]
- Barnett, E.; Schonhoft, J.; Schultz, N.D.; Lee, J.; Zaidi, S.; Abida, W.; Carmichael, T.; Dago, A.E.; Solit, D.B.; Wenstrup, R.; et al. Prevalence and Tissue Concordance of BRCA2 Copy Number Loss Evaluated by Single-Cell, Shallow Whole Genome Sequencing of Circulating Tumor Cells (CTCs) in Castration-Resistant Prostate Cancer (CRPC). J. Clin. Oncol. 2020, 38, 5531. [Google Scholar] [CrossRef]
- Franken, A.; Kraemer, A.; Sicking, A.; Watolla, M.; Rivandi, M.; Yang, L.; Warfsmann, J.; Polzer, B.M.; Friedl, T.W.P.; Meier-Stiegen, F.; et al. Comparative Analysis of EpCAM High-Expressing and Low-Expressing Circulating Tumour Cells with Regard to Their Clonal Relationship and Clinical Value. Br. J. Cancer 2023, 128, 1742–1752. [Google Scholar] [CrossRef]
- Di Donato, M.; Ostacolo, C.; Giovannelli, P.; Di Sarno, V.; Monterrey, I.M.G.; Campiglia, P.; Migliaccio, A.; Bertamino, A.; Castoria, G. Therapeutic Potential of TRPM8 Antagonists in Prostate Cancer. Sci. Rep. 2021, 11, 23232. [Google Scholar] [CrossRef]
- Ouderkirk, J.L.; Krendel, M. Non-Muscle Myosins in Tumor Progression, Cancer Cell Invasion, and Metastasis. Cytoskeleton 2014, 71, 447–463. [Google Scholar] [CrossRef]
- Sperger, J.M.; Emamekhoo, H.; McKay, R.R.; Stahlfeld, C.N.; Singh, A.; Chen, X.E.; Kwak, L.; Gilsdorf, C.S.; Wolfe, S.K.; Wei, X.X.; et al. Prospective Evaluation of Clinical Outcomes Using a Multiplex Liquid Biopsy Targeting Diverse Resistance Mechanisms in Metastatic Prostate Cancer. JCO 2021, 39, 2926–2937. [Google Scholar] [CrossRef]
- Wang, Z.; Shen, H.; Liang, Z.; Mao, Y.; Wang, C.; Xie, L. The Characteristics of Androgen Receptor Splice Variant 7 in the Treatment of Hormonal Sensitive Prostate Cancer: A Systematic Review and Meta-Analysis. Cancer Cell Int. 2020, 20, 149. [Google Scholar] [CrossRef]
- Dahiya, V.; Bagchi, G. Non-Canonical Androgen Signaling Pathways and Implications in Prostate Cancer. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119357. [Google Scholar] [CrossRef]
- Lassche, G.; Tada, Y.; van Herpen, C.M.L.; Jonker, M.A.; Nagao, T.; Saotome, T.; Hirai, H.; Saigusa, N.; Takahashi, H.; Ojiri, H.; et al. Predictive and Prognostic Biomarker Identification in a Large Cohort of Androgen Receptor-Positive Salivary Duct Carcinoma Patients Scheduled for Combined Androgen Blockade. Cancers 2021, 13, 3527. [Google Scholar] [CrossRef] [PubMed]
- Enna, S.J.; Bylund, D.B. Dihydrotestosterone. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–2. [Google Scholar] [CrossRef]
- He, Y.; Wei, T.; Ye, Z.; Orme, J.J.; Lin, D.; Sheng, H.; Fazli, L.; Jeffrey Karnes, R.; Jimenez, R.; Wang, L.; et al. A Noncanonical AR Addiction Drives Enzalutamide Resistance in Prostate Cancer. Nat. Commun. 2021, 12, 1521. [Google Scholar] [CrossRef] [PubMed]
- Tien, A.H.; Sadar, M.D. Keys to Unlock Androgen Receptor Translocation. J. Biol. Chem. 2019, 294, 8711–8712. [Google Scholar] [CrossRef] [PubMed]
- Matuszczak, M.; Schalken, J.A.; Salagierski, M. Prostate Cancer Liquid Biopsy Biomarkers’ Clinical Utility in Diagnosis and Prognosis. Cancers 2021, 13, 3373. [Google Scholar] [CrossRef] [PubMed]
- Hamid, A.R.A.H.; Hoogland, A.M.; Smit, F.; Jannink, S.; Van Rijt-Van De Westerlo, C.; Jansen, C.F.J.; Van Leenders, G.J.L.H.; Verhaegh, G.W.; Schalken, J.A. The Role of HOXC6 in Prostate Cancer Development. Prostate 2015, 75, 1868–1876. [Google Scholar] [CrossRef]
- Luo, Z.; Farnham, P.J. Genome-Wide Analysis of HOXC4 and HOXC6 Regulated Genes and Binding Sites in Prostate Cancer Cells. PLoS ONE 2020, 15, e0228590. [Google Scholar] [CrossRef]
- Goel, S.; Bhatia, V.; Kundu, S.; Biswas, T.; Carskadon, S.; Gupta, N.; Asim, M.; Morrissey, C.; Palanisamy, N.; Ateeq, B. Transcriptional Network Involving ERG and AR Orchestrates Distal-Less Homeobox-1 Mediated Prostate Cancer Progression. Nat. Commun. 2021, 12, 5325. [Google Scholar] [CrossRef]
- Leyten, G.H.J.M.; Hessels, D.; Smit, F.P.; Jannink, S.A.; De Jong, H.; Melchers, W.J.G.; Cornel, E.B.; De Reijke, T.M.; Vergunst, H.; Kil, P.; et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin. Cancer Res. 2015, 21, 3061–3070. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Gu, Y.; Hui, K.; Huang, J.; Xu, S.; Wu, S.; Li, L.; Fan, J.; Wang, X.; Hsieh, J.T.; et al. AKR1C3, a Crucial Androgenic Enzyme in Prostate Cancer, Promotes Epithelial-Mesenchymal Transition and Metastasis through Activating ERK Signaling. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 472.e11–472.e20. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wang, Z.; Li, J.; Wu, X.; Fan, N.; Li, D.; Liu, F.; Plum, P.S.; Hoppe, S.; Hillmer, A.M.; et al. Aldo-Keto Reductase 1c3 Mediates Chemotherapy Resistance in Esophageal Adenocarcinoma via Ros Detoxification. Cancers 2021, 13, 2403. [Google Scholar] [CrossRef] [PubMed]
- Majewski, Ł.; Sobczak, M.; Wasik, A.; Skowronek, K.; Rędowicz, M.J. Myosin VI in PC12 Cells Plays Important Roles in Cell Migration and Proliferation but Not in Catecholamine Secretion. J. Muscle Res. Cell Motil. 2011, 32, 291–302. [Google Scholar] [CrossRef]
- Wang, D.; Zhu, L.; Liao, M.; Zeng, T.; Zhuo, W.; Yang, S.; Wu, W. MYO6 Knockdown Inhibits the Growth and Induces the Apoptosis of Prostate Cancer Cells by Decreasing the Phosphorylation of ERK1/2 and PRAS40. Oncol. Rep. 2016, 36, 1285–1292. [Google Scholar] [CrossRef]
- Dunn, T.A.; Chen, S.; Faith, D.A.; Hicks, J.L.; Platz, E.A.; Chen, Y.; Ewing, C.M.; Sauvageot, J.; Isaacs, W.B.; De Marzo, A.M.; et al. A Novel Role of Myosin VI in Human Prostate Cancer. Am. J. Pathol. 2006, 169, 1843–1854. [Google Scholar] [CrossRef]
- Asim, M.; Tarish, F.; Zecchini, H.I.; Sanjiv, K.; Gelali, E.; Massie, C.E.; Baridi, A.; Warren, A.Y.; Zhao, W.; Ogris, C.; et al. Synthetic Lethality between Androgen Receptor Signalling and the PARP Pathway in Prostate Cancer. Nat. Commun. 2017, 8, 374. [Google Scholar] [CrossRef]
- Nizialek, E.; Haffner, M.; Bhamidipati, A.; Yegnasubramanian, S. The Effect of PARP Inhibition on Androgen Receptor Localization and Activity in Castration Resistant Prostate Cancer. J. Clin. Oncol. 2022, 40, e17037. [Google Scholar] [CrossRef]
- Congregado, B.; Rivero, I.; Osmán, I.; Sáez, C.; López, R.M. PARP Inhibitors: A New Horizon for Patients with Prostate Cancer. Biomedicines 2022, 10, 1416. [Google Scholar] [CrossRef]
- Ehsani, M.; David, F.O.; Baniahmad, A. Androgen Receptor-Dependent Mechanisms Mediating Drug Resistance in Prostate Cancer. Cancers 2021, 13, 1534. [Google Scholar] [CrossRef]
- Au, S.H.; Edd, J.; Haber, D.A.; Maheswaran, S.; Stott, S.L.; Toner, M. Clusters of Circulating Tumor Cells: A Biophysical and Technological Perspective. Curr. Opin. Biomed. Eng. 2017, 3, 13–19. [Google Scholar] [CrossRef]
- Castro-Giner, F.; Aceto, N. Tracking Cancer Progression: From Circulating Tumor Cells to Metastasis. Genome Med. 2020, 12, 31. [Google Scholar] [CrossRef]
- Bilandzic, M.; Rainczuk, A.; Green, E.; Fairweather, N.; Jobling, T.W.; Plebanski, M.; Stephens, A.N. Keratin-14 (KRT14) Positive Leader Cells Mediate Mesothelial Clearance and Invasion by Ovarian Cancer Cells. Cancers 2019, 11, 1228. [Google Scholar] [CrossRef]
- Simper, N.B.; Jones, C.L.; MacLennan, G.T.; Montironi, R.; Williamson, S.R.; Osunkoya, A.O.; Wang, M.; Zhang, S.; Grignon, D.J.; Eble, J.N.; et al. Basal Cell Carcinoma of the Prostate Is an Aggressive Tumor with Frequent Loss of PTEN Expression and Overexpression of EGFR. Hum. Pathol. 2015, 46, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Fels, B.; Bulk, E.; Pethő, Z.; Schwab, A. The Role of TRP Channels in the Metastatic Cascade. Pharmaceuticals 2018, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.J.; Wang, R.; Kuang, X.R.; Zhou, J.Y.; Hu, X.L. Elevated Expression of Myosin VI Contributes to Breast Cancer Progression via MAPK/ERK Signaling Pathway. Cell. Signal. 2023, 106, 110633. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Quinn, M.C.J.; Wilson, D.J.; Young, F.; Dempsey, A.A.; Arcand, S.L.; Birch, A.H.; Wojnarowicz, P.M.; Provencher, D.; Mes-Masson, A.M.; Englert, D.; et al. The Chemiluminescence Based Ziplex® Automated Workstation Focus Array Reproduces Ovarian Cancer Affymetrix GeneChip® Expression Profiles. J. Transl. Med. 2009, 7, 55. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Patil, I. Visualizations with Statistical Details: The “ggstatsplot” Approach. J. Open Source Softw. 2021, 6, 3167. [Google Scholar] [CrossRef]



| Number of patients | 40 |
| Age, median (range) | 65 (45–85) |
| Years since CRPC, the median (range) | 2 (0.8–5.5) |
| PSA (μg/L) at inclusion, median (range) | 31.5 (1.2–1600) |
| De novo metastatic diseases, Num. (%) | |
| Yes | 29 (72.5%) |
| No | 11 (27.5%) |
| Alkaline phosphatase (U/L), median (range) | 106 (54–1866) |
| Type of therapy following inclusion, Num. (%) | |
| ARSI | 22 (54%) |
| Chemotherapy | 9 (22%) |
| Immunotherapy | 2 (5%) |
| Radioligand therapy | 2 (5%) |
| PARPi therapy | 2 (5%) |
| Active surveillance | 3 (7%) |
| 1-year survival per therapy, percentage (95% CI) | |
| Therapy-agnostic | 82% (72–95%) |
| ARSI | 86% (73–100%) |
| Chemotherapy | 89% (71–100%) |
| Immunotherapy | 50% (13–100%) |
| Radioligand | 50% (13–100%) |
| PARPi | 100% |
| Active surveillance | 33% (6.7–100%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Groen, L.; Kloots, I.; Englert, D.; Seto, K.; Estafanos, L.; Smith, P.; Verhaegh, G.W.; Mehra, N.; Schalken, J.A. Transcriptome Profiling of Circulating Tumor Cells to Predict Clinical Outcomes in Metastatic Castration-Resistant Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 9002. https://doi.org/10.3390/ijms24109002
Groen L, Kloots I, Englert D, Seto K, Estafanos L, Smith P, Verhaegh GW, Mehra N, Schalken JA. Transcriptome Profiling of Circulating Tumor Cells to Predict Clinical Outcomes in Metastatic Castration-Resistant Prostate Cancer. International Journal of Molecular Sciences. 2023; 24(10):9002. https://doi.org/10.3390/ijms24109002
Chicago/Turabian StyleGroen, Levi, Iris Kloots, David Englert, Kelly Seto, Lana Estafanos, Paul Smith, Gerald W. Verhaegh, Niven Mehra, and Jack A. Schalken. 2023. "Transcriptome Profiling of Circulating Tumor Cells to Predict Clinical Outcomes in Metastatic Castration-Resistant Prostate Cancer" International Journal of Molecular Sciences 24, no. 10: 9002. https://doi.org/10.3390/ijms24109002
APA StyleGroen, L., Kloots, I., Englert, D., Seto, K., Estafanos, L., Smith, P., Verhaegh, G. W., Mehra, N., & Schalken, J. A. (2023). Transcriptome Profiling of Circulating Tumor Cells to Predict Clinical Outcomes in Metastatic Castration-Resistant Prostate Cancer. International Journal of Molecular Sciences, 24(10), 9002. https://doi.org/10.3390/ijms24109002