Circulating Tumor DNA in Gastric Adenocarcinoma: Future Clinical Applications and Perspectives
Abstract
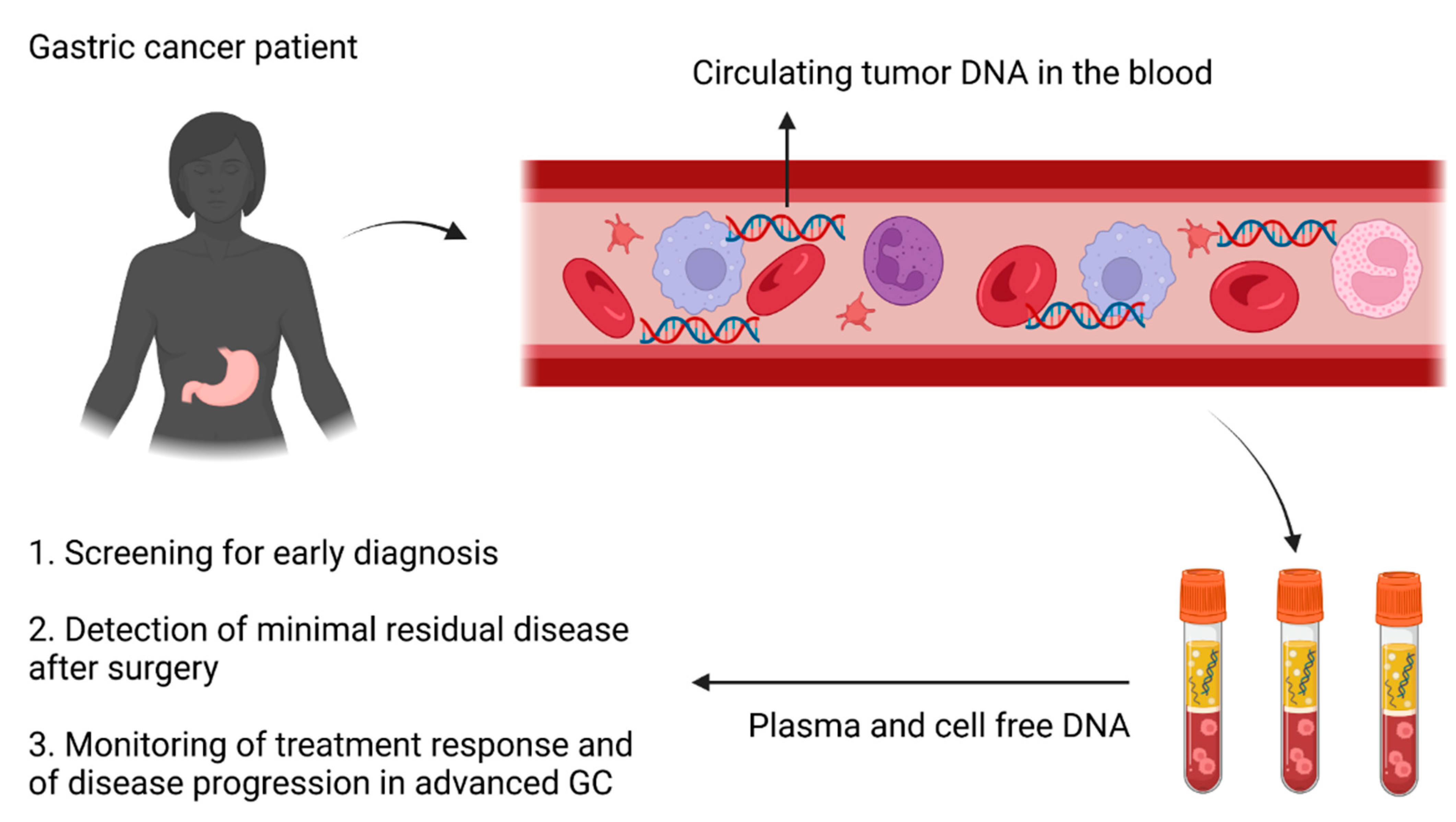
1. Introduction
2. Characteristics of ctDNA and Detection Methods
- (1)
- A targeted approach that identifies specific mutations previously detected in the primary tumor. It is fast, highly specific, and can detect very low allele frequencies. It can be used for early disease detection, monitoring of early-stage tumors, or MRD analysis. The difficulty of this approach is that it requires abundant information about the tumor genome [17,18,19,20];
- (2)
- An untargeted approach that implies a wide genome or exome sequencing and does not require a prior characterization of the primary tumor. This approach is more useful in advanced or metastatic settings in order to identify actionable molecular alterations or mutations related to therapeutic resistance. The disadvantage is that high concentrations of ctDNA are required with an overall low sensitivity [17,18,19,20]. Implemented methodologies include NGS, digital PCR, quantitative PCR (qPCR), and mass spectrometry [17]. As ctDNA levels are influenced by multiple tumor- and host-related variables, choosing the appropriate ct-DNA assay and its timing to address a specific clinical or scientific issue is essential. At present, no assay can be considered fit for all purposes. The CfDNA fraction fluctuates according to tumor stage, type, and site. In the case of higher tumor burden and/or extra-cranial progression, solid tumors will more likely shed ct-DNA into the bloodstream if compared to tumors with limited burden and or intracranial progression only [17]. Moreover, detectable ct-DNA levels have been found in >75% of patients with advanced pancreatic, bladder, ovarian colorectal, breast, melanoma, hepatocellular, head and neck, and gastroesophageal cancers, but in less than 50% of patients with brain, renal, thyroid, and prostate cancers [21].
3. Clinical Applications of ctDNA in Gastric Cancer
3.1. Early Diagnosis
3.2. Minimal Residual Disease
3.3. Advanced Disease
4. Main Challenges and Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Sexton, R.E.; Al Hallak, M.N.; Diab, M.; Azmi, A.S. Gastric cancer: A comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020, 39, 1179–1203. [Google Scholar] [CrossRef] [PubMed]
- Alsina, M.; Arrazubi, V.; Diez, M.; Tabernero, J. Current developments in gastric cancer: From molecular profiling to treatment strategy. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 155–170. [Google Scholar] [CrossRef]
- Lordick, F.; Carneiro, F.; Cascinu, S.; Fleitas, T.; Haustermans, K.; Piessen, G.; Vogel, A.; Smyth, E.C. Gastric cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 1005–1020. [Google Scholar] [CrossRef]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Shitara, K.; Moehler, M.; Garrido, M.; Salman, P.; Shen, L.; Wyrwicz, L.; Yamaguchi, K.; Skoczylas, T.; Campos Bragagnoli, A.; et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet 2021, 398, 27–40. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Miceli, R.; Raimondi, A.; Kim, Y.W.; Kang, W.K.; Langley, R.E.; Choi, Y.Y.; Kim, K.M.; Nankivell, M.G.; Morano, F.; et al. Individual Patient Data Meta-Analysis of the Value of Microsatellite Instability As a Biomarker in Gastric Cancer. J. Clin. Oncol. 2019, 37, 3392–3400. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Fuchs, C.S.; Shitara, K.; Tabernero, J.; Muro, K.; Van Cutsem, E.; Bang, Y.J.; De Vita, F.; Landers, G.; Yen, C.J.; et al. Assessment of Pembrolizumab Therapy for the Treatment of Microsatellite Instability-High Gastric or Gastroesophageal Junction Cancer Among Patients in the KEYNOTE-059, KEYNOTE-061, and KEYNOTE-062 Clinical Trials. JAMA Oncol. 2021, 7, 895–902. [Google Scholar] [CrossRef]
- Shitara, K.; Lordick, F.; Bang, Y.J.; Enzinger, P.; Ilson, D.; Shah, M.A.; Van Cutsem, E.; Xu, R.H.; Aprile, G.; Xu, J.; et al. Zolbetuximab plus mFOLFOX6 in patients with CLDN18.2-positive, HER2-negative, untreated, locally advanced unresectable or metastatic gastric or gastro-oesophageal junction adenocarcinoma (SPOTLIGHT): A multicentre, randomised, double-blind, phase 3 trial. Lancet 2023, 401, 1655–1668. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Ma, X.; Ou, K.; Liu, X.; Yang, L. Application progress of liquid biopsy in gastric cancer. Front. Oncol. 2022, 12, 969866. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, C.G.; Hussain, S.; Trapani, D.; El Bairi, K.; Altuna, S.C.; Seeber, A.; Odhiambo, A.; Habeeb, B.S.; Seid, F. The Emerging Role of Liquid Biopsy in Gastric Cancer. J. Clin. Med. 2021, 10, 2108. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zhou, M.; Xu, Y.; Gu, X.; Zou, M.; Abudushalamu, G.; Yao, Y.; Fan, X.; Wu, G. Clinical application and detection techniques of liquid biopsy in gastric cancer. Mol. Cancer 2023, 22, 7. [Google Scholar] [CrossRef]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef]
- Rolfo, C.; Cardona, A.F.; Cristofanilli, M.; Paz-Ares, L.; Diaz Mochon, J.J.; Duran, I.; Raez, L.E.; Russo, A.; Lorente, J.A.; Malapelle, U.; et al. Challenges and opportunities of cfDNA analysis implementation in clinical practice: Perspective of the International Society of Liquid Biopsy (ISLB). Crit. Rev. Oncol. Hematol. 2020, 151, 102978. [Google Scholar] [CrossRef]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
- Rodriguez, J.; Avila, J.; Rolfo, C.; Ruiz-Patino, A.; Russo, A.; Ricaurte, L.; Ordonez-Reyes, C.; Arrieta, O.; Zatarain-Barron, Z.L.; Recondo, G.; et al. When Tissue is an Issue the Liquid Biopsy is Nonissue: A Review. Oncol. Ther. 2021, 9, 89–110. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ji, H.; Yang, W.; Zhang, M.; Guo, Y.; Li, B.; Wang, J.; Chen, R.; Chen, Y.; Wang, X. Liquid biopsy using ascitic fluid and pleural effusion supernatants for genomic profiling in gastrointestinal and lung cancers. BMC Cancer 2022, 22, 1020. [Google Scholar] [CrossRef] [PubMed]
- Connors, D.; Allen, J.; Alvarez, J.D.; Boyle, J.; Cristofanilli, M.; Hiller, C.; Keating, S.; Kelloff, G.; Leiman, L.; McCormack, R.; et al. International liquid biopsy standardization alliance white paper. Crit. Rev. Oncol. Hematol. 2020, 156, 103112. [Google Scholar] [CrossRef] [PubMed]
- Ofman, J.; Raza, A. Taking Early Cancer Detection to the Next Level. Scientific American, 1 November 2020. [Google Scholar]
- Clarke, C.A.; Hubbell, E.; Kurian, A.W.; Colditz, G.A.; Hartman, A.R.; Gomez, S.L. Projected Reductions in Absolute Cancer-Related Deaths from Diagnosing Cancers Before Metastasis, 2006–2015. Cancer Epidemiol. Biomark. Prev. 2020, 29, 895–902. [Google Scholar] [CrossRef]
- Shimada, H.; Noie, T.; Ohashi, M.; Oba, K.; Takahashi, Y. Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer 2014, 17, 26–33. [Google Scholar] [CrossRef]
- Kim, K.; Shin, D.G.; Park, M.K.; Baik, S.H.; Kim, T.H.; Kim, S.; Lee, S. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: Diagnostic validity and significant reduction of cfDNA after surgical resection. Ann. Surg. Treat. Res. 2014, 86, 136–142. [Google Scholar] [CrossRef]
- Park, J.L.; Kim, H.J.; Choi, B.Y.; Lee, H.C.; Jang, H.R.; Song, K.S.; Noh, S.M.; Kim, S.Y.; Han, D.S.; Kim, Y.S. Quantitative analysis of cell-free DNA in the plasma of gastric cancer patients. Oncol. Lett. 2012, 3, 921–926. [Google Scholar] [CrossRef]
- Sai, S.; Ichikawa, D.; Tomita, H.; Ikoma, D.; Tani, N.; Ikoma, H.; Kikuchi, S.; Fujiwara, H.; Ueda, Y.; Otsuji, E. Quantification of plasma cell-free DNA in patients with gastric cancer. Anticancer Res. 2007, 27, 2747–2751. [Google Scholar]
- Qian, C.; Ju, S.; Qi, J.; Zhao, J.; Shen, X.; Jing, R.; Yu, J.; Li, L.; Shi, Y.; Zhang, L.; et al. Alu-based cell-free DNA: A novel biomarker for screening of gastric cancer. Oncotarget 2017, 8, 54037–54045. [Google Scholar] [CrossRef]
- Lan, Y.T.; Chen, M.H.; Fang, W.L.; Hsieh, C.C.; Lin, C.H.; Jhang, F.Y.; Yang, S.H.; Lin, J.K.; Chen, W.S.; Jiang, J.K.; et al. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget 2017, 8, 3009–3017. [Google Scholar] [CrossRef]
- Grenda, A.; Wojas-Krawczyk, K.; Skoczylas, T.; Krawczyk, P.; Sierocinska-Sawa, J.; Wallner, G.; Milanowski, J. HER2 gene assessment in liquid biopsy of gastric and esophagogastric junction cancer patients qualified for surgery. BMC Gastroenterol. 2020, 20, 382. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.; Chakrabarty, M.; Cohn, E.M.; Leon, S.A. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer 1983, 51, 2116–2120. [Google Scholar] [CrossRef] [PubMed]
- Abbaszadegan, M.R.; Moaven, O.; Sima, H.R.; Ghafarzadegan, K.; A’Rabi, A.; Forghani, M.N.; Raziee, H.R.; Mashhadinejad, A.; Jafarzadeh, M.; Esmaili-Shandiz, E.; et al. p16 promoter hypermethylation: A useful serum marker for early detection of gastric cancer. World J. Gastroenterol. 2008, 14, 2055–2060. [Google Scholar] [CrossRef]
- Bernal, C.; Aguayo, F.; Villarroel, C.; Vargas, M.; Diaz, I.; Ossandon, F.J.; Santibanez, E.; Palma, M.; Aravena, E.; Barrientos, C.; et al. Reprimo as a potential biomarker for early detection in gastric cancer. Clin. Cancer Res. 2008, 14, 6264–6269. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, D.; Koike, H.; Ikoma, H.; Ikoma, D.; Tani, N.; Otsuji, E.; Kitamura, K.; Yamagishi, H. Detection of aberrant methylation as a tumor marker in serum of patients with gastric cancer. Anticancer Res. 2004, 24, 2477–2481. [Google Scholar]
- Kanyama, Y.; Hibi, K.; Nakayama, H.; Kodera, Y.; Ito, K.; Akiyama, S.; Nakao, A. Detection of p16 promoter hypermethylation in serum of gastric cancer patients. Cancer Sci. 2003, 94, 418–420. [Google Scholar] [CrossRef]
- Kim, T.Y.; Lee, H.J.; Hwang, K.S.; Lee, M.; Kim, J.W.; Bang, Y.J.; Kang, G.H. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab. Investig. 2004, 84, 479–484. [Google Scholar] [CrossRef]
- Ren, J.; Lu, P.; Zhou, X.; Liao, Y.; Liu, X.; Li, J.; Wang, W.; Wang, J.; Wen, L.; Fu, W.; et al. Genome-Scale Methylation Analysis of Circulating Cell-Free DNA in Gastric Cancer Patients. Clin. Chem. 2022, 68, 354–364. [Google Scholar] [CrossRef]
- Anderson, B.W.; Suh, Y.S.; Choi, B.; Lee, H.J.; Yab, T.C.; Taylor, W.R.; Dukek, B.A.; Berger, C.K.; Cao, X.; Foote, P.H.; et al. Detection of Gastric Cancer with Novel Methylated DNA Markers: Discovery, Tissue Validation, and Pilot Testing in Plasma. Clin. Cancer Res. 2018, 24, 5724–5734. [Google Scholar] [CrossRef]
- Kandimalla, R.; Xu, J.; Link, A.; Matsuyama, T.; Yamamura, K.; Parker, M.I.; Uetake, H.; Balaguer, F.; Borazanci, E.; Tsai, S.; et al. EpiPanGI Dx: A Cell-free DNA Methylation Fingerprint for the Early Detection of Gastrointestinal Cancers. Clin. Cancer Res. 2021, 27, 6135–6144. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Ueberroth, B.E.; Jones, J.C.; Bekaii-Saab, T. The role of circulating tumor DNA in evaluating minimal residual disease in luminal gastrointestinal malignancies. Clin. Adv. Hematol. Oncol. 2022, 20, 444–455. [Google Scholar]
- Tarazona, N.; Gimeno-Valiente, F.; Cervantes, A. Minimal residual disease in gastroesophageal adenocarcinoma: The search for the invisible. ESMO Open 2022, 7, 100547. [Google Scholar] [CrossRef] [PubMed]
- Cmero, M.; Yuan, K.; Ong, C.S.; Schroder, J.; PCAWG Evolution and Heterogeneity Working Group; Corcoran, N.M.; Papenfuss, T.; Hovens, C.M.; Markowetz, F.; Macintyre, G.; et al. Inferring structural variant cancer cell fraction. Nat. Commun. 2020, 11, 730. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Stanislaw, S.; Spain, L.; Gallegos, L.L.; Rowan, A.; Schnidrig, D.; Rosenbaum, H.; Harle, A.; Au, L.; Hill, S.M.; et al. Representative Sequencing: Unbiased Sampling of Solid Tumor Tissue. Cell Rep. 2020, 31, 107550. [Google Scholar] [CrossRef]
- Yang, J.; Gong, Y.; Lam, V.K.; Shi, Y.; Guan, Y.; Zhang, Y.; Ji, L.; Chen, Y.; Zhao, Y.; Qian, F.; et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Leal, A.; van Grieken, N.C.T.; Palsgrove, D.N.; Phallen, J.; Medina, J.E.; Hruban, C.; Broeckaert, M.A.M.; Anagnostou, V.; Adleff, V.; Bruhm, D.C.; et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat. Commun. 2020, 11, 525. [Google Scholar] [CrossRef]
- Kim, Y.W.; Kim, Y.H.; Song, Y.; Kim, H.S.; Sim, H.W.; Poojan, S.; Eom, B.W.; Kook, M.C.; Joo, J.; Hong, K.M. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Ko, K.; Kananazawa, Y.; Yamada, T.; Kakinuma, D.; Matsuno, K.; Ando, F.; Kuriyama, S.; Matsuda, A.; Yoshida, H. Methylation status and long-fragment cell-free DNA are prognostic biomarkers for gastric cancer. Cancer Med. 2021, 10, 2003–2012. [Google Scholar] [CrossRef]
- Shoda, K.; Ichikawa, D.; Fujita, Y.; Masuda, K.; Hiramoto, H.; Hamada, J.; Arita, T.; Konishi, H.; Komatsu, S.; Shiozaki, A.; et al. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer 2017, 20, 126–135. [Google Scholar] [CrossRef]
- Ling, Z.Q.; Lv, P.; Lu, X.X.; Yu, J.L.; Han, J.; Ying, L.S.; Zhu, X.; Zhu, W.Y.; Fang, X.H.; Wang, S.; et al. Circulating Methylated XAF1 DNA Indicates Poor Prognosis for Gastric Cancer. PLoS ONE 2013, 8, e67195. [Google Scholar] [CrossRef]
- Maron, S.B.; Chase, L.M.; Lomnicki, S.; Kochanny, S.; Moore, K.L.; Joshi, S.S.; Landron, S.; Johnson, J.; Kiedrowski, L.A.; Nagy, R.J.; et al. Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma. Clin. Cancer Res. 2019, 25, 7098–7112. [Google Scholar] [CrossRef] [PubMed]
- Davidson, M.; Barber, L.J.; Woolston, A.; Cafferkey, C.; Mansukhani, S.; Griffiths, B.; Moorcraft, S.Y.; Rana, I.; Begum, R.; Assiotis, I.; et al. Detecting and Tracking Circulating Tumour DNA Copy Number Profiles during First Line Chemotherapy in Oesophagogastric Adenocarcinoma. Cancers 2019, 11, 736. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, C.; Zhang, M.; Li, B.; Niu, Y.; Chen, L.; Yang, J.; Lu, S.; Gao, J.; Shen, L. Chromosomal instability of circulating tumor DNA reflect therapeutic responses in advanced gastric cancer. Cell Death Dis. 2019, 10, 697. [Google Scholar] [CrossRef]
- Shitara, K.; Bang, Y.J.; Iwasa, S.; Sugimoto, N.; Ryu, M.H.; Sakai, D.; Chung, H.C.; Kawakami, H.; Yabusaki, H.; Lee, J.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Gastric Cancer. N. Engl. J. Med. 2020, 382, 2419–2430. [Google Scholar] [CrossRef]
- Wang, H.; Li, B.; Liu, Z.; Gong, J.; Shao, L.; Ren, J.; Niu, Y.; Bo, S.; Li, Z.; Lai, Y.; et al. HER2 copy number of circulating tumour DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer. Eur. J. Cancer 2018, 88, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.S.; Liu, Z.X.; Lu, Y.X.; Bao, H.; Wu, X.; Zeng, Z.L.; Liu, Z.; Zhao, Q.; He, C.Y.; Lu, J.H.; et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 2019, 68, 1152–1161. [Google Scholar] [CrossRef]
- Shitara, K.; Bang, Y.J.; Iwasa, S.; Sugimoto, N.; Ryu, M.H.; Sakai, D.; Chung, H.; Omuro, H.; Kawakami, H.; Yabusaki, H.; et al. Exploratory biomarker analysis of trastuzumab deruxtecan in DESTINY-Gastric01, a randomized, phase 2, multicenter, open-label study in patients with HER2-positive or -low advanced gastric or gastroesophageal junction adenocarcinoma. In Proceedings of the ESMO World Congress on Gastrointestinal Cancer, virtual event, 30 June–3 July 2021; p. S224. [Google Scholar]
- Jin, Y.; Chen, D.L.; Wang, F.; Yang, C.P.; Chen, X.X.; You, J.Q.; Huang, J.S.; Shao, Y.; Zhu, D.Q.; Ouyang, Y.M.; et al. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol. Cancer 2020, 19, 154. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef]
- Xie, T.; Liu, Y.; Zhang, Z.; Zhang, X.; Gong, J.; Qi, C.; Li, J.; Shen, L.; Peng, Z. Positive Status of Epstein-Barr Virus as a Biomarker for Gastric Cancer Immunotherapy: A Prospective Observational Study. J. Immunother. 2020, 43, 139–144. [Google Scholar] [CrossRef]
- Qiu, M.Z.; He, C.Y.; Lu, S.X.; Guan, W.L.; Wang, F.; Wang, X.J.; Jin, Y.; Wang, F.H.; Li, Y.H.; Shao, J.Y.; et al. Prospective observation: Clinical utility of plasma Epstein-Barr virus DNA load in EBV-associated gastric carcinoma patients. Int. J. Cancer 2020, 146, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Boldrin, E.; Piano, M.A.; Alfieri, R.; Mazza, M.; Vassallo, L.; Scapinello, A.; Pilati, P.; Curtarello, M. MSI Analysis in Solid and Liquid Biopsies of Gastroesophageal Adenocarcinoma Patients: A Molecular Approach. Int. J. Mol. Sci. 2021, 22, 7244. [Google Scholar] [CrossRef]
- Deng, N.; Goh, L.K.; Wang, H.; Das, K.; Tao, J.; Tan, I.B.; Zhang, S.; Lee, M.; Wu, J.; Lim, K.H.; et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut 2012, 61, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, Z.A.; Enzinger, P.C.; Kang, Y.K.; Qin, S.; Yamaguchi, K.; Kim, I.H.; Saeed, A.; Oh, S.C.; Li, J.; Turk, H.M.; et al. Bemarituzumab in patients with FGFR2b-selected gastric or gastro-oesophageal junction adenocarcinoma (FIGHT): A randomised, double-blind, placebo-controlled, phase 2 study. Lancet Oncol. 2022, 23, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, S.T.; Kim, K.; Lee, H.; Kozarewa, I.; Mortimer, P.G.S.; Odegaard, J.I.; Harrington, E.A.; Lee, J.; Lee, T.; et al. Tumor Genomic Profiling Guides Patients with Metastatic Gastric Cancer to Targeted Treatment: The VIKTORY Umbrella Trial. Cancer Discov. 2019, 9, 1388–1405. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Liu, H.; Yu, J.; Shi, G.Y.; Zhao, L.Y.; Li, G.X. Prognostic and predictive blood biomarkers in gastric cancer and the potential application of circulating tumor cells. World J. Gastroenterol. 2018, 24, 2236–2246. [Google Scholar] [CrossRef]
- Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life 2021, 11, 890. [Google Scholar] [CrossRef]
- Salvianti, F.; Gelmini, S.; Costanza, F.; Mancini, I.; Sonnati, G.; Simi, L.; Pazzagli, M.; Pinzani, P. The pre-analytical phase of the liquid biopsy. New Biotechnol. 2020, 55, 19–29. [Google Scholar] [CrossRef]
- Brito-Rocha, T.; Constancio, V.; Henrique, R.; Jeronimo, C. Shifting the Cancer Screening Paradigm: The Rising Potential of Blood-Based Multi-Cancer Early Detection Tests. Cells 2023, 12, 935. [Google Scholar] [CrossRef]
- Chivu-Economescu, M.; Necula, L.; Matei, L.; Dragu, D.; Bleotu, C.; Diaconu, C.C. Clinical Applications of Liquid Biopsy in Gastric Cancer. Front. Med. 2021, 8, 749250. [Google Scholar] [CrossRef]
- Fernandez-Lazaro, D.; Garcia Hernandez, J.L.; Garcia, A.C.; Cordova Martinez, A.; Mielgo-Ayuso, J.; Cruz-Hernandez, J.J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, H.; Chong, W.; Shang, L.; Jing, C.; Li, L. Liquid biopsy in gastric cancer: Predictive and prognostic biomarkers. Cell Death Dis. 2022, 13, 903. [Google Scholar] [CrossRef]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: The future of cancer early detection. J. Transl. Med. 2023, 21, 118. [Google Scholar] [CrossRef]
- Armakolas, A.; Kotsari, M.; Koskinas, J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers 2023, 15, 1579. [Google Scholar] [CrossRef]
- Fu, M.; Gu, J.; Jiang, P.; Qian, H.; Xu, W.; Zhang, X. Exosomes in gastric cancer: Roles, mechanisms, and applications. Mol. Cancer 2019, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Qian, H.; Shen, L.; Zhang, X.; Zhu, W.; Huang, L.; Yan, Y.; Mao, F.; Zhao, C.; Shi, Y.; et al. Gastric cancer exosomes trigger differentiation of umbilical cord derived mesenchymal stem cells to carcinoma-associated fibroblasts through TGF-beta/Smad pathway. PLoS ONE 2012, 7, e52465. [Google Scholar] [CrossRef] [PubMed]
- Yen, E.Y.; Miaw, S.C.; Yu, J.S.; Lai, I.R. Exosomal TGF-beta1 is correlated with lymphatic metastasis of gastric cancers. Am. J. Cancer Res. 2017, 7, 2199–2208. [Google Scholar] [PubMed]
- Zhang, H.; Deng, T.; Liu, R.; Bai, M.; Zhou, L.; Wang, X.; Li, S.; Wang, X.; Yang, H.; Li, J.; et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017, 8, 15016. [Google Scholar] [CrossRef]
- Ren, J.; Zhou, Q.; Li, H.; Li, J.; Pang, L.; Su, L.; Gu, Q.; Zhu, Z.; Liu, B. Characterization of exosomal RNAs derived from human gastric cancer cells by deep sequencing. Tumour Biol. 2017, 39, 1010428317695012. [Google Scholar] [CrossRef]
- Tang, X.H.; Guo, T.; Gao, X.Y.; Wu, X.L.; Xing, X.F.; Ji, J.F.; Li, Z.Y. Exosome-derived noncoding RNAs in gastric cancer: Functions and clinical applications. Mol. Cancer 2021, 20, 99. [Google Scholar] [CrossRef]
- Fan, Y.; Che, X.; Qu, J.; Hou, K.; Wen, T.; Li, Z.; Li, C.; Wang, S.; Xu, L.; Liu, Y.; et al. Exosomal PD-L1 Retains Immunosuppressive Activity and is Associated with Gastric Cancer Prognosis. Ann. Surg. Oncol. 2019, 26, 3745–3755. [Google Scholar] [CrossRef] [PubMed]
- Varkey, J.; Nicolaides, T. Tumor-Educated Platelets: A Review of Current and Potential Applications in Solid Tumors. Cureus 2021, 13, e19189. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [PubMed]
| Author, Year | Number of pts | Study Design | Results |
|---|---|---|---|
| Kim, 2019 [49] | 25 | WGS → Tumor tissue qPCR → ctDNA testing in serum (before surgery, at months 1, 3, 6, 9, and 12 after surgery) | - Median lead time ctDNA + vs. imaging relapse: 4.05 months. - ctDNA + associated with relapse in 12-month postop period (p = 0.0294). |
| Ko, 2021 [50] | 49 | qPCR → ctDNA evaluation for LINE1 fragments, ligation-mediated PCR → methylation proportion of LINE1 postop | High postop concentration of LINE1 associated with: - Shorter RFS (p = 0.009); - Shorter OS (p = 0.04). |
| Shoda, 2017 [51] | 4 | RT-PCR and dd-PCR → Postop levels of HER2 + RPPH1 (control gene) copy number | Four pts relapsed, all with HER2-to-RPPH1 ratio above the cutoff. |
| Ling, 2013 [52] | 72 | RT-PCR → XAF1 hypermethylation (matched tumor-ctDNA) | Overall, 12 pts relapsed, 10 pts with negative to positive XAF1 methylation status on ctDNA. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grizzi, G.; Salati, M.; Bonomi, M.; Ratti, M.; Holladay, L.; De Grandis, M.C.; Spada, D.; Baiocchi, G.L.; Ghidini, M. Circulating Tumor DNA in Gastric Adenocarcinoma: Future Clinical Applications and Perspectives. Int. J. Mol. Sci. 2023, 24, 9421. https://doi.org/10.3390/ijms24119421
Grizzi G, Salati M, Bonomi M, Ratti M, Holladay L, De Grandis MC, Spada D, Baiocchi GL, Ghidini M. Circulating Tumor DNA in Gastric Adenocarcinoma: Future Clinical Applications and Perspectives. International Journal of Molecular Sciences. 2023; 24(11):9421. https://doi.org/10.3390/ijms24119421
Chicago/Turabian StyleGrizzi, Giulia, Massimiliano Salati, Maria Bonomi, Margherita Ratti, Lauren Holladay, Maria Caterina De Grandis, Daniele Spada, Gian Luca Baiocchi, and Michele Ghidini. 2023. "Circulating Tumor DNA in Gastric Adenocarcinoma: Future Clinical Applications and Perspectives" International Journal of Molecular Sciences 24, no. 11: 9421. https://doi.org/10.3390/ijms24119421
APA StyleGrizzi, G., Salati, M., Bonomi, M., Ratti, M., Holladay, L., De Grandis, M. C., Spada, D., Baiocchi, G. L., & Ghidini, M. (2023). Circulating Tumor DNA in Gastric Adenocarcinoma: Future Clinical Applications and Perspectives. International Journal of Molecular Sciences, 24(11), 9421. https://doi.org/10.3390/ijms24119421







