Inhibition of Replication Fork Formation and Progression: Targeting the Replication Initiation and Primosomal Proteins
Abstract
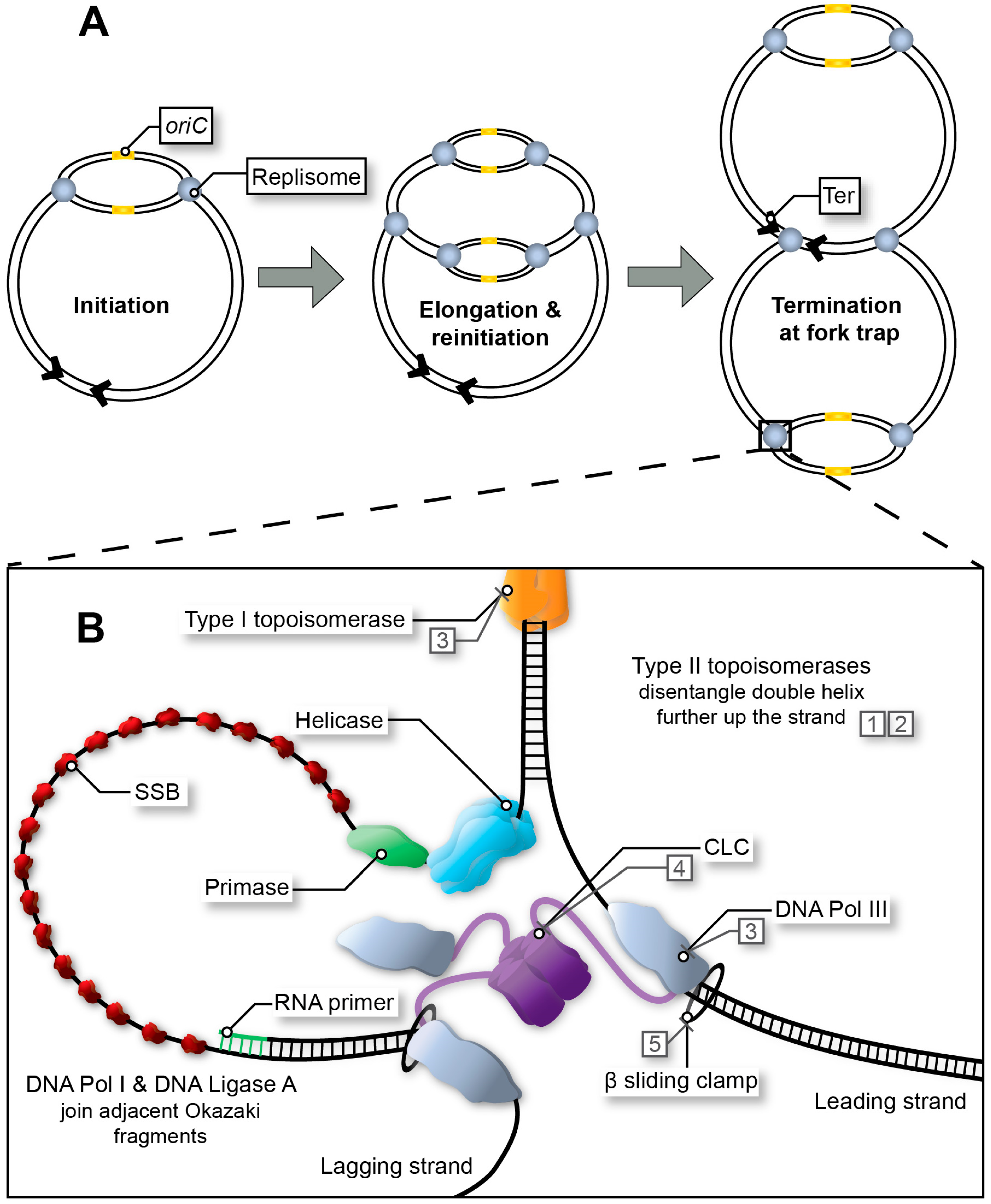
1. DNA Replication: A Valid Antibacterial Drug Target
2. Targeting DNA Replication Initiation
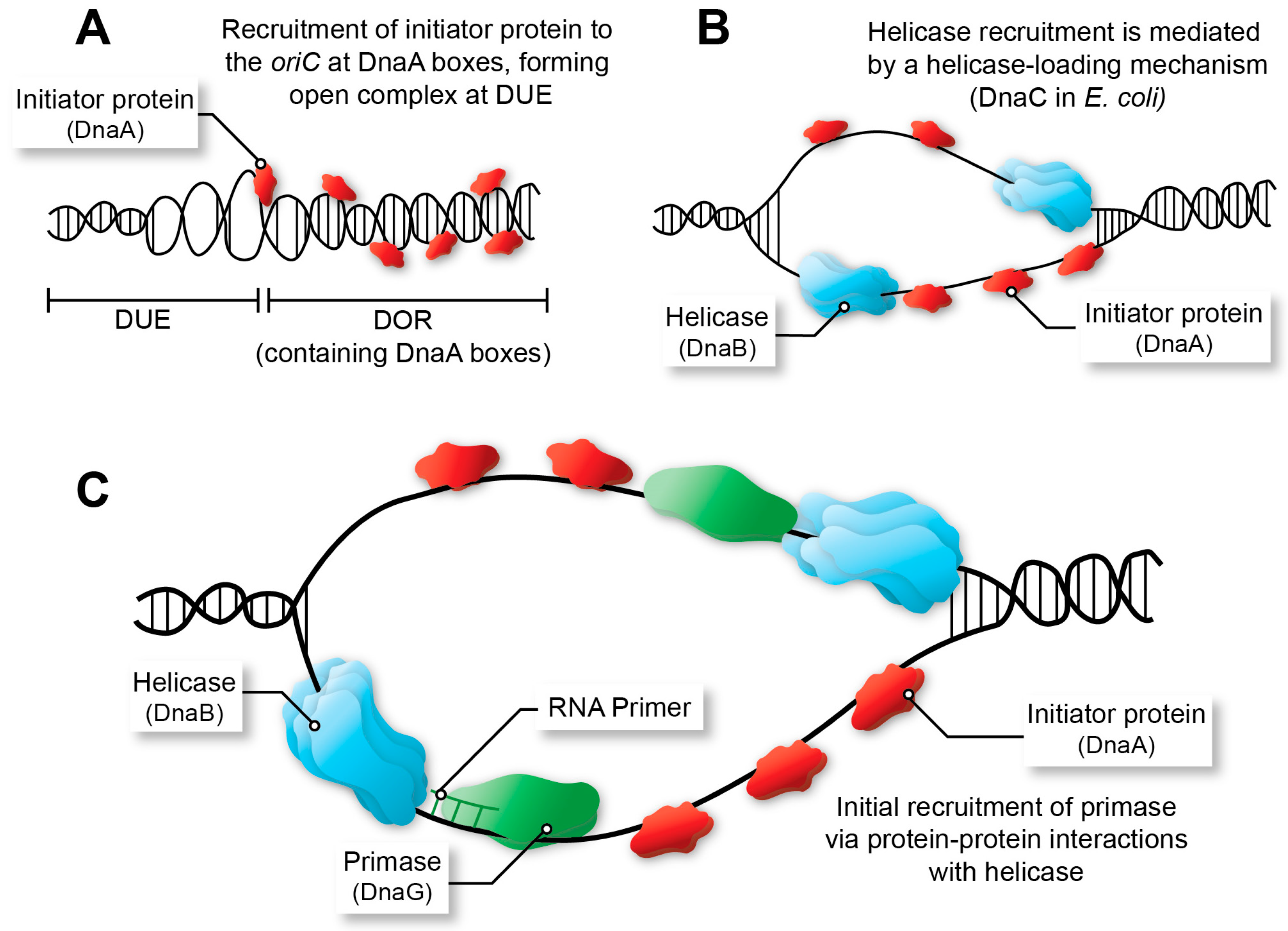
2.1. The Initiator Protein
2.2. Other Factors Associated with the Origin and Initiation
2.3. The Helicase
2.4. The Primase
2.5. Alternative Helicase Loading
3. Replication Initiation Inhibitors
3.1. DnaA Inhibitors
3.2. Helicase Inhibitors
3.3. Primase Inhibitors
3.4. Synopsis
4. Advantages and Limitations of Current Methods
5. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schaeffer, P.M.; Headlam, M.J.; Dixon, N.E. Protein—Protein Interactions in the Eubacterial Replisome. IUBMB Life 2005, 57, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Neylon, C.; Kralicek, A.V.; Hill, T.M.; Dixon, N.E. Replication termination in Escherichia coli: Structure and antihelicase activity of the Tus-Ter complex. Microbiol. Mol. Biol. Rev. 2005, 69, 501–526. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Q.; Dixon, N.E. Bacterial replisomes. Curr. Opin. Struct. Biol. 2018, 53, 159–168. [Google Scholar] [CrossRef]
- Yao, N.Y.; O’Donnell, M.E. The DNA Replication Machine: Structure and Dynamic Function. In Subcellular Biochemistry; Springer: Cham, Switzerland, 2021; Volume 96, pp. 233–258. [Google Scholar] [CrossRef]
- Cooper, S.; Helmstetter, C.E. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 1968, 31, 519–540. [Google Scholar] [CrossRef] [PubMed]
- Fossum, S.; Crooke, E.; Skarstad, K. Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli. EMBO J. 2007, 26, 4514–4522. [Google Scholar] [CrossRef]
- Wolański, M.; Donczew, R.; Zawilak-Pawlik, A.; Zakrzewska-Czerwińska, J. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 2015, 5, 735. [Google Scholar] [CrossRef]
- Lin, Y.S.; Kieser, H.M.; Hopwood, D.A.; Chen, C.W. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 1993, 10, 923–933. [Google Scholar] [CrossRef]
- Reiche, M.A.; Warner, D.F.; Mizrahi, V. Targeting DNA Replication and Repair for the Development of Novel Therapeutics against Tuberculosis. Front. Mol. Biosci. 2017, 4, 75. [Google Scholar] [CrossRef]
- Oakley, A.J. A structural view of bacterial DNA replication. Protein Sci. 2019, 28, 990–1004. [Google Scholar] [CrossRef]
- Beattie, T.R.; Reyes-Lamothe, R. A Replisome’s journey through the bacterial chromosome. Front. Microbiol. 2015, 6, 562. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Wieczorek, K.; Dec, M.; Stepien-Pysniak, D.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria-A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Sanyal, G.; Doig, P. Bacterial DNA replication enzymes as targets for antibacterial drug discovery. Expert Opin. Drug Discov. 2012, 7, 327–339. [Google Scholar] [CrossRef]
- Robinson, A.; Causer, R.J.; Dixon, N.E. Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr. Drug Targets 2012, 13, 352–372. [Google Scholar] [CrossRef]
- Kaguni, J.M. The Macromolecular Machines that Duplicate the Escherichia coli Chromosome as Targets for Drug Discovery. Antibiotics 2018, 7, 23. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Hooper, D.C. Review of the Quinolone Family. In Antibiotic Discovery and Development; Dougherty, T.J., Pucci, M.J., Eds.; Springer: Boston, MA, USA, 2012; pp. 119–146. [Google Scholar] [CrossRef]
- Badshah, S.L.; Ullah, A. New developments in non-quinolone-based antibiotics for the inhibiton of bacterial gyrase and topoisomerase IV. Eur. J. Med. Chem. 2018, 152, 393–400. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Reyes-Lamothe, R.; Sherratt, D.J.; Leake, M.C. Stoichiometry and Architecture of Active DNA Replication Machinery in Escherichia coli. Science 2010, 328, 498–501. [Google Scholar] [CrossRef]
- Graham, J.E.; Marians, K.J.; Kowalczykowski, S.C. Independent and Stochastic Action of DNA Polymerases in the Replisome. Cell 2017, 169, 1201–1213.E17. [Google Scholar] [CrossRef]
- Duggin, I.G.; Wake, R.G.; Bell, S.D.; Hill, T.M. The replication fork trap and termination of chromosome replication. Mol. Microbiol. 2008, 70, 1323–1333. [Google Scholar] [CrossRef]
- Toft, C.J.; Sorenson, A.E.; Schaeffer, P.M. A soft Tus-Ter interaction is hiding a fail-safe lock in the replication fork trap of Dickeya paradisiaca. Microbiol. Res. 2022, 263, 127147. [Google Scholar] [CrossRef]
- Toft, C.J.; Sorenson, A.E.; Schaeffer, P.M. Rise of the terminator protein tus: A versatile tool in the biotechnologist’s toolbox. Anal. Chim. Acta 2022, 1213, 339946. [Google Scholar] [CrossRef] [PubMed]
- Toft, C.J.; Moreau, M.J.J.; Perutka, J.; Mandapati, S.; Enyeart, P.; Sorenson, A.E.; Ellington, A.D.; Schaeffer, P.M. Delineation of the Ancestral Tus-Dependent Replication Fork Trap. Int. J. Mol. Sci. 2021, 22, 13533. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T. Initiation of DNA Replication at the Chromosomal Origin of E. coli, oriC. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; Volume 1042, pp. 79–98. [Google Scholar] [CrossRef]
- Mansfield, R.E.; Dixon, N.E. Priming the engine of DNA synthesis. Structure 2012, 20, 1447–1448. [Google Scholar] [CrossRef] [PubMed]
- Trojanowski, D.; Holowka, J.; Zakrzewska-Czerwinska, J. Where and When Bacterial Chromosome Replication Starts: A Single Cell Perspective. Front. Microbiol. 2018, 9, 2819. [Google Scholar] [CrossRef]
- Chodavarapu, S.; Kaguni, J. Replication initiation in bacteria. Enzymes 2016, 39, 1–30. [Google Scholar]
- Kaguni, J.M. Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 2011, 15, 606–613. [Google Scholar] [CrossRef]
- Van Eijk, E.; Wittekoek, B.; Kuijper, E.J.; Smits, W.K. DNA replication proteins as potential targets for antimicrobials in drug-resistant bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 1275–1284. [Google Scholar] [CrossRef]
- Windgassen, T.A.; Wessel, S.R.; Bhattacharyya, B.; Keck, J.L. Mechanisms of bacterial DNA replication restart. Nucleic Acids Res. 2018, 46, 504–519. [Google Scholar] [CrossRef]
- Naue, N.; Beerbaum, M.; Bogutzki, A.; Schmieder, P.; Curth, U. The helicase-binding domain of Escherichia coli DnaG primase interacts with the highly conserved C-terminal region of single-stranded DNA-binding protein. Nucleic Acids Res. 2013, 41, 4507–4517. [Google Scholar] [CrossRef]
- Bianco, P.R. DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication forks in Escherichia coli. Genes 2020, 11, 471. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, C.C.; Chen, C.C.; Yang, K.J.; Huang, C.Y. Inhibition of Staphylococcus aureus PriA Helicase by Flavonol Kaempferol. Protein J. 2015, 34, 169–172. [Google Scholar] [CrossRef]
- Costa, A.; Diffley, J.F.X. The Initiation of Eukaryotic DNA Replication. Annu. Rev. Biochem. 2022, 91, 107–131. [Google Scholar] [CrossRef]
- Akbari, M.; Nilsen, H.L.; Montaldo, N.P. Dynamic features of human mitochondrial DNA maintenance and transcription. Front. Cell Dev. Biol. 2022, 10, 984245. [Google Scholar] [CrossRef]
- Periago, J.; Mason, C.; Griep, M.A. Theoretical Development of DnaG Primase as a Novel Narrow-Spectrum Antibiotic Target. ACS Omega 2022, 7, 8420–8428. [Google Scholar] [CrossRef]
- Blaine, H.C.; Simmons, L.A.; Stallings, C.L. Diverse Mechanisms of Helicase Loading during DNA Replication Initiation in Bacteria. J. Bacteriol. 2023, 205, e0048722. [Google Scholar] [CrossRef]
- Biswas, T.; Resto-Roldán, E.; Sawyer, S.K.; Artsimovitch, I.; Tsodikov, O.V. A novel non-radioactive primase-pyrophosphatase activity assay and its application to the discovery of inhibitors of Mycobacterium tuberculosis primase DnaG. Nucleic Acids Res. 2013, 41, e56. [Google Scholar] [CrossRef]
- Lin, H.H.; Huang, C.Y. Characterization of flavonol inhibition of DnaB helicase: Real-time monitoring, structural modeling, and proposed mechanism. J. Biomed. Biotechnol. 2012, 2012, 735368. [Google Scholar] [CrossRef]
- Mizushima, T.; Sasaki, S.; Ohishi, H.; Kobayashi, M.; Katayama, T.; Miki, T.; Maeda, M.; Sekimizu, K. Molecular design of inhibitors of in vitro oriC DNA replication based on the potential to block the ATP binding of DnaA protein. J. Biol. Chem. 1996, 271, 25178–25183. [Google Scholar] [CrossRef]
- Catazaro, J.; Periago, J.; Shortridge, M.D.; Worley, B.; Kirchner, A.; Powers, R.; Griep, M.A. Identification of a Ligand-Binding Site on the Staphylococcus aureus DnaG Primase C-Terminal Domain. Biochemistry 2017, 56, 932–943. [Google Scholar] [CrossRef]
- Anderson, A.C. The process of structure-based drug design. Chem. Biol. 2003, 10, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Jameson, K.H.; Wilkinson, A.J. Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli. Genes 2017, 8, 22. [Google Scholar] [CrossRef]
- Leonard, A.C.; Grimwade, J.E. The orisome: Structure and function. Front. Microbiol. 2015, 6, 545. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, J.E.; Leonard, A.C. Targeting the Bacterial Orisome in the Search for New Antibiotics. Front. Microbiol. 2017, 8, 2352. [Google Scholar] [CrossRef] [PubMed]
- Hansen, F.G.; Atlung, T. The DnaA Tale. Front. Microbiol. 2018, 9, 319. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Kasho, K.; Kawakami, H. The DnaA Cycle in Escherichia coli: Activation, Function and Inactivation of the Initiator Protein. Front. Microbiol. 2017, 8, 2496. [Google Scholar] [CrossRef]
- Kohiyama, M. Research on DnaA in the early days. Res. Microbiol. 2020, 171, 287–289. [Google Scholar] [CrossRef]
- Menikpurage, I.P.; Woo, K.; Mera, P.E. Transcriptional Activity of the Bacterial Replication Initiator DnaA. Front. Microbiol. 2021, 12, 662317. [Google Scholar] [CrossRef]
- Ozaki, S.; Katayama, T. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid 2009, 62, 71–82. [Google Scholar] [CrossRef]
- Katayama, T.; Ozaki, S.; Keyamura, K.; Fujimitsu, K. Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010, 8, 163–170. [Google Scholar] [CrossRef]
- Sekimizu, K.; Yung, B.Y.; Kornberg, A. The dnaA protein of Escherichia coli. Abundance, improved purification, and membrane binding. J. Biol. Chem. 1988, 263, 7136–7140. [Google Scholar] [CrossRef]
- Messer, W.; Weigel, C. DnaA initiator—Also a transcription factor. Mol. Microbiol. 1997, 24, 1–6. [Google Scholar] [CrossRef]
- Leonard, A.C.; Rao, P.; Kadam, R.P.; Grimwade, J.E. Changing Perspectives on the Role of DnaA-ATP in Orisome Function and Timing Regulation. Front. Microbiol. 2019, 10, 2009. [Google Scholar] [CrossRef]
- Mackiewicz, P.; Zakrzewska-Czerwińska, J.; Zawilak, A.; Dudek, M.R.; Cebrat, S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 2004, 32, 3781–3791. [Google Scholar] [CrossRef]
- Rao, P.; Rozgaja, T.A.; Alqahtani, A.; Grimwade, J.E.; Leonard, A.C. Low Affinity DnaA-ATP Recognition Sites in E. coli oriC Make Non-equivalent and Growth Rate-Dependent Contributions to the Regulated Timing of Chromosome Replication. Front. Microbiol. 2018, 9, 1673. [Google Scholar] [CrossRef]
- Ozaki, S.; Katayama, T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012, 40, 1648–1665. [Google Scholar] [CrossRef]
- Grimwade, J.E.; Rozgaja, T.A.; Gupta, R.; Dyson, K.; Rao, P.; Leonard, A.C. Origin recognition is the predominant role for DnaA-ATP in initiation of chromosome replication. Nucleic Acids Res. 2018, 46, 6140–6151. [Google Scholar] [CrossRef]
- Sakiyama, Y.; Nishimura, M.; Hayashi, C.; Akama, Y.; Ozaki, S.; Katayama, T. The DnaA AAA plus Domain His136 Residue Directs DnaB Replicative Helicase to the Unwound Region of the Replication Origin, oriC. Front. Microbiol. 2018, 9, 2017. [Google Scholar] [CrossRef]
- Ozaki, S.; Kawakami, H.; Nakamura, K.; Fujikawa, N.; Kagawa, W.; Park, S.-Y.; Yokoyama, S.; Kurumizaka, H.; Katayama, T. A Common Mechanism for the ATP-DnaA-dependent Formation of Open Complexes at the Replication Origin. J. Biol. Chem. 2008, 283, 8351–8362. [Google Scholar] [CrossRef]
- Skarstad, K.; Katayama, T. Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 2013, 5, a012922. [Google Scholar] [CrossRef]
- Camara, J.E.; Breier, A.M.; Brendler, T.; Austin, S.; Cozzarelli, N.R.; Crooke, E. Hda inactivation of DnaA is the predominant mechanism preventing hyperinitiation of Escherichia coli DNA replication. EMBO Rep. 2005, 6, 736–741. [Google Scholar] [CrossRef]
- Kasho, K.; Katayama, T. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. USA 2013, 110, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Molt, K.L.; Sutera, V.A., Jr.; Moore, K.K.; Lovett, S.T. A Role for Nonessential Domain II of Initiator Protein, DnaA, in Replication Control. Genetics 2009, 183, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Chodavarapu, S.; Felczak, M.M.; Rouviere-Yaniv, J.; Kaguni, J.M. Escherichia coli DnaA interacts with HU in initiation at the E-coli replication origin. Mol. Microbiol. 2008, 67, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Kasho, K.; Tanaka, H.; Sakai, R.; Katayama, T. Cooperative DnaA Binding to the Negatively Supercoiled datA Locus Stimulates DnaA-ATP Hydrolysis. J. Biol. Chem. 2017, 292, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Tatsumoto, Y.; Ozaki, S.; Katayama, T. Negative feedback for DARS2-Fis complex by ATP-DnaA supports the cell cycle-coordinated regulation for chromosome replication. Nucleic Acids Res. 2021, 49, 12820–12835. [Google Scholar] [CrossRef]
- Fujimitsu, K.; Senriuchi, T.; Katayama, T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009, 23, 1221–1233. [Google Scholar] [CrossRef]
- Abe, Y.; Jo, T.; Matsuda, Y.; Matsunaga, C.; Katayama, T.; Ueda, T. Structure and Function of DnaA N-terminal Domains. J. Biol. Chem. 2007, 282, 17816–17827. [Google Scholar] [CrossRef]
- Sugiyama, R.; Kasho, K.; Miyoshi, K.; Ozaki, S.; Kagawa, W.; Kurumizaka, H.; Katayama, T. A novel mode of DnaA-DnaA interaction promotes ADP dissociation for reactivation of replication initiation activity. Nucleic Acids Res. 2019, 47, 11209–11224. [Google Scholar] [CrossRef]
- Biswas, S.B.; Chen, P.H.; Biswas, E.E. Structure and Function of Escherichia-Coli Dnab Protein—Role of the N-Terminal Domain in Helicase Activity. Biochemistry 1994, 33, 11307–11314. [Google Scholar] [CrossRef]
- Lewis, J.S.; Jergic, S.; Dixon, N.E. Chapter Two—The E. coli DNA Replication Fork. Enzymes 2016, 39, 31–88. [Google Scholar]
- Bell, S.P.; Kaguni, J.M. Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010124. [Google Scholar] [CrossRef] [PubMed]
- Felczak, M.M.; Chodavarapu, S.; Kaguni, J.M. DnaC, the indispensable companion of DnaB helicase, controls the accessibility of DnaB helicase by primase. J. Biol. Chem. 2017, 292, 20871–20882. [Google Scholar] [CrossRef] [PubMed]
- Spinks, R.R.; Spenkelink, L.M.; Stratmann, S.A.; Xu, Z.Q.; Stamford, N.P.J.; Brown, S.E.; Dixon, N.E.; Jergic, S.; Van Oijen, A.M. DnaB helicase dynamics in bacterial DNA replication resolved by single-molecule studies. Nucleic Acids Res. 2021, 49, 6804–6816. [Google Scholar] [CrossRef]
- Thirlway, J.; Turner, I.J.; Gibson, C.T.; Gardiner, L.; Brady, K.; Allen, S.; Roberts, C.J.; Soultanas, P. DnaG interacts with a linker region that joins the N- and C-domains of DnaB and induces the formation of 3-fold symmetric rings. Nucleic Acids Res. 2004, 32, 2977–2986. [Google Scholar] [CrossRef] [PubMed]
- Oakley, A.J.; Loscha, K.V.; Schaeffer, P.M.; Liepinsh, E.; Pintacuda, G.; Wilce, M.C.; Otting, G.; Dixon, N.E. Crystal and solution structures of the helicase-binding domain of Escherichia coli primase. J. Biol. Chem. 2005, 280, 11495–11504. [Google Scholar] [CrossRef]
- Chodavarapu, S.; Jones, A.D.; Feig, M.; Kaguni, J.M. DnaC traps DnaB as an open ring and remodels the domain that binds primase. Nucleic Acids Res. 2016, 44, 210–220. [Google Scholar] [CrossRef]
- Bárcena, M.; Ruiz, T.; Donate, L.E.; Brown, S.E.; Dixon, N.E.; Radermacher, M.; Carazo, J.M. The DnaB.DnaC complex: A structure based on dimers assembled around an occluded channel. EMBO J. 2001, 20, 1462–1468. [Google Scholar] [CrossRef]
- Fairman-Williams, M.E.; Guenther, U.-P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Wiegand, T.; Cadalbert, R.; Lacabanne, D.; Timmins, J.; Terradot, L.; Böckmann, A.; Meier, B.H. The conformational changes coupling ATP hydrolysis and translocation in a bacterial DnaB helicase. Nat. Commun. 2019, 10, 31. [Google Scholar] [CrossRef]
- Syson, K.; Thirlway, J.; Hounslow, A.M.; Soultanas, P.; Waltho, J.P. Solution structure of the helicase-interaction domain of the primase DnaG: A model for helicase activation. Structure 2005, 13, 609–616. [Google Scholar] [CrossRef]
- Chintakayala, K.; Larson, M.A.; Grainger, W.H.; Scott, D.J.; Griep, M.A.; Hinrichs, S.H.; Soultanas, P. Domain swapping reveals that the C- and N-terminal domains of DnaG and DnaB, respectively, are functional homologues. Mol. Microbiol. 2007, 63, 1629–1639. [Google Scholar] [CrossRef]
- van Eijk, E.; Paschalis, V.; Green, M.; Friggen, A.H.; Larson, M.A.; Spriggs, K.; Briggs, G.S.; Soultanas, P.; Smits, W.K. Primase is required for helicase activity and helicase alters the specificity of primase in the enteropathogen Clostridium difficile. Open Biol. 2016, 6, 160272. [Google Scholar] [CrossRef]
- Ma, J.B.; Chen, Z.; Xu, C.H.; Huang, X.Y.; Jia, Q.; Zou, Z.Y.; Mi, C.Y.; Ma, D.F.; Lu, Y.; Zhang, H.D.; et al. Dynamic structural insights into the molecular mechanism of DNA unwinding by the bacteriophage T7 helicase. Nucleic Acids Res. 2020, 48, 3156–3164. [Google Scholar] [CrossRef]
- Ilic, S.; Cohen, S.; Singh, M.; Tam, B.; Dayan, A.; Akabayov, B. DnaG Primase-A Target for the Development of Novel Antibacterial Agents. Antibiotics 2018, 7, 72. [Google Scholar] [CrossRef]
- Corn, J.E.; Berger, J.M. Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions. Nucleic Acids Res. 2006, 34, 4082–4088. [Google Scholar] [CrossRef]
- Zechner, E.L.; Wu, C.A.; Marians, K.J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. III. A polymerase-primase interaction governs primer size. J. Biol. Chem. 1992, 267, 4054–4063. [Google Scholar] [CrossRef]
- Tougu, K.; Marians, K.J. The Interaction between Helicase and Primase Sets the Replication Fork Clock. J. Biol. Chem. 1996, 271, 21398–21405. [Google Scholar] [CrossRef]
- Van Oijen, A.M.; Hamdan, S.M.; Tanner, N.A.; Schaeffer, P.M.; Loscha, K.V.; Dixon, N.E.; Jergic, S. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat. Struct. Mol. Biol. 2008, 15, 170–176. [Google Scholar] [CrossRef]
- Monachino, E.; Jergic, S.; Lewis, J.S.; Xu, Z.Q.; Lo, A.T.Y.; O’Shea, V.L.; Berger, J.M.; Dixon, N.E.; van Oijen, A.M. A Primase-Induced Conformational Switch Controls the Stability of the Bacterial Replisome. Mol. Cell 2020, 79, 140–154.E7. [Google Scholar] [CrossRef]
- Lee, S.J.; Zhu, B.; Akabayov, B.; Richardson, C.C. Zinc-binding domain of the bacteriophage T7 DNA primase modulates binding to the DNA template. J. Biol. Chem. 2012, 287, 39030–39040. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Wigley, D.B. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure 2000, 8, 231–239. [Google Scholar] [CrossRef]
- Hou, C.X.; Biswas, T.; Tsodikov, O.V. Structures of the Catalytic Domain of Bacterial Primase DnaG in Complexes with DNA Provide Insight into Key Priming Events. Biochemistry 2018, 57, 2084–2093. [Google Scholar] [CrossRef] [PubMed]
- Spenkelink, L.M.; Lewis, J.S.; Jergic, S.; Xu, Z.-Q.; Robinson, A.; Dixon, N.E.; van Oijen, A.M. Recycling of single-stranded DNA-binding protein by the bacterial replisome. Nucleic Acids Res. 2019, 47, 4111–4123. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R. The tale of SSB. Prog. Biophys. Mol. Biol. 2017, 127, 111–118. [Google Scholar] [CrossRef]
- Yuzhakov, A.; Kelman, Z.; O’Donnell, M. Trading Places on DNA—A Three-Point Switch Underlies Primer Handoff from Primase to the Replicative DNA Polymerase. Cell 1999, 96, 153–163. [Google Scholar] [CrossRef]
- Velten, M.; McGovern, S.; Marsin, S.; Ehrlich, S.D.; Noirot, P.; Polard, P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell 2003, 11, 1009–1020. [Google Scholar] [CrossRef]
- Ioannou, C.; Schaeffer, P.M.; Dixon, N.E.; Soultanas, P. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 2006, 34, 5247–5258. [Google Scholar] [CrossRef]
- Loscha, K.V.; Jaudzems, K.; Ioannou, C.; Su, X.C.; Hill, F.R.; Otting, G.; Dixon, N.E.; Liepinsh, E. A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader. Nucleic Acids Res. 2009, 37, 2395–2404. [Google Scholar] [CrossRef]
- Brézellec, P.; Vallet-Gely, I.; Possoz, C.; Quevillon-Cheruel, S.; Ferat, J.-L. DciA is an ancestral replicative helicase operator essential for bacterial replication initiation. Nat. Commun. 2016, 7, 13271. [Google Scholar] [CrossRef]
- Mann, K.M.; Huang, D.L.; Hooppaw, A.J.; Logsdon, M.M.; Richardson, K.; Lee, H.J.; Kimmey, J.M.; Aldridge, B.B.; Stallings, C.L. Rv0004 is a new essential member of the mycobacterial DNA replication machinery. PLoS Genet. 2017, 13, e1007115. [Google Scholar] [CrossRef]
- Ozaki, S.; Wang, D.; Wakasugi, Y.; Itani, N.; Katayama, T. The Caulobacter crescentus DciA promotes chromosome replication through topological loading of the DnaB replicative helicase at replication forks. Nucleic Acids Res. 2022, 50, 12896–12912. [Google Scholar] [CrossRef]
- Jensen, R.B.; Wang, S.C.; Shapiro, L. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 2001, 20, 4952–4963. [Google Scholar] [CrossRef]
- Marsin, S.; Adam, Y.; Cargemel, C.; Andreani, J.; Baconnais, S.; Legrand, P.; Li de la Sierra-Gallay, I.; Humbert, A.; Aumont-Nicaise, M.; Velours, C.; et al. Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase. Nucleic Acids Res. 2021, 49, 6569–6586. [Google Scholar] [CrossRef]
- Zawilak-Pawlik, A.; Zakrzewska-Czerwińska, J. Recent Advances in Helicobacter pylori Replication: Possible Implications in Adaptation to a Pathogenic Lifestyle and Perspectives for Drug Design. In Current Topics in Microbiology and Immunology; Springer: Cham, Switzerland, 2017; Volume 400, pp. 73–103. [Google Scholar] [CrossRef]
- Nitharwal, R.G.; Verma, V.; Dasgupta, S.; Dhar, S.K. Helicobacter pylori chromosomal DNA replication: Current status and future perspectives. FEBS Lett. 2011, 585, 7–17. [Google Scholar] [CrossRef]
- Stelter, M.; Gutsche, I.; Kapp, U.; Bazin, A.; Bajic, G.; Goret, G.; Jamin, M.; Timmins, J.; Terradot, L. Architecture of a Dodecameric Bacterial Replicative Helicase. Structure 2012, 20, 554–564. [Google Scholar] [CrossRef]
- Bazin, A.; Cherrier, M.V.; Gutsche, I.; Timmins, J.; Terradot, L. Structure and primase-mediated activation of a bacterial dodecameric replicative helicase. Nucleic Acids Res. 2015, 43, 8564–8576. [Google Scholar] [CrossRef]
- Sasaki, S.; Mizushima, T.; Hashimoto, T.; Maeda, M.; Kazuhisa, S. 3-Acetoxy-2,2′-bi-1H-indol as a novel inhibitor of ATP binding to DnaA, the protein initiating chromosomal replication in Escherichia coli. Bioorg. Med. Chem. Lett. 1994, 4, 1771–1774. [Google Scholar] [CrossRef]
- Griep, M.A.; Blood, S.; Larson, M.A.; Koepsell, S.A.; Hinrichs, S.H. Myricetin inhibits Escherichia coli DnaB helicase but not primase. Bioorganic Med. Chem. 2007, 15, 7203–7208. [Google Scholar] [CrossRef]
- Chen, C.C.; Huang, C.Y. Inhibition of Klebsiella pneumoniae DnaB helicase by the flavonol galangin. Protein J. 2011, 30, 59–65. [Google Scholar] [CrossRef]
- Li, B.; Pai, R.; Di, M.; Aiello, D.; Barnes, M.H.; Butler, M.M.; Tashjian, T.F.; Peet, N.P.; Bowlin, T.L.; Moir, D.T. Coumarin-based inhibitors of Bacillus anthracis and Staphylococcus aureus replicative DNA helicase: Chemical optimization, biological evaluation, and antibacterial activities. J. Med. Chem. 2012, 55, 10896–10908. [Google Scholar] [CrossRef] [PubMed]
- McKay, G.A.; Reddy, R.; Arhin, F.; Belley, A.; Lehoux, D.; Moeck, G.; Sarmiento, I.; Parr, T.R.; Gros, P.; Pelletier, J.; et al. Triaminotriazine DNA helicase inhibitors with antibacterial activity. Bioorg. Med. Chem. Lett. 2006, 16, 1286–1290. [Google Scholar] [CrossRef] [PubMed]
- Chilingaryan, Z.; Headey, S.J.; Lo, A.T.Y.; Xu, Z.Q.; Otting, G.; Dixon, N.E.; Scanlon, M.J.; Oakley, A.J. Fragment-Based Discovery of Inhibitors of the Bacterial DnaG-SSB Interaction. Antibiotics 2018, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Louise-May, S.; Thanassi, J.A.; Podos, S.D.; Cheng, J.; Thoma, C.; Liu, C.; Wiles, J.A.; Nelson, D.M.; Phadke, A.S.; et al. Small molecule inhibitors of E. coli primase, a novel bacterial target. Bioorg. Med. Chem. Lett. 2007, 17, 2807–2810. [Google Scholar] [CrossRef] [PubMed]
- Biswas, T.; Green, K.D.; Garneau-Tsodikova, S.; Tsodikov, O.V. Discovery of inhibitors of Bacillus anthracis primase DnaG. Biochemistry 2013, 52, 6905–6910. [Google Scholar] [CrossRef]
- Gajadeera, C.; Willby, M.J.; Green, K.D.; Shaul, P.; Fridman, M.; Garneau-Tsodikova, S.; Posey, J.E.; Tsodikov, O.V. Antimycobacterial activity of DNA intercalator inhibitors of Mycobacterium tuberculosis primase DnaG. J. Antibiot. 2015, 68, 153–157. [Google Scholar] [CrossRef]
- Hakeem, S.; Singh, I.; Sharma, P.; Verma, V.; Chandra, R. in silico screening and molecular dynamics simulations study to identify novel potent inhibitors against Mycobacterium tuberculosis DnaG primase. Acta Trop. 2019, 199, 105154. [Google Scholar] [CrossRef]
- Ilic, S.; Akabayov, S.R.; Arthanari, H.; Wagner, G.; Richardson, C.C.; Akabayov, B. Identification of DNA primase inhibitors via a combined fragment-based and virtual screening. Sci. Rep. 2016, 6, 36322. [Google Scholar] [CrossRef]
- Singh, M.; Ilic, S.; Tam, B.; Ben-Ishay, Y.; Sherf, D.; Pappo, D.; Akabayov, B. Dual-Acting Small-Molecule Inhibitors Targeting Mycobacterial DNA Replication. Chemistry 2020, 26, 10849–10860. [Google Scholar] [CrossRef]
- Choi, S.R.; Larson, M.A.; Hinrichs, S.H.; Narayanasamy, P. Development of potential broad spectrum antimicrobials using C2-symmetric 9-fluorenone alkyl amine. Bioorg. Med. Chem. Lett. 2016, 26, 1997–1999. [Google Scholar] [CrossRef]
- Green, K.D.; Punetha, A.; Chandrika, N.T.; Hou, C.; Garneau-Tsodikova, S.; Tsodikov, O.V. Development of Single-Stranded DNA Bisintercalating Inhibitors of Primase DnaG as Antibiotics. ChemMedChem 2021, 16, 1986–1995. [Google Scholar] [CrossRef]
- Hegde, V.R.; Pu, H.; Patel, M.; Black, T.; Soriano, A.; Zhao, W.; Gullo, V.P.; Chan, T.M. Two new bacterial DNA primase inhibitors from the plant Polygonum cuspidatum. Bioorg. Med. Chem. Lett. 2004, 14, 2275–2277. [Google Scholar] [CrossRef]
- Mehta, G.; Shinde, H.M. Enantioselective total synthesis of bioactive natural product (+)-Sch 642305: A structurally novel inhibitor of bacterial DNA primase and HIV-1 Tat transactivation. Chem. Commun. 2005, 29, 3703–3705. [Google Scholar] [CrossRef]
- Chu, M.; Mierzwa, R.; Xu, L.; He, L.; Terracciano, J.; Patel, M.; Gullo, V.; Black, T.; Zhao, W.; Chan, T.M.; et al. Isolation and structure elucidation of Sch 642305, a novel bacterial DNA primase inhibitor produced by Penicillium verrucosum. J. Nat. Prod. 2003, 66, 1527–1530. [Google Scholar] [CrossRef]
- Wilson, E.M.; Trauner, D. Concise synthesis of the bacterial DNA primase inhibitor (+) -Sch 642305. Org. Lett. 2007, 9, 1327–1329. [Google Scholar] [CrossRef]
- Haystead, T.A. The purinome, a complex mix of drug and toxicity targets. Curr. Top. Med. Chem. 2006, 6, 1117–1127. [Google Scholar] [CrossRef]
- Xu, H.; Ziegelin, G.; Schroder, W.; Frank, J.; Ayora, S.; Alonso, J.C.; Lanka, E.; Saenger, W. Flavones inhibit the hexameric replicative helicase RepA. Nucleic Acids Res. 2001, 29, 5058–5066. [Google Scholar] [CrossRef]
- Bird, L.E.; Pan, H.; Soultanas, P.; Wigley, D.B. Mapping protein-protein interactions within a stable complex of DNA primase and DnaB helicase from Bacillus stearothermophilus. Biochemistry 2000, 39, 171–182. [Google Scholar] [CrossRef]
- Fossum, S.; De Pascale, G.; Weigel, C.; Messer, W.; Donadio, S.; Skarstad, K. A robust screen for novel antibiotics: Specific knockout of the initiator of bacterial DNA replication. FEMS Microbiol. Lett. 2008, 281, 210–214. [Google Scholar] [CrossRef]
- Klitgaard, R.N.; Lobner-Olesen, A. A Novel Fluorescence-Based Screen for Inhibitors of the Initiation of DNA Replication in Bacteria. Curr. Drug Discov. Technol. 2019, 16, 272–277. [Google Scholar] [CrossRef]
- Charbon, G.; Klitgaard, R.N.; Liboriussen, C.D.; Thulstrup, P.W.; Maffioli, S.I.; Donadio, S.; Lobner-Olesen, A. Iron chelation increases the tolerance of Escherichia coli to hyper-replication stress. Sci. Rep. 2018, 8, 10550. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Wang, X.; Ahmad, S.; Shahid, F.; Aslam, S.; Ashfaq, U.A.; Alrumaihi, F.; Qasim, M.; Hashem, A.; Al-Hazzani, A.A.; et al. In Silico Core Proteomics and Molecular Docking Approaches for the Identification of Novel Inhibitors against Streptococcus pyogenes. Int. J. Environ. Res. Public Health 2021, 18, 11355. [Google Scholar] [CrossRef]
- Johnsen, L.; Weigel, C.; von Kries, J.; Moller, M.; Skarstad, K. A novel DNA gyrase inhibitor rescues Escherichia coli dnaAcos mutant cells from lethal hyperinitiation. J. Antimicrob. Chemother. 2010, 65, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, F.; Kao, Y.C.; Kurilla, M.G.; Pompliano, D.L.; Dicker, I.B. Homogenous assays for Escherichia coli DnaB-stimulated DnaG primase and DnaB helicase and their use in screening for chemical inhibitors. Anal. Biochem. 2002, 304, 174–179. [Google Scholar] [CrossRef]
- Aiello, D.; Barnes, M.H.; Biswas, E.E.; Biswas, S.B.; Gu, S.; Williams, J.D.; Bowlin, T.L.; Moir, D.T. Discovery, characterization and comparison of inhibitors of Bacillus anthracis and Staphylococcus aureus replicative DNA helicases. Bioorganic Med. Chem. 2009, 17, 4466–4476. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Pai, R.; Aiello, D.; Di, M.; Barnes, M.H.; Peet, N.P.; Bowlin, T.L.; Moir, D.T. Optimization of a novel potent and selective bacterial DNA helicase inhibitor scaffold from a high throughput screening hit. Bioorg. Med. Chem. Lett. 2013, 23, 3481–3486. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, D.L.; Moore, K.J.; Greenwood, C.J.; Djaballah, H.; Jurewicz, A.J.; Murray, K.J.; Pope, A.J. Time-Resolved Fluorescence Energy Transfer DNA Helicase Assays for High Throughput Screening. J. Biomol. Screen 1999, 4, 239–248. [Google Scholar] [CrossRef]
- Mueller, S.H.; Fitschen, L.J.; Shirbini, A.; Hamdan, S.M.; Spenkelink, L.M.; van Oijen, A.M. Rapid single-molecule characterisation of enzymes involved in nucleic-acid metabolism. Nucleic Acids Res. 2023, 51, e5. [Google Scholar] [CrossRef]
- Gardiner, L.; Coyle, B.J.; Chan, W.C.; Soultanas, P. Discovery of antagonist peptides against bacterial helicase-primase interaction in B. stearothermophilus by reverse yeast three-hybrid. Chem. Biol. 2005, 12, 595–604. [Google Scholar] [CrossRef]
- Koepsell, S.; Bastola, D.; Hinrichs, S.H.; Griep, M.A. Thermally denaturing high-performance liquid chromatography analysis of primase activity. Anal. Biochem. 2004, 332, 330–336. [Google Scholar] [CrossRef]
- Dallmann, H.G.; Fackelmayer, O.J.; Tomer, G.; Chen, J.; Wiktor-Becker, A.; Ferrara, T.; Pope, C.; Oliveira, M.T.; Burgers, P.M.; Kaguni, L.S.; et al. Parallel multiplicative target screening against divergent bacterial replicases: Identification of specific inhibitors with broad spectrum potential. Biochemistry 2010, 49, 2551–2562. [Google Scholar] [CrossRef]
- Koepsell, S.A.; Hanson, S.; Hinrichs, S.H.; Griep, M.A. Fluorometric assay for bacterial primases. Anal. Biochem. 2005, 339, 353–355. [Google Scholar] [CrossRef] [PubMed]
- Blay, V.; Tolani, B.; Ho, S.P.; Arkin, M.R. High-Throughput Screening: Today’s biochemical and cell-based approaches. Drug Discov. Today 2020, 25, 1807–1821. [Google Scholar] [CrossRef] [PubMed]
- Olaru, A.; Bala, C.; Jaffrezic-Renault, N.; Aboul-Enein, H.Y. Surface plasmon resonance (SPR) biosensors in pharmaceutical analysis. Crit. Rev. Anal. Chem. 2015, 45, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Cuozzo, J. Review article: High-throughput affinity-based technologies for small-molecule drug discovery. J. Biomol. Screen 2009, 14, 1157–1164. [Google Scholar] [CrossRef]
- Cummings, M.D.; Farnum, M.A.; Nelen, M.I. Universal screening methods and applications of ThermoFluor. J. Biomol. Screen 2006, 11, 854–863. [Google Scholar] [CrossRef]
- Moreau, M.J.J.; Morin, I.; Askin, S.; Cooper, A.E.; Moreland, N.J.; Vasudevan, S.G.; Schaeffer, P.M. Rapid determination of protein stability and ligand binding by differential scanning fluorimetry of GFP-tagged proteins. RSC Adv. 2012, 2, 11892–11900. [Google Scholar] [CrossRef]
- Askin, S.; Bond, T.E.H.; Sorenson, A.E.; Moreau, M.J.J.; Antony, H.; Davis, R.A.; Schaeffer, P.M. Selective protein unfolding: A universal mechanism of action for the development of irreversible inhibitors. Chem. Commun. 2018, 54, 1738–1741. [Google Scholar] [CrossRef]
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013, 65, 157–170. [Google Scholar] [CrossRef]
- Renaud, J.P.; Chung, C.W.; Danielson, U.H.; Egner, U.; Hennig, M.; Hubbard, R.E.; Nar, H. Biophysics in drug discovery: Impact, challenges and opportunities. Nat. Rev. Drug Discov. 2016, 15, 679–698. [Google Scholar] [CrossRef]
- Chavanieu, A.; Pugniere, M. Developments in SPR Fragment Screening. Expert Opin. Drug Discov. 2016, 11, 489–499. [Google Scholar] [CrossRef]
- Chen, G.; Fan, M.; Liu, Y.; Sun, B.; Liu, M.; Wu, J.; Li, N.; Guo, M. Advances in MS Based Strategies for Probing Ligand-Target Interactions: Focus on Soft Ionization Mass Spectrometric Techniques. Front. Chem. 2019, 7, 703. [Google Scholar] [CrossRef]
- Ishii, K.; Noda, M.; Uchiyama, S. Mass spectrometric analysis of protein-ligand interactions. Biophys. Phys. 2016, 13, 87–95. [Google Scholar] [CrossRef]
- Schermann, S.M.; Simmons, D.A.; Konermann, L. Mass spectrometry-based approaches to protein-ligand interactions. Expert Rev. Proteom. 2005, 2, 475–485. [Google Scholar] [CrossRef]
- Muchiri, R.N.; van Breemen, R.B. Affinity selection-mass spectrometry for the discovery of pharmacologically active compounds from combinatorial libraries and natural products. J. Mass Spectrom. 2021, 56, e4647. [Google Scholar] [CrossRef]
- Johnson, B.M.; Nikolic, D.; van Breemen, R.B. Applications of pulsed ultrafiltration-mass spectrometry. Mass Spectrom. Rev. 2002, 21, 76–86. [Google Scholar] [CrossRef]
- O’Connell, T.N.; Ramsay, J.; Rieth, S.F.; Shapiro, M.J.; Stroh, J.G. Solution-based indirect affinity selection mass spectrometry--a general tool for high-throughput screening of pharmaceutical compound libraries. Anal. Chem. 2014, 86, 7413–7420. [Google Scholar] [CrossRef]
- Van Breemen, R.B.; Huang, C.R.; Nikolic, D.; Woodbury, C.P.; Zhao, Y.Z.; Venton, D.L. Pulsed ultrafiltration mass spectrometry: A new method for screening combinatorial libraries. Anal. Chem. 1997, 69, 2159–2164. [Google Scholar] [CrossRef]
- Beck, J.L.; Urathamakul, T.; Watt, S.J.; Sheil, M.M.; Schaeffer, P.M.; Dixon, N.E. Proteomic dissection of DNA polymerization. Expert Rev. Proteom. 2006, 3, 197–211. [Google Scholar] [CrossRef]
- Watt, S.J.; Urathamakul, T.; Schaeffer, P.M.; Williams, N.K.; Sheil, M.M.; Dixon, N.E.; Beck, J.L. Multiple oligomeric forms of Escherichia coli DnaB helicase revealed by electrospray ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Pantoliano, M.W.; Petrella, E.C.; Kwasnoski, J.D.; Lobanov, V.S.; Myslik, J.; Graf, E.; Carver, T.; Asel, E.; Springer, B.A.; Lane, P.; et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen 2001, 6, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.J.; Schaeffer, P.M. Dissecting the salt dependence of the Tus-Ter protein-DNA complexes by high-throughput differential scanning fluorimetry of a GFP-tagged Tus. Mol. Biosyst. 2013, 9, 3146–3154. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.J.; Schaeffer, P.M. Differential Tus-Ter binding and lock formation: Implications for DNA replication termination in Escherichia coli. Mol. Biosyst. 2012, 8, 2783–2791. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, J.E.; Leonard, A.C. Blocking the Trigger: Inhibition of the Initiation of Bacterial Chromosome Replication as an Antimicrobial Strategy. Antibiotics 2019, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Koonin, E.V. Archaeology of eukaryotic DNA replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a012963. [Google Scholar] [CrossRef]
- Altieri, A.S.; Kelman, Z. DNA Sliding Clamps as Therapeutic Targets. Front. Mol. Biosci. 2018, 5, 87. [Google Scholar] [CrossRef]
- Georgescu, R.E.; Yurieva, O.; Kim, S.S.; Kuriyan, J.; Kong, X.P.; O’Donnell, M. Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp. Proc. Natl. Acad. Sci. USA 2008, 105, 11116–11121. [Google Scholar] [CrossRef]
- Young, M.J. Off-Target Effects of Drugs that Disrupt Human Mitochondrial DNA Maintenance. Front. Mol. Biosci. 2017, 4, 74. [Google Scholar] [CrossRef]
- Datta, A.; Brosh, R.M., Jr. New Insights Into DNA Helicases as Druggable Targets for Cancer Therapy. Front. Mol. Biosci. 2018, 5, 59. [Google Scholar] [CrossRef]
- Seo, Y.S.; Kang, Y.H. The Human Replicative Helicase, the CMG Complex, as a Target for Anti-cancer Therapy. Front. Mol. Biosci. 2018, 5, 26. [Google Scholar] [CrossRef]
| E. coli | B. subtilis | H. pylori | M. tuberculosis | Phage T4 | Phage T7 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Initiation accessory proteins | IHF, HU, Fis, DiaA | DnaD, YabA, SirA | HobA, HU | - | - | - | Initiation | ||
| Initiator protein | DnaA | DnaA | DnaA | DnaA | - | T7 RNAP | |||
| Helicase loader | DnaC | DnaI, DnaB | HP0897, - | DciA | gp59 | - | |||
| Replicative helicase | DnaB6 | DnaC6 | DnaB6,12 | DnaB6 | gp41 | gp4 | Primosome | ||
| Primase | DnaG | DnaG | DnaG | DnaG | gp61 | gp4 | |||
| SSB 1 | SSB4 | SSB4 | SSB4 | SSB4 | gp32 | gp2.52 | |||
| CLC 2 | gp444gp62 | - | Elongation | ||||||
| Sliding clamp | gp453 | Trx (E. coli) | |||||||
| DNA Pol III core | PolC, DnaE | DnaE1 () | gp43 | gp5 | |||||
| Terminator | Tus | RTP2 | - | - | - | - | Termination | ||
| Inhibitor | Status | Target | Access. No. | |
|---|---|---|---|---|
| 1 | Quinolones/Fluoroquinolones (Nalidixic acid derivatives) | Clinical use | Type II topoisomerases | DB00779 |
| 2 | Aminocoumarins (Novobiocin) | Withdrawn | Type II topoisomerase (specifically Gyrase B) | DB01051 |
| 3 | Thymidine monophosphate | Experimental | DNA topoisomerase I DNA Pol III ( subunit) | DB01643 |
| 4 | Adenosine 5′-[gamma-thio]triphosphate | Experimental | CLC 1 ( subunit) | DB02930 |
| 5 | [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | Experimental | Sliding clamp () | DB06998 |
| Protein Target | Inhibitor | Species * | Ref. |
|---|---|---|---|
| DnaA | Bisindole derivatives | Eco | [41,112] |
| DnaB | Myricetin | Eco | [113] |
| Galangin | Kpn | [114] | |
| Coumarin-based | Ban Sau | [115] | |
| Triaminotriazine derivatives | Eco Sau Pau | [116] | |
| DnaG | Para-phenyl substituted tetrazoles | Eco | [117] |
| Benzo[d]imidazo[2,1-b]imidazoles Benzo[d]pyrimido[5,4-b]furans Pyrido[3’,2’:4,5]thieno[3,2-d]pyrimidines | Eco | [118] | |
| Tilorone Doxorubicin Suramin | Ban | [119] | |
| Doxorubicin Suramin Ellagic acid | Mtb | [39] | |
| Anthracyclines and aloe-emodin | Mtb | [120] | |
| Daunorubicin derivatives | Mtb | [121] | |
| 3-[1-benzyl-5-chloro-2-(ethoxycarbonyl)-4-(trifluoromethyl)-1H-indol-3-yl]propanoic acid 3-[1-benzyl-7-chloro-2-(ethoxycarbonyl)-5-(trifluoromethyl)-1H-indol-3-yl]propanoic acid | Mtb | [122,123] | |
| 9-fluorenone-based derivatives | Sau Ban Bth | [124] | |
| Dequalinium analogues | Sau | [125] | |
| Phenolic monosaccharides derived from Polygonum cuspidatum | Eco | [126] | |
| Bicyclic 10-membered macrolide (Sch 642305) from P. verrucosum | Eco | [127,128,129] |
| Target | Species * | Type | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| DnaA | Eco | Cell-based (dnaA219rnhA reporter strain) | Compounds can cross bacterial membrane | Cannot distinguish compounds with specificity for DnaA | [133] |
| Eco | Minichromosome-based (GFP reporter) | Compounds can cross bacterial membrane | Inhibition of plasmid-based oriC not always repeatable with chromosomal oriC | [134] | |
| Eco | Cell-based (pBR322-DARS2 and hda mutant reporter strains) | Compounds can cross bacterial membrane | Cannot distinguish compounds with specificity for DnaA | [135] | |
| Eco | Filter binding assay ([α-32P]ATP) | Could be converted for HTS | Requires radioactive labelling and scintillation counter | [41] | |
| Spy | In silico (molecular dynamics simulation) | Inexpensive, can be used for pre-screening | Requires additional in vivo efficacy conformation | [136] | |
| Eco | Cell-based (SF53 reporter strain) | HTS format (384-well plates) | Cannot distinguish compounds with specificity for DnaA | [137] | |
| DnaB | Kpn | ATPase assay (molybdophosphoric acid complex) | Could be converted for HTS | Low throughput | [40] |
| Kpn | Helicase activity assay (FRET 1) | Could be converted for HTS | Low throughput | [40] | |
| Sau | ATPase assay (molybdophosphoric acid complex) | HTS format (96-well plates) | Requires helicase activity assay for confirmation | [115] | |
| Eco Sau Ban Pau | Helicase activity assay (FRET 1) | HTS format (96-well plates) | Cannot distinguish compounds with specificity for DnaB | [115,116,138,139,140] | |
| Kpn | dNTP dissociation (fluorescence) | Could be converted for HTS | Requires helicase activity assay for confirmation | [114] | |
| viral and bacterial | Time-resolved FRET 1 (Tb3+, Eu3+) | HTS format (Up to 1536-well plates) | Optical interference from compounds | [141] | |
| Eco Bst | ATPase assay (NADH) | Could be converted for higher throughput | Low throughput | [113,132] | |
| Bst | Helicase activity (radioactive label not specified) | Can be used for inhibitors of DnaB/G interaction | Low-throughput, requires radioactive labelling | [132] | |
| UvrD | Eco | Helicase activity (SYTOX stain) | Microfluidic flowcell format, could be used for DnaB | Low throughput | [142] |
| DnaB/ DnaG | Bst | Reverse yeast three-hybrid (β-galactosidase) | Allows screening of potential antimicrobial peptides | Low-throughput, for screening of peptides only | [143] |
| Eco | SPA 2 ([3H]CTP) | HTS format (96-well plates) | Requires radioactive labelling and scintillation counter | [118,138] | |
| DnaG | Eco | Thermally denaturing HPLC (260 nm) | Can distinguish between de novo synthesis and elongation | Low throughput | [113,144] |
| Mtb Ban Sau | Pyrophosphatase assay (molybdophosphoric acid complex) | HTS format (384-well plates) | Cannot distinguish compounds with specificity for DnaG | [39,119,120,125] | |
| Eco Mtb | Primase activity ([3H]NTP, [α-32P]ATP) | Can be used for inhibitors of DnaB/G interaction | Low throughput, requires radioactive labelling, cannot distinguish compounds with specificity for DnaG | [123,126,128] | |
| Eco | SPR 3 competition assay | Can be used for inhibitors of DnaG/SSB interaction | Cannot distinguish compounds with specificity for DnaG | [117] | |
| Eco T7 | STD 4 and 2D NMR (Imax, 15N-1H HSQC 5) | Could be used for inhibitors of other proteins | Requires pooling of compounds for initial screens | [117,122] | |
| Eco | Primase/Replicase activity assay (PicoGreen) | Can be used for inhibitors of DnaG/SSB interaction, HTS format (384-well plates) | Cannot distinguish compounds with specificity for DnaG | [145,146] | |
| All | Any species | Molecular docking (AutoDock Vina, Glide, Molecular Operating Environment) | Can give indication of mechanism of action | Requires additional in vivo efficacy conformation | [117,118,121,122,136] |
| Any species | SPR 3 | Sensitive, can determine binding kinetics | Expensive, need to control for buffer effects | [147,148] | |
| Any species | Mass spectrometry (AS-MS 6, LC-ESI-MS 7) | Sensitive, can pool compounds to increase throughput | Requires multiple rounds for inhibitor ranking | [147,149] | |
| Any species | Thermofluor (SYPRO Orange & ANS stain) | HTS format (384-well plates) | Optical interference from compounds | [147,150] | |
| Any species | DSF-GTP 8 (GFP) | HTS format (96-well plates), can be used in mixed samples, can test target access | Optical interference from compounds | [151,152] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radford, H.M.; Toft, C.J.; Sorenson, A.E.; Schaeffer, P.M. Inhibition of Replication Fork Formation and Progression: Targeting the Replication Initiation and Primosomal Proteins. Int. J. Mol. Sci. 2023, 24, 8802. https://doi.org/10.3390/ijms24108802
Radford HM, Toft CJ, Sorenson AE, Schaeffer PM. Inhibition of Replication Fork Formation and Progression: Targeting the Replication Initiation and Primosomal Proteins. International Journal of Molecular Sciences. 2023; 24(10):8802. https://doi.org/10.3390/ijms24108802
Chicago/Turabian StyleRadford, Holly M., Casey J. Toft, Alanna E. Sorenson, and Patrick M. Schaeffer. 2023. "Inhibition of Replication Fork Formation and Progression: Targeting the Replication Initiation and Primosomal Proteins" International Journal of Molecular Sciences 24, no. 10: 8802. https://doi.org/10.3390/ijms24108802
APA StyleRadford, H. M., Toft, C. J., Sorenson, A. E., & Schaeffer, P. M. (2023). Inhibition of Replication Fork Formation and Progression: Targeting the Replication Initiation and Primosomal Proteins. International Journal of Molecular Sciences, 24(10), 8802. https://doi.org/10.3390/ijms24108802







