Abstract
The medaka (Oryzias latipes) is an excellent vertebrate model for studying the development of the retina. Its genome database is complete, and the number of opsin genes is relatively small compared to zebrafish. Short wavelength sensitive 2 (sws2), a G-protein-coupled receptor expressed in the retina, has been lost in mammals, but its role in eye development in fish is still poorly understood. In this study, we established a sws2a and sws2b knockout medaka model by CRISPR/Cas9 technology. We discovered that medaka sws2a and sws2b are mainly expressed in the eyes and may be regulated by growth differentiation factor 6a (gdf6a). Compared with the WT, sws2a−/− and sws2b−/− mutant larvae displayed an increase in swimming speed during the changes from light to dark. We also observed that sws2a−/− and sws2b−/− larvae both swam faster than WT in the first 10 s of the 2 min light period. The enhanced vision-guided behavior in sws2a−/− and sws2b−/− medaka larvae may be related to the upregulation of phototransduction-related genes. Additionally, we also found that sws2b affects the expression of eye development genes, while sws2a is unaffected. Together, these findings indicate that sws2a and sws2b knockouts increase vision-guided behavior and phototransduction, but on the other hand, sws2b plays an important role in regulating eye development genes. This study provides data for further understanding of the role of sws2a and sws2b in medaka retina development.
1. Introduction
Visual pigments in photoreceptor cells are very important in light detection and signal transduction [1,2]. Visual pigments consist of opsin receptor proteins that are covalently bound to vitamin A-derived photochromophores, and opsins mediate light absorption in cones and rods [3]. In vertebrates, rod cells responsible for dark or low-light vision express only rhodopsin (rh1), while cone cells for bright or well-lit vision fall into four main categories: short- (sws1, sws2), medium- (rh2), and long- (lws) wavelength-sensitive opsins [4]. Several studies have shown that opsins play a crucial part in feeding, behavior, and color sensitivity in fish [5,6,7,8]. In the process of fish evolution, some copies of the opsin gene may be lost, but all opsins are basically present, while sws2 and rh2 are lost in most mammals [9,10]. Therefore, the function of sws2 and rh2 in fish is worth studying. RH2 mainly performs its visual function in fish that are active at night or dusk [11]. Much less is known about sws2, with only Percomorpha fish having received extensive research [12].
Visual pigment proteins show great diversity under natural selection, and many fish genomes contain one to three copies of sws2 genes [13,14,15]. In rainbow trout (Oncorhynchus mykiss), sws2 is switched from sws1, a process that begins before the yolk sac is fully absorbed and continues throughout the juvenile period [16,17]. Atlantic Cod (Gadus morhua) have lost sws1 and lws opsins in evolution, and only uses sws2 and rh2 opsins to detect prey, avoid predators, and adapt to light response [18]. Similarly, the threespine stickleback (Gasterosteus aculeatus) adapts its visual perception to blackwater habitats by tuning spectra at sws2 sites [14]. There is a group of weakly electric teleost fishes in South America (Gymnotiforms) that are nocturnal, many living in muddy streams or deep rivers, and which have lost the sws1 and sws2 opsin genes through evolution [19]. In addition, changes in opsin expression patterns and gene expression regulation also affect opsin function.
The expression of opsin varies greatly among different fish species, but each species may change according to ontogeny and changes in the light environment [20]. On the other hand, the regulation of sws2 gene expression is also controversial. Both gdf6a and forkhead box Q2 (foxq2) have been proven to regulate the expression and differentiation of sws2 [21,22]. In addition, some studies have shown that changes in the opsin genes affected by transcriptional regulators or drug exposure can also cause changes in visual guidance behavior [23,24].
The medaka (Oryzias latipes) is an ideal model organism for studying visual development, and its retina is rich in various types of cone photoreceptors that mediate color perception [25,26]. Medaka and zebrafish (Danio rerio) retinas exhibit distinct patterns of photoreceptor cell mosaics, with each blue-sensitive cone (sws2) surrounded by four double cones (lws and rh2) [27]. Despite the complexity of the spatiotemporal pattern of cone opsin expression in fish, recent genome-editing technologies have made the use of mutants in genome studies possible [28,29].
In this study, we generated sws2a and sws2b knockout medaka models using CRISPR/Cas9. We discovered that medaka sws2a and sws2b are mainly expressed in the eyes and may be regulated by gdf6a. The enhanced vision-guided behavior in sws2a−/− and sws2b−/− medaka larvae may be related to the enhanced photoconductive signaling. Additionally, we also found that sws2b affects the expression of eye development genes, while sws2a does not. These findings indicate that sws2a and sws2b knockouts increase vision-guided behavior and upregulated phototransduction-related genes, but on the other hand, sws2b plays an important role in regulating eye development genes.
2. Results
2.1. Expression of sws2a and sws2b in Medaka
There are two orthologous genes of sws2 in medaka, namely sws2a and sws2b, and sequence alignment showed the amino acid residues of medaka sws2a and sws2b had 77.27% homology (Figure 1A). In addition, the expression of sws2a and sws2b mRNA in different tissues was detected by RT-PCR. Both sws2a and sws2b were expressed mainly in the eyes among the adult tissues (Figure 1B). The expression levels of sws2a and sws2b were low in larval fish, while sws2b expression was increased in adult fish (Figure 1C).
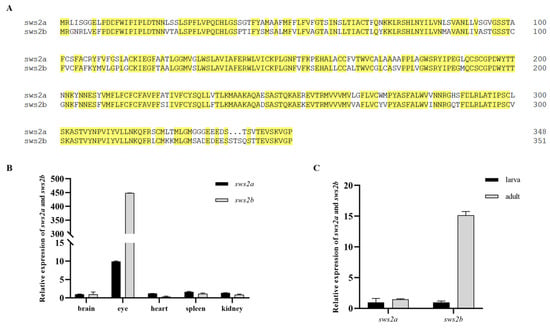
Figure 1.
Conservation analysis and expression characterization of sws2a and sws2b in medaka. (A) Sequence alignment of amino acids in sws2a and sws2b (n = 3). (B) Differences of sws2a and sws2b mRNA expression in different tissues of medaka adult. (C) Expression of sws2a and sws2b in larva and adults (n = 6).
2.2. Establishment of sws2a and sws2b Mutant Medaka
To understand the importance of sws2a and sws2b in visual performance, we generated sws2a and sws2b mutant medaka by CRISPR/Cas9 technology. The CRISPR/Cas9 guide-RNAs close to the translation start codon were selected to change the base sequences of sws2a and sws2b, thereby blocking protein translation. Compared with the WT medaka, sws2a and sws2b homozygous mutants showed 4 bp and 274 bp deletions, respectively, and the deletions both resulted in the early termination of translation of the entire seven-transmembrane domain (7tm_1) in SWS2 (Figure 2A–D). Protein structure prediction further elucidated the knockout results (Figure 2E,F).
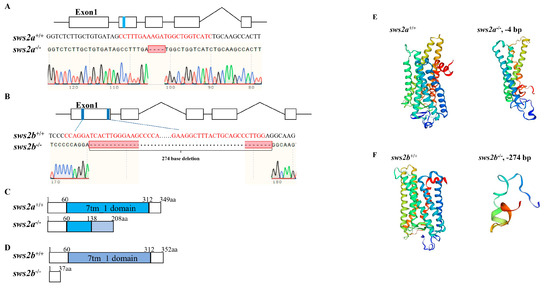
Figure 2.
Generation and characterization of sws2a and sws2b mutant medaka. (A,B) Design target sites of sws2a and sws2b based on CRISPR/Cas9 technology. Exons are represented by boxes, and single-guide RNAs (sgRNAs) are labeled with blue color in exon. The sgRNA sequences are highlighted in red, and the −4 bp and −274 bp deletions are indicated by sequencing validation. (C,D) Illustration of deduced protein structure of WT and sws2a and sws2b mutants. These numbers represent amino acid positions from the initiation codon. (E,F) The sws2a and sws2b protein tertiary structure prediction of WT and sws2a and sws2b mutants.
In order to determine whether the sws2a−/− and sws2b−/− mRNA was decayed, we analyzed the transcript levels of sws2a and sws2b in WT, sws2a−/−, and sws2b−/− medaka by RT-qPCR. The sws2a−/− and sws2b−/− medaka both had no significant decrease in sws2a and sws2b mRNA levels, respectively, while transcriptional compensation of sws2b deletion was observed sws2a−/− (Figure 3A,B). Moreover, we designed two pairs of total-length primers to distinguish the expression differences between the two transcripts of medaka sws2 (Figure S1). Next, we sequenced the total lengths of sws2a and sws2b in sws2a−/− and sws2b−/− medaka, which showed 4 bp and 274 bp deletions in sws2a and sws2b mRNA levels in sws2a−/− and sws2b−/− mutants (Figure S2). sws2a−/− and sws2b−/− showed no morphological difference from the WT medaka (Figure S3).
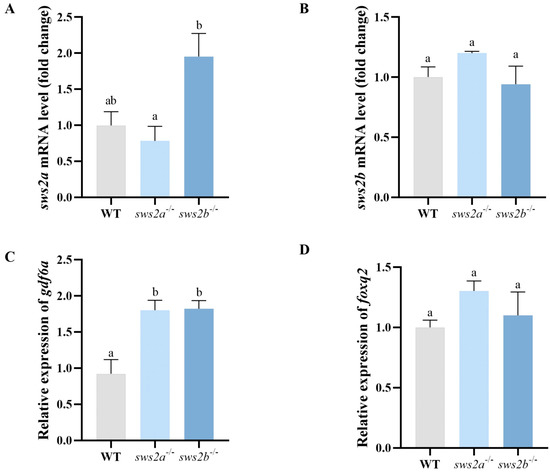
Figure 3.
The mRNA levels of sws2a and sws2b, and the transcription factors gdf6a and foxq2 in the larval eyes at 6 dph. (A) sws2a. (B) sws2b. (C) gdf6a. (D) foxq2. All data are expressed as the mean ± SEM (n = 6). Vertical bars not sharing the same letter are significantly different (p < 0.05).
Because gdf6a and foxq2 were reported to determine the expression of sws2 [21,22], we analyzed the mRNA expression levels of gdf6a and foxq2 in sws2a−/− and sws2b−/− mutants. GDF6A mRNA expression levels were significantly increased in both sws2a−/− and sws2b−/− compared to WT (Figure 3C, p < 0.05). However, the mRNA expression levels of foxq2 exhibited no significant difference in the fish (Figure 3D, p > 0.05).
2.3. Feeding and the Behavioral Tests
The food intake of Artemia in sws2a−/− medaka larvae was significantly higher than that of WT and sws2b−/− (Figure 4A, p < 0.05), but there was no significant difference in growth (Figure 4B, p > 0.05).
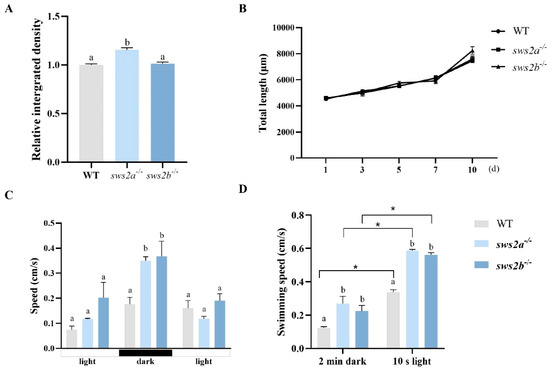
Figure 4.
Analysis of feeding and the behavioral tests among WT, sws2a−/−, and sws2b−/− medaka at the larval stage. (A) Relative levels of ingested Artemia shown in the digestive tract (n = 60). (B) Total length of medaka from 1 to 10 days from first feeding (n = 15/group). (C) The statistical analysis of swimming speed during photoperiod stimulation period in WT, sws2a−/−, and sws2b−/− larvae. (D) The swimming speed during the last 2 min dark and the first 10 s of the 2 min light period (* p < 0.05 by Student’s t-test). Vertical bars not sharing the same letter are significantly different (p < 0.05).
We next examined the visual responsiveness in sws2a−/− and sws2b−/− mutants at 6 dph. Compared with the WT, sws2a−/− and sws2b−/− mutant larvae displayed an increase in swimming speed during the changes from light to dark (Figure 4C, p < 0.05). We also observed that sws2a−/− and sws2b−/− larvae both swam faster than WT in the dark and the first 10 s of the 2 min light period (Figure 4D, p < 0.05). They all had a peak in the first 10 s of the 2 min light period, while the swimming speed was detected to increase by 48.1% and 57.3% in sws2a−/− and sws2b−/− larvae, respectively.
2.4. Transcript Levels of Phototransduction-Related Genes in Larvae of sws2a−/− and sws2b−/− Mutants
We examined the mRNA levels of phototransduction-related genes in larvae of the WT, sws2a−/−, and sws2b−/− medaka by RT-qPCR. Surprisingly, the deletion of sws2a or sws2b did not affect the expression of other cone and rod genes (Figure 5A, p > 0.05). We further measured the mRNA levels of cone phototransduction genes in WT, sws2a−/−, and sws2b−/− medaka at 6 dph. Expression of guanine nucleotide-binding protein (G protein), beta polypeptide 3b (gnb3a), and G protein-coupled receptor kinase 7a (grk7a) were increased significantly in the sws2b−/− medaka, while only grk7a was elevated in sws2a−/− larvae (Figure 5B, p < 0.05).
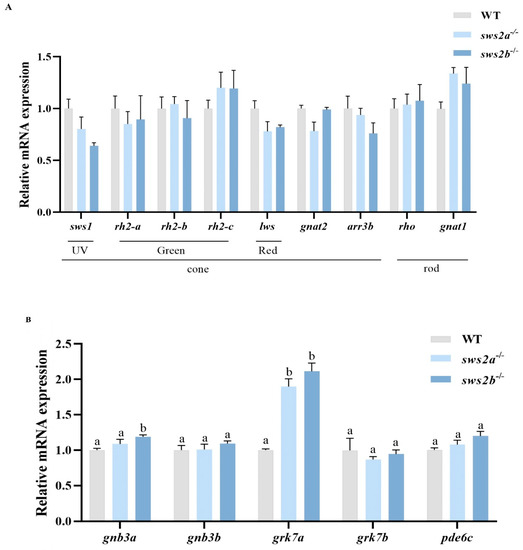
Figure 5.
The transcriptional levels of opsin- and phototransduction-related genes in the larval eyes at 6 dph. (A) Genes involved in opsin (sws1, rh2-a, rh2-b, rh2-c, lws, gnat2, arr3b, rho, and gnat1). (B) Genes involved in phototransduction (gnb3a, gnb3b, grk7a, and grk7b, and pde6c). The data are represented by mean ± SEM (n = 6). Vertical bars not sharing the same letter are significantly different (p < 0.05).
2.5. Transcript Levels of Eye Development Gene in Larval sws2a and sws2b Knockout Medaka
We next tested whether sws2a and sws2b knockout affected the expression of the eye development genes in the larvae. Compared to WT, sws2a−/− larvae displayed an increased mRNA expression pattern of SIX homeobox 7 (six7) (Figure 6A, p < 0.05), while no alteration was observed in the sws2b−/−. In addition, the transcriptional expression levels of paired box 6 (pax6), SIX homeobox 3a (six3a), and six3b were significantly decreased in the sws2b−/− larvae (Figure 6B–D, p < 0.05).
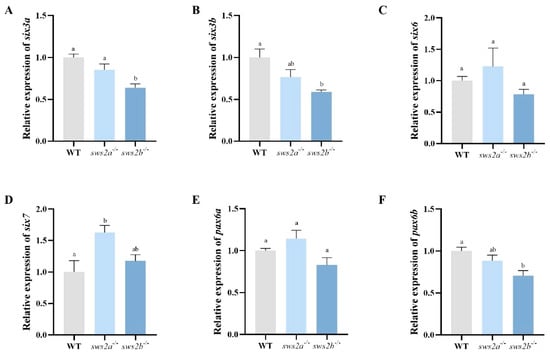
Figure 6.
The results of the qRT-PCR analysis of eye development genes in larval eyes at 6 dph. (A) six3a. (B) sin3b. (C) six6. (D) six7. (E) pax6a. (F) pax6b. All data are expressed as the mean ± SEM (n = 6). Vertical bars not sharing the same letter are significantly different (p < 0.05).
3. Discussion
In contrast to most mammals that have lost sws2 and rh2 genes over evolutionary time, all four opsin gene types are present in whole-genome duplication in teleost fish, although there is a phenomenon of gene copy loss [30,31]. SWS2A and sws2b were developed by the tandem duplication of sws2 in the common ancestor of Neoteleostei fishes [32]. For instance, despite having highly conserved foxl2a and foxl2b domains in zebrafish, these two genes have divergent functions and synergies. Disruption of foxl2a and foxl2b led to premature ovarian failure and partial sex reversal, respectively, and foxl2a and foxl2b jointly regulate the development and maintenance of zebrafish ovaries [33]. The structural domain of sws2a and sws2b proteins in medaka was also highly conserved, and the protein similarity between the two proteins reached 77.27%. Medaka sws2a and sws2b are mainly expressed in the eye during adulthood, while sws2b expression is significantly elevated in adult fish relative to larva. This is consistent with the expression pattern of sws2a and sws2b in bluefin killifish (Lucania goodei) [34].
In this study, we produced and characterized sws2a and sws2b medaka mutant lines with 4 bp and 274 bp deletions that caused the loss of seven transmembrane domains in SWS2A and SWS2B. We detected that the transcription levels of sws2a and sws2b were not affected by the mutation. Although we did not find specific antibodies for medaka SWS2A or SWS2B that confirm them at the protein level, we further amplified the total lengths of the gene using cDNA as a template, and sequenced it. The results showed that sws2a and sws2b mutants had 4 bp and 274 bp deletions of sws2a and sws2b mRNA, respectively. These results indicated that the knockouts of sws2a and sws2b in this study are effective. Our results are in accordance with the recent work on lca5 knockout zebrafish [35]. SWS2 gene and protein expressions are regulated by upstream signals [36]. GDF6A is a member of the bone morphogenetic protein family that induces dorsal retinal differentiation during ocular morphogenesis [37]. GDF6A deletion zebrafish larvae displayed fewer blue cone photoreceptor cells, and gdf6a could develop and maintain the sws2 [21]. Disruption of foxq2 in zebrafish showed the loss of sws2 cone expression in 5-day post-fertilization (dpf) larvae, and further results indicated that foxq2 was an activator of sws2 transcription and inhibited sws1 expression [22]. In addition, the thyroid hormone (TH) may induce the transition of sws1 to sws2 opsin by binding to its receptor thrβ2 [38]. Growth hormone (GH) can accelerate the process of sws1 to sws2 opsin conversion [39]. Here, we observed that gdf6a expression was significantly upregulated in sws2a−/− and sws2b−/− mutants, while foxq2 expression was unaffected. We speculated that sws2a and sws2b are mainly regulated by gdf6a in medaka, mainly because when sws2a or sws2b was knocked out, its upstream gene gdf6a expression increased through negative feedback regulation. Similar to foxl2 and sox9, foxl2 is downstream of sox9, and sox9 transcript level significantly increased in foxl2a−/− and foxl2b−/− zebrafish mutants [33,40].
Feeding assessment showed increased food intake in sws2a−/− mutant zebrafish, while sws2b−/− was unaffected. In a previous report, tbx2b zebrafish mutants (reduced UV (SWS1) cones) showed declined foraging performance [41]. Another study demonstrated that sws2 and rh2 opsins decreased, and visual prey capture was impaired in six6a/six6b/six7 triple-knockout zebrafish at 6 dpf [23]. It has also been proposed that in the case of sws1 regulation, the increase in sws2 may be related to prey search [5,42]. Therefore, we think that the increased intake of sws2a−/− may be related to the deletion of sws2a. Interestingly, the feeding rate and survival rate of larvae of haddock (Mellanogrammus aeglefinus) increased under blue and green light compared with other light colors [43,44]. Light/dark motion tests and responses to light stimuli are effective methods to reflect the integrated function of visual pathways [45,46,47]. In this study, compared with the control group, sws2a−/− and sws2b−/− mutants both significantly increased swimming speed when they changed from light to darkness. When the larvae were suddenly stimulated by light, sws2a−/− and sws2b−/− mutant larvae both reacted strongly within the first 10 s. The swimming speed increased by 48.1% and 57.3% in sws2a−/− and sws2b−/− larvae, respectively. These results are consistent with our finding that cone phototransduction pathway-related genes are further activated in sws2a−/− and sws2b−/− larvae. GRK7A is mainly expressed in the outer cone segment; grk7 knockdown in larval zebrafish caused a delay in dark adaptation and impaired cone response recovery [48]. Moreover, grk7a has been shown to be involved in the recovery of cone light response in larval zebrafish [49]. Additionally, GNB3 is a G-protein beta subunit located in the outer segment of cones and mainly plays a role in the phototransduction cascade of cones [50]. We concluded that the enhanced vision-guided behavior in sws2a−/− and sws2b−/− medaka larvae may be the result of upregulated phototransduction genes.
In vertebrates, opsins are G-protein-coupled receptors expressed in the retina, responsible for promoting eye sensitivity to light. Each cone opsin covers a distinct part of the visible spectrum, with corresponding non-absorption maxima [51]. Our RT-PCR analysis indicated that the loss of sws2a and sws2b did not affect the normal expression of other opsins. In addition to being absorbed by sws2, blue light may also be absorbed by neighboring sws1 and rh2. Thus, the sensitivity of blue light is not controlled by sws2 [52]. Fish such as the southern catfish (silurus meridionalis), which are mainly nocturnal and live in underground or even burrowing freshwater environments, lost sws2 [53,54]. Some recently studied cartilaginous fishes also show no expression of sws2, further supporting the hypothesis that sws2 was lost early in the cartilaginous lineage [55,56,57]. However, sws2 plays an irreplaceable role in some species [14]. The Japanese flounder (Paralichthys olivaceus) adjusted their sws2a and rh2 genes as their light environment changed during development [58]. Therefore, diverse visual systems are adaptive responses to varying environments in different species.
In this study, there was no significant retinal histological damage observed during the retinal histological examination, indicating that its effect may be at the molecular level. SIX3, six6, and six7 are important regulators of fish retinal development and differentiation [23,59]. Knock-down six3b and six7 resulted in the loss of eyes, whereas disruption of six3a and six7 had no significant eye phenotypes [60]. A previous study demonstrated that six6 deletion mice showed severe retinal abnormalities [61]. PAX6 is necessary for the normal development of fish eyes [62]. Deletion of a single copy of pax6 in mice showed microphthalmia, while mutation with a double copy showed anophthalmia [63]. In the present study, we found that six3a, six3b, and pax6b were downregulated in sws2b−/− medaka larvae, suggesting that the loss of sws2b may affect the retinal development of medaka larvae. In addition, the upregulated expression of six7 in sws2a−/− medaka larvae might be due to the regulation of sws2a by six7 [64]. Paradoxically, sws2b−/− medaka larvae showed increased motor capacity, but decreased transcription levels of eye development and regulatory genes. Our explanation is that sws2b may delay the movement of medaka larvae, but on the other hand, sws2b plays an important role in regulating eye development genes. The mechanism of medaka sws2a and sws2b double knockout in regulating eye development needs to be determined in future studies.
In summary, this study established sws2a and sws2b knockout medaka by CRISPR/Cas9 technology. We speculated that the enhanced vision sensitivity in regulating vision-guided behavior in sws2a and sws2b knockout medaka larvae might be related to the upregulation of phototransduction-related genes. Additionally, sws2b affects eye development gene expression, implying different mechanisms of sws2a and sws2b. This study provides data for further understanding of the role of sws2a and sws2b in medaka retina development.
4. Materials and Methods
4.1. Medaka Lines and Maintenance
The wild-type (WT) medaka are an orange strain, and were maintained in an environment of 26~28 °C and 14 light/10 h dark cycle. Medaka embryos were cultured at 27~28 °C in medaka embryo medium (MEM) [65]. The 6 dph (day post-hatching) larvae were fed with live Artemia twice daily when the yolk sac was almost completely consumed. All the fish were anesthetized with tricaine methanesulfonate (MS-222) before the tissue collection.
4.2. Generating sws2a−/− and sws2b−/− Mutants by CRISPR/Cas9 Technology
Medaka sws2a (ENSORLG00000040205) and sws2b (ENSORLG00000028370) genes were targeted using CRISPR/Cas9 technology. The sequencing of single-guide RNAs (sgRNAs) and PCR primers are shown in Supplementary Table S1. sgRNAs were cloned into pMD-18T vector and were synthesized using TranscriptAid T7 High Yield Transcription kit (ThermoFisher Scientific, Waltham, MA, USA). The compounds of sgRNAs (50 ng/µL) and Cas9 protein (New England Biolabs, Ipswich, MA, USA) were co-injected into one- or two-cell stage wild-type embryos. The F0 medaka were raised to adulthood and outcrossed with wild-type to produce F1 medaka. A T7 endonuclease 1 assay (Vazyme, Nanjing, China) was used to detect sws2a heterozygous mutant individuals according to the manufacturer’s instructions. The heterozygous individuals of sws2b mutation with large fragment deletion were distinguished by PCR detection and then sequenced. The F1 heterozygous individuals in-crossed to generate F2 homozygous individuals, and all experiments were conducted with F3 homozygous individuals. Unless otherwise mentioned, the homozygous mutant lines of sws2a and sws2b in subsequent experiments were addressed as sws2a−/− and sws2b−/−, respectively. The SWISS_MODEL (https://swissmodel.expasy.org/interactive) (1 March 2023) was used to predict the protein tertiary structures. The protein tertiary structures of sws2a and sws2b were predicted using the SWISS_MODEL (https://swissmodel.expasy.org/).
4.3. Larvae Feeding Assays
For larvae food intake, 6 dph larvae were fed with Artemia in wells of a 6-well plate with 8 mL MEM (diameter 3.48 cm wells) for 30 min (6 larvae per 6-well plate). The density of the Artemia is adjusted to 150 Artemia per ml. Then, larvae were anesthetized with MS-222 (Argent Chemical Laboratories, Redmond, WA, USA) and fixed with 4% paraformaldehyde (PFA) (Servicebio, Wuhan, China) overnight. Photographs of the orange area of Artemia in the digestive tract were taken by a stereomicroscope and measured with Image J1 software. The amount of food ingested by medaka larvae was developed by the procedure described previously [66].
4.4. Growth Performance and Survival Rate
For growth performance and survival rate, 6 dph larvae were fed with abundant Artemia twice daily for 10 days. Twenty WT, sws2a−/−, and sws2b−/− medaka larvae were randomly selected, anesthetized, and fixed with 4% PFA for total length measurement. The experiment was repeated 3 times.
4.5. Behavioral Tests
Behavioral tests were conducted between 15:00 and 17:00 using the DanioVision Observation Chamber (Noldus Information Technology, Wageningen, The Netherlands) linked to the EthoVision XT13 software. The 6 dph larvae were plated onto 24-well plates (diameter 15.6 mm wells) with 1 mL MEM (individual larvae per 24-well plate). Further analysis was performed using custom Open Office Org 2.4 software.
Light response: The larvae were acclimated for 30 min in the dark at 28 °C and then tracked the movement of larvae for 4 min, with 2 min of the dark period and 10 s of the light stimulation period [46]. The average swimming speed (cm/s) for dark and light were collected every 2 min and 10 s, respectively.
Light/dark behavior analysis: The larvae were acclimated for 10 min at 28 °C, and the larval locomotor activity was tested in response to dark–light conversion (3 min light/3 min dark/3 min light/3 min dark) based on the protocol by Huang et al. [67], with modifications to the transition stimulation time. The average swimming speed (cm/s) for each individual larva was collected every 60 s.
4.6. Hematoxylin-Eosin (H&E) Staining
Medaka at 6 dph were preserved in 4% PFA for 24 h and dehydrated in 70–100% ethanol, embedded in paraffin, and sectioned at thickness of 4 μm (Leica, Heidelberg, Germany). Then, the sections were stained with hematoxylin-eosin (H&E) according to standard protocols. The slides were imaged by slice digital scanning (Pannoramic250, Pannoramic250 MIDI, 3D HISTECH).
4.7. RNA Isolation and Quantitative RT-PCR
All fish were sampled in the light phase of the light/dark cycle. The adult fish tissues (n = 3) and the two larval eyes of the 6 dph medaka (n = 6) were collected and frozen in liquid nitrogen. An equal amount of RNA was extracted from each sample according to the RNAiso instruction, and the cDNA was reversed transcribed by the HiScript® III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). The reaction system (20 μL) contained 1 μL cDNA template, 10 μL SYBR (Vazyme, China), 0.4 μL of each primer, and 8.2 μL ddH2O. The cycling parameters were 95 °C for 30 s, 40 cycles at 95 °C for 10 s, 58 °C for 30 s, and melting curve from 65 °C to 95 °C (gradually increasing 0.5 °C s−1), with data acquired every 6 s. The results were normalized to β-actin, and relative transcript abundances of genes were performed using the 2−ΔΔCt value method [68]. All primers are shown in Supplementary Table S2.
4.8. Statistical Analysis
All results are presented as means ± S.E.M (standard error of the mean), and the normality of the data was first tested by the Shapiro–Wilk test. The differences among three groups were analyzed by one-way ANOVA and Duncan’s multiple-range test, and (p < 0.05) was considered a significant difference. The differences between the two groups were determined with Student’s t-test, and statistical significance was determined at p < 0.05.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms24108786/s1.
Author Contributions
K.L. designed and performed the experiment, analyzed experimental data, and wrote the original draft. J.W., S.T. and X.J. raised fish and participated in some of the experiments. X.-F.L. designed and supervised the experiment, and wrote and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Development Project of Hubei Province (2020BBA035) and the National Natural Science Foundation of China (31972809).
Institutional Review Board Statement
All experimental protocols were approved by the Institutional Animal Care and Use Ethics Committee of Huazhong Agricultural University (an approval reference number HZAUFI-2020-0024).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available from the corresponding author by request.
Acknowledgments
We want to thank Tiansheng Chen at Jimei University for providing us with the orange strain wild-type medaka.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Archer, S. Light and photoreception: Visual pigments and photoreception. In Adaptive Mechanisms in the Ecology of Vision; Springer: Berlin/Heidelberg, Germany, 1999; pp. 25–42. [Google Scholar]
- Palczewski, K.; Kiser, P.D. Shedding new light on the generation of the visual chromophore. Proc. Natl. Acad. Sci. USA 2020, 117, 19629–19638. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S. Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 2000, 19, 385–419. [Google Scholar] [CrossRef] [PubMed]
- Carleton, K. Cichlid fish visual systems: Mechanisms of spectral tuning. Integr. Zool. 2009, 4, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Flamarique, I.N. Opsin switch reveals function of the ultraviolet cone in fish foraging. Proc. Biol. Sci. 2013, 280, 20122490. [Google Scholar]
- Stieb, S.M.; Cortesi, F.; Queiroz, L.J.D.; Carleton, K.L.; Seehausen, O.; Marshall, N.J. Long-wavelength-sensitive (lws) opsin gene expression, foraging and visual communication in coral reef fishes. Mol. Ecol. 2022, 32, 1656–1672. [Google Scholar] [CrossRef]
- Stieb, S.M.; Cortesi, F.; Sueess, L.; Carleton, K.L.; Salzburger, W.; Marshall, N.J. Why UV vision and red vision are important for damselfish (Pomacentridae): Structural and expression variation in opsin genes. Mol. Ecol. 2017, 26, 1323–1342. [Google Scholar] [CrossRef] [PubMed]
- Shimmura, T.; Nakayama, T.; Shinomiya, A.; Fukamachi, S.; Yasugi, M.; Watanabe, E.; Shimo, T.; Senga, T.; Nishimura, T.; Tanaka, M.; et al. Dynamic plasticity in phototransduction regulates seasonal changes in color perception. Nat. Commun. 2017, 8, 412. [Google Scholar] [CrossRef]
- Lin, J.-J.; Wang, F.-Y.; Li, W.-H.; Wang, T.-Z. The rises and falls of opsin genes in 59 ray-finned fish genomes and their implications for environmental adaptation. Sci. Rep. 2017, 7, 15568. [Google Scholar] [CrossRef]
- Trezise, A.E.; Collin, S.P. Opsins: Evolution in waiting. Curr. Biol. 2005, 15, R794–R796. [Google Scholar] [CrossRef]
- Musilova, Z.; Cortesi, F. Multiple ancestral and a plethora of recent gene duplications during the evolution of the green sensitive opsin genes (RH2) in teleost fishes. BioRxiv 2021. [Google Scholar] [CrossRef]
- Cortesi, F.; Musilová, Z.; Stieb, S.M.; Salzburger, W. Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc. Natl. Acad. Sci. USA 2015, 112, 1493–1498. [Google Scholar] [CrossRef] [PubMed]
- Härer, A.; Torres-Dowdall, J.; Meyer, A. Rapid adaptation to a novel light environment: The importance of ontogeny and phenotypic plasticity in shaping the visual system of Nicaraguan Midas cichlid fish (Amphilophus citrinellus spp.). Mol. Ecol. 2017, 26, 5582–5593. [Google Scholar] [CrossRef]
- Marques, D.A.; Taylor, J.S.; Jones, F.C.; Palma, F.D.; Kingsley, D.M.; Reimchen, T.E. Convergent evolution of SWS2 opsin facilitates adaptive radiation of threespine stickleback into different light environments. PLoS Biol. 2017, 15, e2001627. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.-L.; Liang, X.-F.; Li, L.; Wu, J.; Lu, K. Genome-wide identification and expression patterns of opsin genes during larval development in Chinese perch (Siniperca chuatsi). Gene 2022, 825, 146434. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.L.; Gan, K.J.; Flamarique, I.N. The ultraviolet opsin is the first opsin expressed during retinal development of salmonid fishes. Investig. Ophthalmol. Vis. Sci. 2007, 48, 866–873. [Google Scholar] [CrossRef]
- Cheng, C.L.; Flamarique, I.N. Chromatic organization of cone photoreceptors in the retina of rainbow trout: Single cones irreversibly switch from UV (SWS1) to blue (SWS2) light sensitive opsin during natural development. J. Exp. Biol. 2007, 210, 4123–4135. [Google Scholar] [CrossRef]
- Valen, R.; Edvardsen, R.B.; Søviknes, A.M.; Drivenes, Ø.; Helvik, J.V. Molecular evidence that only two opsin subfamilies, the blue light-(SWS2) and green light-sensitive (RH2), drive color vision in Atlantic cod (Gadus morhua). PLoS ONE 2014, 9, e115436. [Google Scholar] [CrossRef]
- Liu, D.W.; Lu, Y.; Yan, H.Y.; Zakon, H.H. South American Weakly Electric Fish (Gymnotiformes) Are Long-Wavelength-Sensitive Cone Monochromats. Brain Behav. Evol. 2016, 88, 204–212. [Google Scholar] [CrossRef]
- Musilova, Z.; Salzburger, W.; Cortesi, F. The visual opsin gene repertoires of teleost fishes: Evolution, ecology, and function. Annu. Rev. Cell Dev. Bi. 2021, 37, 441–468. [Google Scholar] [CrossRef]
- Duval, M.G.; Oel, A.P.; Allison, W.T. gdf6a is required for cone photoreceptor subtype differentiation and for the actions of tbx2b in determining rod versus cone photoreceptor fate. PLoS ONE 2014, 9, e92991. [Google Scholar] [CrossRef]
- Ogawa, Y.; Shiraki, T.; Fukada, Y.; Kojima, D. Foxq2 determines blue cone identity in zebrafish. Sci. Adv. 2021, 7, eabi9784. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Shiraki, T.; Asano, Y.; Fukada, Y. Six6 and Six7 coordinately regulate expression of middle-wavelength opsins in zebrafish. Proc. Natl. Acad. Sci. USA 2019, 116, 4651–4660. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.; Popowitz, J.; Delbridge-Perry, M.; Rowe, C.J.; Connaughton, V.P. The role of estrogen and thyroid hormones in zebrafish visual system function. Front. Pharmacol. 2022, 13, 837687. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Shoji Fukamachi, S.; Mitani, H.; Kawamura, S. Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes). Gene 2006, 371, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Kitambi, S.S.; Malicki, J.J. Spatiotemporal features of neurogenesis in the retina of medaka, Oryzias Latipes. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2008, 237, 3870–3881. [Google Scholar] [CrossRef]
- Tohya, S.; Mochizuki, A.; Iwasa, Y. Difference in the retinal cone mosaic pattern between zebrafish and medaka: Cell-rearrangement model. J. Theor. Biol. 2003, 221, 289–300. [Google Scholar] [CrossRef]
- Harada, Y.; Matsuo, M.; Kamei, Y.; Goto, M.; Fukamachi, S. Evolutionary history of the medaka long-wavelength sensitive genes and effects of artificial regression by gene loss on behavioural photosensitivity. Sci. Rep. 2019, 9, 2726. [Google Scholar] [CrossRef]
- Matsuo, M.; Matsuyama, M.; Kobayashi, T.; Kanda, S.; Ansai, S.; Kawakami, T.; Hosokawa, E.; Daido, Y.; Kusakabe, T.G.; Naruse, K.; et al. Retinal Cone Mosaic in sws1-Mutant Medaka (Oryzias latipes), A Teleost. Investig. Ophthalmol. Vis. Sci. 2022, 63, 21. [Google Scholar] [CrossRef]
- Rennison, D.J.; Owens, G.L.; Taylor, J.S. Opsin gene duplication and divergence in ray-finned fish. Mol. Phylogenet Evol. 2012, 62, 986–1008. [Google Scholar] [CrossRef]
- Musser, J.M.; Arendt, D. Loss and gain of cone types in vertebrate ciliary photoreceptor evolution. Dev. Biol. 2017, 431, 26–35. [Google Scholar] [CrossRef]
- Yokoyama, S. Evolution of dim-light and color vision pigments. Annu. Rev. Genom. Hum. Genet. 2008, 9, 259–282. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-J.; Wang, Y.; Li, Z.; Zhou, L.; Gui, J.-F. Sequential, divergent, and cooperative requirements of Foxl2a and Foxl2b in ovary development and maintenance of zebrafish. Genetics 2017, 205, 1551–1572. [Google Scholar] [CrossRef]
- Chang, C.-H.; Catchen, J.; Moran, R.L.; Rivera-Colón, A.G.; Wang, Y.-C.; Fuller, R.C. Sequence analysis and ontogenetic expression patterns of cone opsin genes in the bluefin killifish (Lucania goodei). J. Hered. 2021, 112, 357–366. [Google Scholar] [CrossRef]
- Qu, Z.; Yimer, T.A.; Xie, S.; Wong, F.; Yu, S.; Liu, X.; Han, S.; Ma, J.; Lu, Z.; Hu, X.; et al. Knocking out lca5 in zebrafish causes cone-rod dystrophy due to impaired outer segment protein trafficking. BBA-Mol. Basis Dis. 2019, 1865, 2694–2705. [Google Scholar] [CrossRef] [PubMed]
- Takechi, M.; Seno, S.; Kawamura, S. Identification of cis-acting elements repressing blue opsin expression in zebrafish UV cones and pineal cells. J. Biol. Chem. 2008, 283, 31625–31632. [Google Scholar] [CrossRef]
- Gosse, N.J.; Baier, H. An essential role for Radar (Gdf6a) in inducing dorsal fate in the zebrafish retina. Proc. Nat. Acad. Sci. USA 2009, 106, 2236–2241. [Google Scholar] [CrossRef] [PubMed]
- Deveau, C.; Jiao, X.; Suzuki, S.C.; Krishnakumar, A.; Yoshimatsu, T.; Hejtmancik, J.F.; Nelson, R.F. Thyroid hormone receptor beta mutations alter photoreceptor development and function in Danio rerio (Zebrafish). PLoS Genet. 2020, 16, e1008869. [Google Scholar] [CrossRef]
- Novales Flamarique, I.; Ahmed, A.S.; Cheng, C.L.; Molday, R.S.; Devlin, R.H. Growth hormone regulates opsin expression in the retina of a salmonid fish. J. Neuroendocrinol. 2019, 31, e12804. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Treier, M. Foxl2 function in ovarian development. Mol. Genet. Metab. 2006, 88, 225–234. [Google Scholar] [CrossRef]
- Flamarique, N.I. Diminished foraging performance of a mutant zebrafish with reduced population of ultraviolet cones. Proc. Biol. Sci. 2016, 283, 20160058. [Google Scholar]
- Viken, E.N. Expression of Visual Opsins in the Retina of Atlantic salmon (Salmo salar L.) during smoltification. Master’s Thesis, The University of Bergen, Bergen, Norway, 2020. [Google Scholar]
- Downing, G. Impact of spectral composition on larval haddock, Melanogrammus aeglefinus L., growth and survival. Aquac. Res. 2002, 33, 251–259. [Google Scholar] [CrossRef]
- Villamizar, N.; Blanco-Vives, B.; Migaud, H.; Davie, A.; Davie, A.; Carboni, S.; Sánchez-Vázquez, F.J. Effects of light during early larval development of some aquacultured teleosts: A review. Aquaculture 2011, 315, 86–94. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Chen, M.; He, S.; Fang, C.; Chen, M.; Li, D.; Wu, D.; Chernick, M.; Hinton, D.E.; Bo, J.; et al. Microplastics decrease the toxicity of triphenyl phosphate (TPhP) in the marine medaka (Oryzias melastigma) larvae. Sci. Total Environ. 2021, 763, 143040. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Chen, Y.; Shang, Y.; Cui, J.; Li, Z.; Li, Y. Knockout of zebrafish interleukin 7 receptor (IL7R) by the CRISPR/Cas9 system delays retinal neurodevelopment. Cell Death Dis. 2018, 9, 273. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Li, F.; Lu, M.; Ge, K.; Gan, L.; Shang, D. LIM Homeobox 9 knockdown by morpholino does not affect zebrafish retinal development. Biol. Open 2021, 10, bio056382. [Google Scholar] [CrossRef]
- Rinner, O.; Makhankov, Y.V.; Biehlmaier, O.; Neuhauss, S.C.F. Knockdown of cone-specific kinase GRK7 in larval zebrafish leads to impaired cone response recovery and delayed dark adaptation. Neuron 2005, 47, 231–242. [Google Scholar] [CrossRef]
- Chrispell, J.D.; Dong, E.; Osawa, S.; Liu, J.; Cameron, D.J.; Weiss, E.R. Grk1b and Grk7a both contribute to the recovery of the isolated cone photoresponse in larval zebrafish. Investig. Ophth. Vis. Sci. 2018, 59, 5116–5124. [Google Scholar] [CrossRef]
- Nikon, S.S.; Lyubarsky, A.; Fina, M.E.; Nikonova, E.S.; Sengupta, A.; Chinniah, C.; Ding, X.-J.; Smith, R.G.; Pugh Jr, E.N.; Vardi, N.; et al. Cones respond to light in the absence of transducin β subunit. J. Neurosci. 2013, 33, 5182–5194. [Google Scholar] [CrossRef]
- Berry, M.H.; Holt, A.; Salari, A.; Veit, J.; Visel, M.; Levitz, J.; Aghi, K.; Gaub, B.M.; Sivyer, B.; Flannery, J.G.; et al. Restoration of high-sensitivity and adapting vision with a cone opsin. Nat. Commun. 2019, 10, 1221. [Google Scholar] [CrossRef]
- Kanazawa, N.; Goto, M.; Harada, Y.; Takimoto, C.; Sasaki, Y.; Uchikawa, T.; Kamei, Y.; Matsuo, M.; Fukamachi, S. Changes in a cone opsin repertoire affect color-dependent social behavior in medaka but not behavioral photosensitivity. Front Genet. 2020, 11, 801. [Google Scholar] [CrossRef]
- Zheng, S.; Shao, F.; Tao, W.; Liu, Z.; Long, J.; Wang, X.; Zhang, S.; Zhao, Q.; Carleton, K.L.; Kocher, T.D.; et al. Chromosome-level assembly of southern catfish (Silurus meridionalis) provides insights into visual adaptation to nocturnal and benthic lifestyles. Mol. Ecol. Resour. 2021, 21, 1575–1592. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, D.A.; Krejca, J.K.; Martinez, J.M.R. Mexican blindcats genus Prietella (Siluriformes: Ictaluridae): An overview of recent explorations. Environ. Biol. Fish. 2001, 62, 315–337. [Google Scholar] [CrossRef]
- Schluessel, V.; Rick, I.P.; Seifert, F.T.; Baumann, C.; Davies, W.I.L. Not just shades of grey: Life is full of colour for the ocellate river stingray (Potamotrygon motoro). J. Exp. Biol. 2021, 224, 9. [Google Scholar] [CrossRef] [PubMed]
- Delroisse, J.; Duchatelet, L.; Flammang, P.; Mallefet, J. De novo transcriptome analyses provide insights into opsin-based photoreception in the lanternshark Etmopterus spinax. PLoS ONE 2018, 13, e0209767. [Google Scholar] [CrossRef]
- Fasick, J.I.; Algrain, H.; Serba, K.M.; Robinson, P.R. The retinal pigments of the whale shark (Rhincodon typus) and their role in visual foraging ecology. Vis. Neurosci. 2019, 36, E011. [Google Scholar] [CrossRef]
- Shi, Y.; Shi, Y.; Ji, W.; Li, X.; Shi, Z.; Hou, J.; Li, W.; Fu, Y. Thyroid Hormone Signaling Is Required for Dynamic Variation in Opsins in the Retina during Metamorphosis of the Japanese Flounder (Paralichthys olivaceus). Biology. 2023, 12, 397. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Poth, E.M.; Zhu, H.; Blackshaw, S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural. Dev. 2011, 6, 32. [Google Scholar] [CrossRef]
- Inbal, A.; Kim, S.; Shin, J.; Solnica-Krezel, L. Six3 represses nodal activity to establish early brain asymmetry in zebrafish. Neuron 2007, 55, 407–415. [Google Scholar] [CrossRef]
- Li, X.; Perissi, V.; Liu, F.; Rose, D.W.; Rosenfeld, M.G. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science 2002, 297, 1180–1183. [Google Scholar] [CrossRef]
- Chen, Y.; Slack, J. Identifying the precursor zone of muscle satellite cells in Xenopus laevis embryos. Dev. Biol. 2008, 319, 557–558. [Google Scholar] [CrossRef]
- Favor, J.; Gloeckner, C.J.; Neuhäuser-Klaus, A.; Pretsch, W.; Sandulache, R.; Saule, S.; Zaus, I. Relationship of Pax6 activity levels to the extent of eye development in the mouse, Mus musculus. Genet. 2008, 179, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Shiraki, T.; Kojima, D.; Fukada, Y. Homeobox transcription factor Six7 governs expression of green opsin genes in zebrafish. Proc. Biol. Sci. 2015, 282, 20150659. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Hong, N.; Hong, Y. Derivation and characterization of haploid embryonic stem cell cultures in medaka fish. Nat. Protoc. 2010, 5, 1418–1430. [Google Scholar] [CrossRef]
- Shimada, Y.; Hirano, M.; Nishimura, Y.; Tanaka, T. A high-throughput fluorescence-based assay system for appetite-regulating gene and drug screening. PLoS ONE 2012, 7, e52549. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Huang, C.; Wang, L.; Ye, X.; Bai, C.; Simonich, M.T.; Tanguay, R.L.; Dong, X. Toxicity, uptake kinetics and behavior assessment in zebrafish embryos following exposure to perfluorooctanesulphonicacid (PFOS). Aquat. Toxicol. 2010, 98, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).