Evolution and Expression of the Meprin and TRAF Homology Domain-Containing Gene Family in Solanaceae
Abstract
1. Introduction
2. Results
2.1. Identification of MATH Genes in Solanaceae Genomes
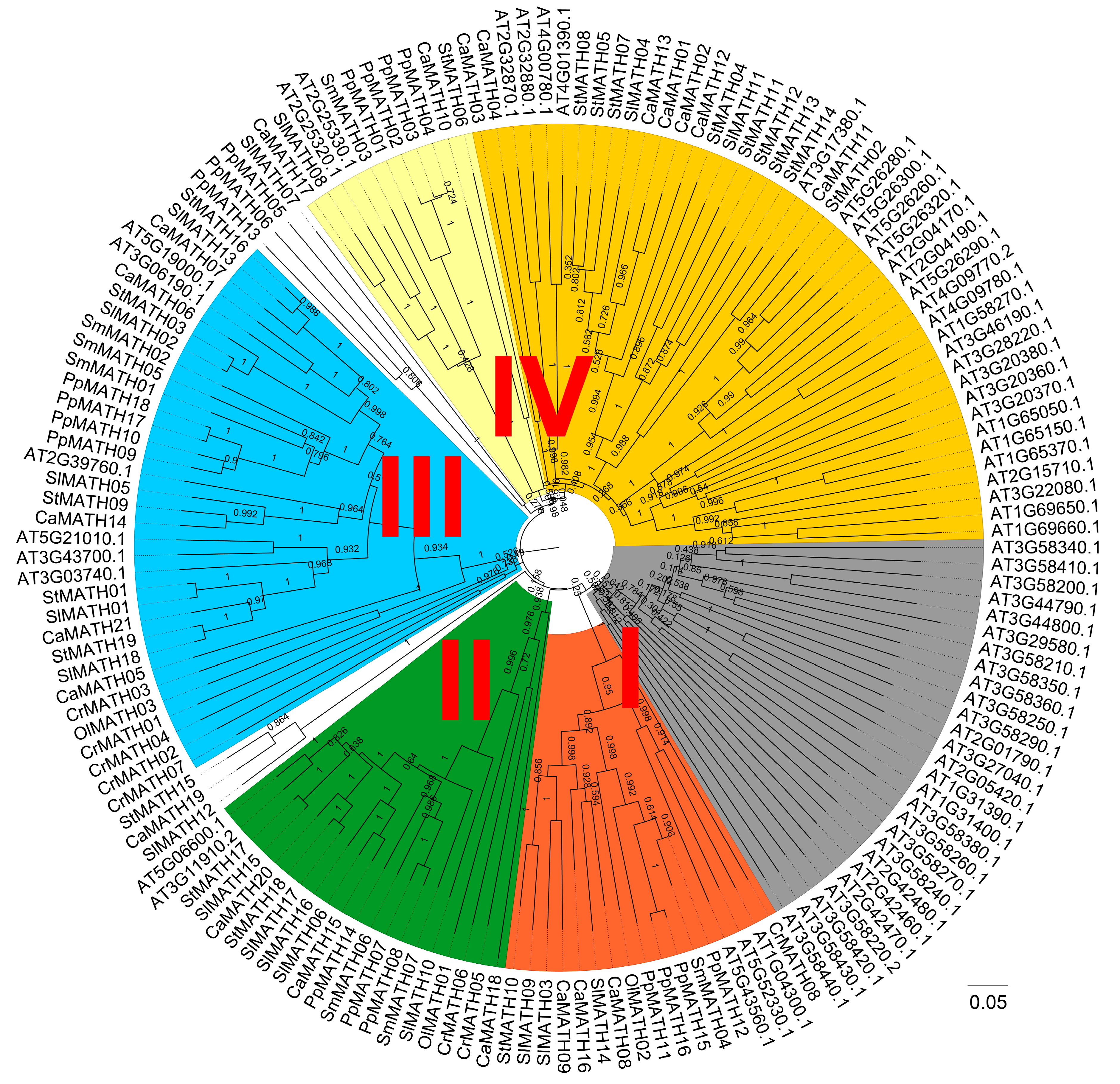
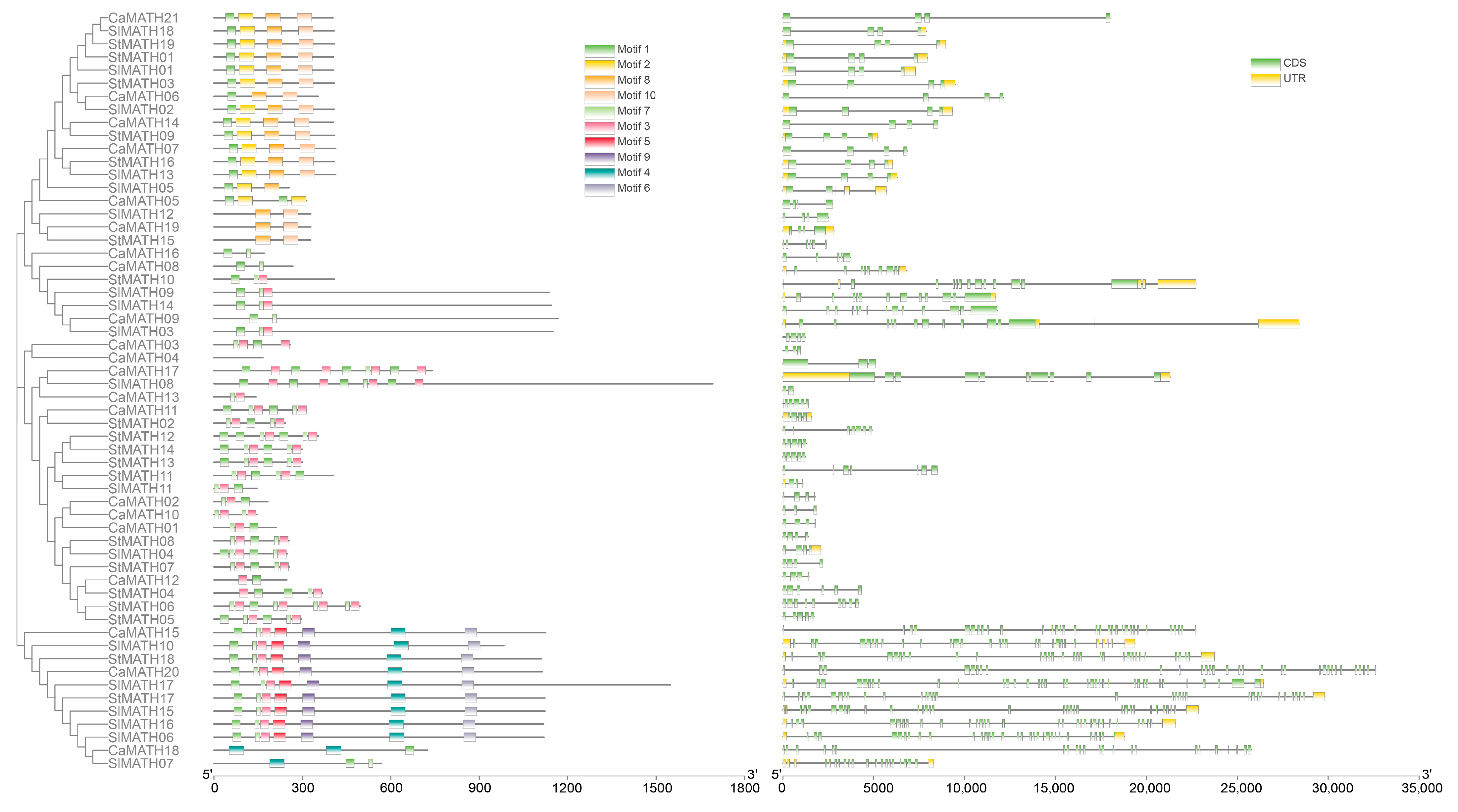
2.2. Phylogenetic Relationship, Conserved Motifs/Domains, and Exon–Intron Organization of Solanaceae MATHs
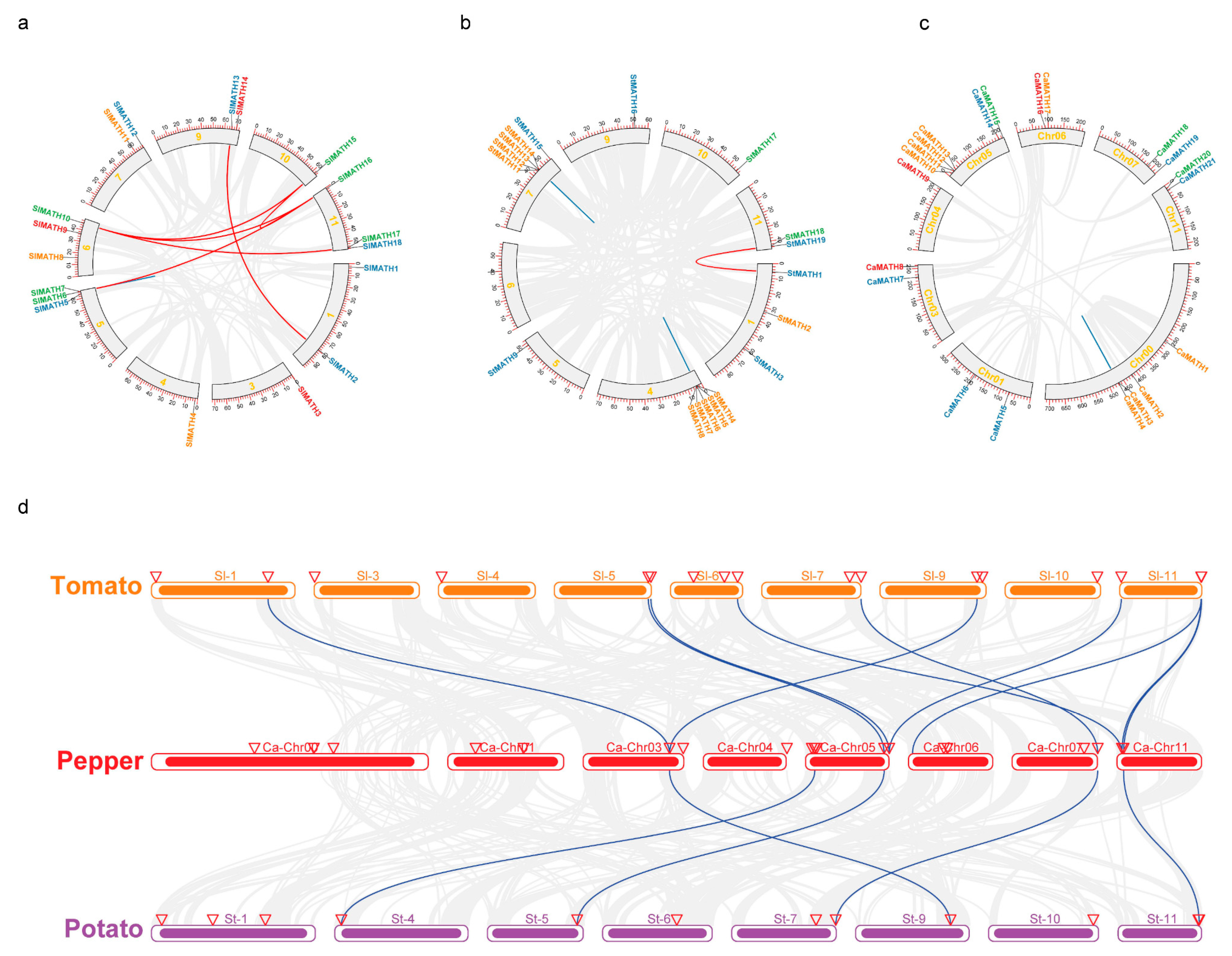
2.3. Gene Duplication and Synteny Analysis of the MATH Gene Family in Solanaceae
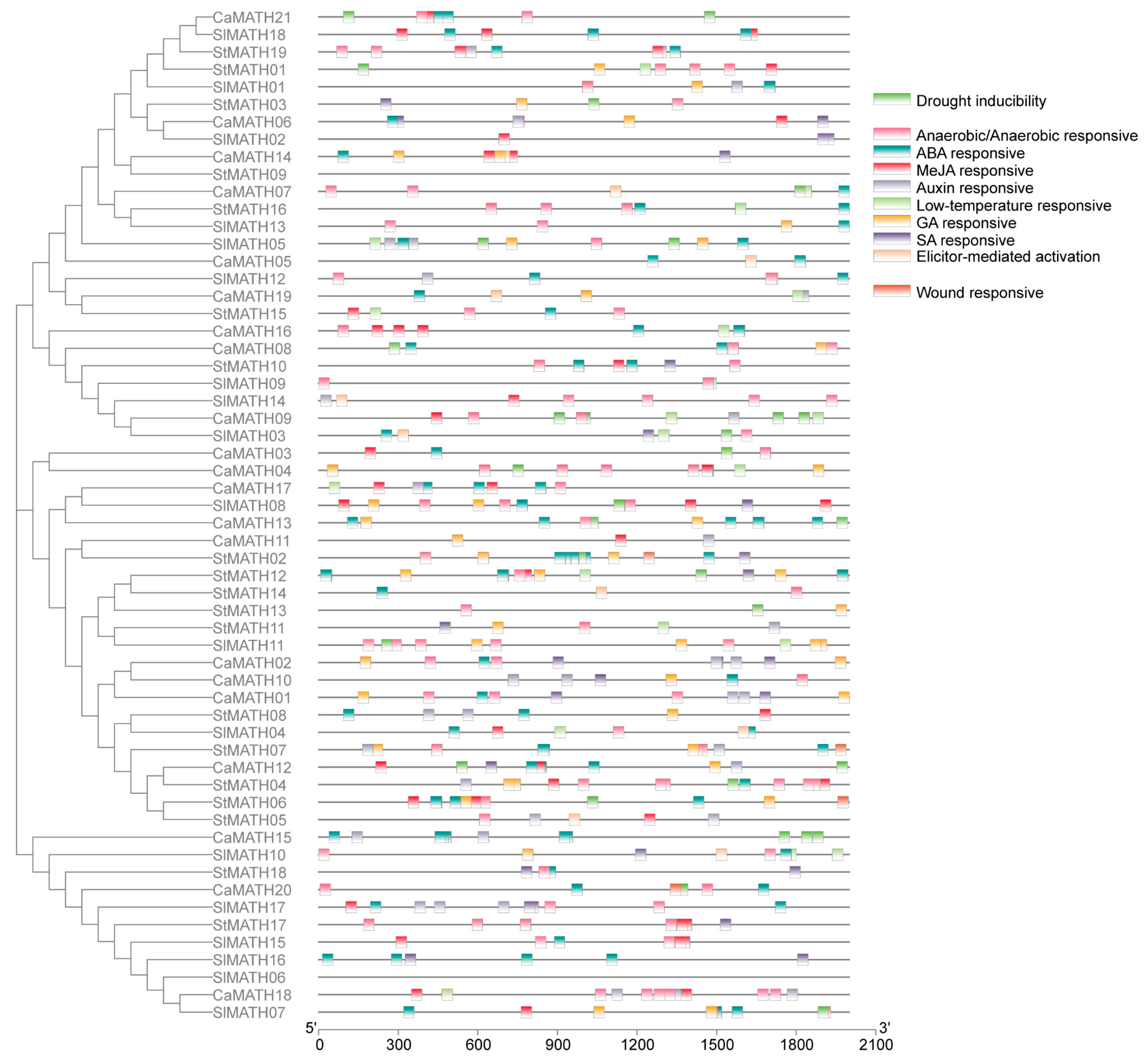
2.4. Cis-Regulatory Elements in MATH Promoters
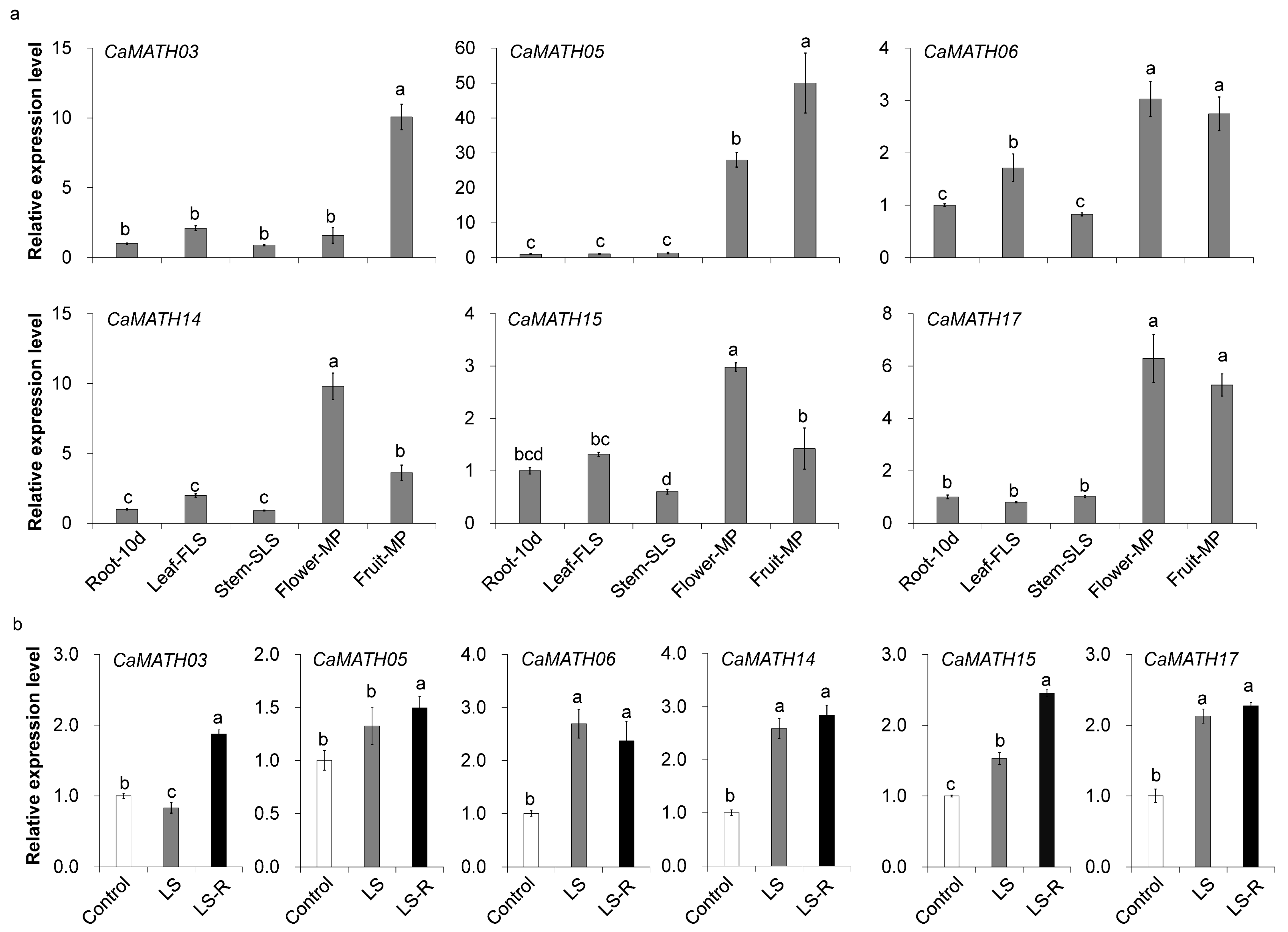
2.5. Tissue-Specific Expression Patterns of Solanaceae MATH Genes
2.6. Expression of MATH Genes in Solanaceae Plants in Response to Phytohormone Treatment and Various Stresses
2.7. Expression of Pepper MATH Genes during Development and in Response to Flooding Treatment
3. Discussion
3.1. Classification and Conserved Nature of MATH Gene Family in Solanaceae
3.2. Gene Duplications Contributed to MATH Gene Expansion in Solanaceae
3.3. MATH Predicted Functions and Gene Expression
4. Materials and Methods
4.1. Identification of MATH Genes
4.2. Protein Property Predictions
4.3. Phylogenetic Analysis
4.4. Duplication Events and Synteny Analysis
4.5. Analysis of Protein Motif and Domain Combinations, Gene Exon–Intron Architecture, and Promoter Cis-Elements in Solanaceae MATH Members
4.6. Analysis of Gene Expression during Different Developmental Stages and under Different Stress and Phytohormone Treatments
4.7. RNA Extraction and Quantitative RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rothe, M.; Wong, S.C.; Henzel, W.J.; Goeddel, D.V. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell 1994, 78, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, B.; Aleithan, F.; Abdul-Sater, Z.; Abdul-Sater, A.A. The Evolving Role of TRAFs in Mediating Inflammatory Responses. Front Immunol. 2019, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Park, H.H. Structure of TRAF family: Current understanding of receptor recognition. Front. Immunol. 2018, 9, 1999. [Google Scholar] [CrossRef]
- Sunnerhagen, M.; Pursglove, S.; Fladvad, M. The new MATH: Homology suggests shared binding surfaces in meprin tetramers and TRAF trimers. FEBS Lett. 2002, 530, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Zapata, J.M.; Martínez-García, V.; Lefebvre, S. Phylogeny of the TRAF/MATH domain. Adv. Exp. Med. Biol. 2007, 597, 1–24. [Google Scholar]
- Uren, A.G.; Vaux, D.L. TRAF proteins and meprins share a conserved domain. Trends Biochem. Sci. 1996, 21, 244–245. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Huang, Y.; Hu, Y.; He, X.; Shen, W.; Liu, C.; Ruan, Y. Phylogenetic Analysis of Brassica rapa MATH-Domain Proteins. Curr. Genom. 2013, 14, 214–223. [Google Scholar] [CrossRef]
- Qi, H.; Xia, F.N.; Xiao, S.; Li, J. TRAF proteins as key regulators of plant development and stress responses. J. Integr. Plant Biol. 2022, 64, 431–448. [Google Scholar] [CrossRef]
- Cosson, P.; Sofer, L.; Le, Q.H.; Léger, V.; Schurdi-Levraud, V.; Whitham, S.A.; Yamamoto, M.L.; Gopalan, S.; Le Gall, O.; Candresse, T.; et al. RTM3, which controls long-distance movement of potyviruses, is a member of a new plant gene family encoding a meprin and TRAF homology domain containing protein. Plant Physiol. 2010, 154, 222–232. [Google Scholar] [CrossRef]
- Kushwaha, H.R.; Joshi, R.; Pareek, A.; Singla-Pareek, S.L. MATH-Domain Family Shows Response toward Abiotic Stress in Arabidopsis and Rice. Front. Plant Sci. 2016, 7, 923. [Google Scholar] [CrossRef]
- Juranić, M.; Srilunchang, K.O.; Krohn, N.G.; Leljak-Levanic, D.; Sprunck, S.; Dresselhaus, T. Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell 2012, 24, 4974–4991. [Google Scholar] [CrossRef] [PubMed]
- Juranić, M.; Dresselhaus, T. Phylogenetic analysis of the expansion of the MATH-BTB gene family in the grasses. Plant Signal. Behav. 2014, 9, e28242. [Google Scholar] [CrossRef]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.J.; Shinde, B.; Cardenas, P.D.; Bocobza, S.E.; Unger, T.; Malitsky, S.; Finkers, R.; et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef]
- The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA. 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Huang, S.; Chen, X.; Zhong, X.; Li, M.; Ao, K.; Huang, J.; Li, X. Plant TRAF proteins regulate NLR immune receptor turnover. Cell Host Microbe 2016, 19, 204–215. [Google Scholar] [CrossRef]
- Qi, H.; Xia, F.N.; Xie, L.J.; Yu, L.J.; Chen, Q.F.; Zhuang, X.H.; Wang, Q.; Li, F.; Jiang, L.; Xie, Q.; et al. TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 2017, 29, 890–911. [Google Scholar] [CrossRef]
- Linden, K.J.; Callis, J. The ubiquitin system affects agronomic plant traits. J. Biol. Chem. 2020, 295, 13940–13955. [Google Scholar] [CrossRef]
- Norizuki, T.; Minamino, N.; Ueda, T. Role of Autophagy in Male Reproductive Processes in Land Plants. Front. Plant Sci. 2020, 11, 756. [Google Scholar] [CrossRef] [PubMed]
- March, E.; Farrona, S. Plant Deubiquitinases and Their Role in the Control of Gene Expression Through Modification of Histones. Front. Plant Sci. 2018, 8, 2274. [Google Scholar] [CrossRef] [PubMed]
- Vanhaeren, H.; Chen, Y.; Vermeersch, M.; De Milde, L.; De Vleeschhauwer, V.; Natran, A.; Persiau, G.; Eeckhout, D.; De Jaeger, G.; Gevaert, K.; et al. UBP12 and UBP13 negatively regulate the activity of the ubiquitin-dependent peptidases DA1, DAR1 and DAR2. eLife 2020, 9, e52276. [Google Scholar] [CrossRef] [PubMed]
- Skelly, M.J. The emerging roles of deubiquitinases in plant proteostasis. Essays Biochem. 2022, 66, 147–154. [Google Scholar]
- Lee, C.M.; Li, M.W.; Feke, A.; Liu, W.; Saffer, A.M.; Gendron, J.M. GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat. Commun. 2019, 10, 3750. [Google Scholar] [CrossRef]
- Ewan, R.; Pangestuti, R.; Thornber, S.; Craig, A.; Carr, C.; O'Donnell, L.; Zhang, C.; Sadanandom, A. Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 2011, 191, 92–106. [Google Scholar] [CrossRef]
- Park, S.H.; Jeong, J.S.; Seo, J.S.; Park, B.S.; Chua, N.H. Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol. 2019, 223, 1447–1460. [Google Scholar] [CrossRef]
- Sakuraba, Y. Molecular basis of nitrogen starvation-induced leaf senescence. Front. Plant Sci. 2022, 13, 1013304. [Google Scholar] [CrossRef]
- Jeong, J.S.; Jung, C.; Seo, J.S.; Kim, J.K.; Chua, N.H. The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 2017, 29, 1406–1424. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Zhou, L.; You, S.; Deng, H.; Chen, Y.; Alseekh, S.; Yuan, Y.; Fu, R.; Zhang, Z.; et al. MicroTom Metabolic Network: Rewiring Tomato Metabolic Regulatory Network throughout the Growth Cycle. Mol. Plant. 2020, 13, 1203–1218. [Google Scholar] [CrossRef]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant. 2017, 10, 1129–1132. [Google Scholar] [CrossRef]
- Gingerich, D.J.; Hanada, K.; Shiu, S.H.; Vierstra, R.D. Largescale, lineage-specific expansion of a brica-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 2007, 19, 2329–2348. [Google Scholar] [CrossRef] [PubMed]
- Kersey, P.J.; Allen, J.E.; Christensen, M.; Davis, P.; Falin, L.J.; Grabmueller, C.; Hughes, D.S.; Humphrey, J.; Kerhornou, A.; Khobova, J.; et al. Ensembl Genomes 2013: Scaling up access to genome-wide data. Nucleic Acids Res. 2014, 42, D546–D552. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Mistry, J.; Schuster-Böckler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, D247–D251. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Liu, D.; Chen, S.; Dai, Y.; Guo, W.; Zhang, X.; Wang, L.; Ma, S.; Xiao, M.; Qi, H.; et al. Evolution and Expression of the Membrane Attack Complex and Perforin Gene Family in the Poaceae. Int. J. Mol. Sci. 2020, 21, 5736. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Sigrist, C.J.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef]
- Maheshwari, S.; Brylinski, M. Predicting protein interface residues using easily accessible on-line resources. Brief Bioinform. 2015, 16, 1025–1034. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Schauser, L.; Wieloch, W.; Stougaard, J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 2005, 60, 229–237. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, L.; Liu, D.; Ma, S.; Dai, Y.; Zhang, X.; Wang, Y.; Hu, T.; Xiao, M.; Zhou, Y.; et al. Identification and Expression of the Multidrug and Toxic Compound Extrusion (MATE) Gene Family in Capsicum annuum and Solanum tuberosum. Plants 2020, 9, 1448. [Google Scholar] [CrossRef] [PubMed]
- Hartman, S.; Sasidharan, R.; Voesenek, L.A.C.J. The role of ethylene in metabolic acclimations to low oxygen. New Phytol. 2021, 229, 64–70. [Google Scholar] [CrossRef]
- Zhou, Y.; Tan, W.J.; Xie, L.J.; Qi, H.; Yang, Y.C.; Huang, L.P.; Lai, Y.X.; Tan, Y.F.; Zhou, D.M.; Yu, L.J.; et al. Polyunsaturated linolenoyl-CoA modulates ERF-VII-mediated hypoxia signaling in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 330–348. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.S.; Liu, D.; Guo, W.; Liu, Z.X.; Zhang, X.; Shi, L.L.; Zhou, D.M.; Wang, L.N.; Kang, K.; Wang, F.Z.; et al. Poaceae-specific β-1,3;1,4-D-glucans link jasmonate signalling to OsLecRK1-mediated defence response during rice-brown planthopper interactions. Plant Biotechnol. J. 2023. [Google Scholar] [CrossRef]
- Jeon, H.; Bae, J.; Hwang, S.H.; Whang, K.Y.; Lee, H.S.; Kim, H.; Kim, M.S. MRPrimerW2: An enhanced tool for rapid design of valid high-quality primers with multiple search modes for qPCR experiments. Nucleic Acids Res. 2019, 47, W614–W622. [Google Scholar] [CrossRef] [PubMed]


| Plant Species | Clade | Genome Size (Mb) | Total | One MATH | Two MATH | Three MATH | Four MATH | MATH-BTB | MATH-USP7 |
|---|---|---|---|---|---|---|---|---|---|
| Ostreococcus lucimarinus | Chlorophytes | 12.56 | 3 | 1 | - | - | - | 1 | 1 |
| Chlamydomonas reinhardtii | Chlorophytes | 120 | 8 | 3 | - | - | - | 4 | 1 |
| Physcomitrium patens | Bryophytes | 472 | 18 | 6 | 1 | - | 4 | 4 | 3 |
| Selaginella moellendorffii | Lycophytes | 100 | 7 | 1 | - | - | 1 | 3 | 2 |
| Arabidopsis thaliana | Eudicots | 135 | 67 | 32 | 25 | - | 2 | 6 | 2 |
| Solanum lycopersicum | Eudicots | 844 | 18 | 5 | 1 | - | 1 | 6 | 5 |
| Solanum tuberosum | Eudicots | 900 | 19 | 1 | 7 | 2 | 1 | 6 | 2 |
| Capsicum annuum-Zunla | Eudicots | 3260 | 21 | 6 | 5 | - | 1 | 6 | 3 |
| Total | 161 | 55 | 39 | 2 | 10 | 36 | 19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Y.; Ma, S.; Guo, Y.; Zhang, X.; Liu, D.; Gao, Y.; Zhai, C.; Chen, Q.; Xiao, S.; Zhang, Z.; et al. Evolution and Expression of the Meprin and TRAF Homology Domain-Containing Gene Family in Solanaceae. Int. J. Mol. Sci. 2023, 24, 8782. https://doi.org/10.3390/ijms24108782
Dai Y, Ma S, Guo Y, Zhang X, Liu D, Gao Y, Zhai C, Chen Q, Xiao S, Zhang Z, et al. Evolution and Expression of the Meprin and TRAF Homology Domain-Containing Gene Family in Solanaceae. International Journal of Molecular Sciences. 2023; 24(10):8782. https://doi.org/10.3390/ijms24108782
Chicago/Turabian StyleDai, Yangshuo, Sirui Ma, Yixian Guo, Xue Zhang, Di Liu, Yan Gao, Chendong Zhai, Qinfang Chen, Shi Xiao, Zhenfei Zhang, and et al. 2023. "Evolution and Expression of the Meprin and TRAF Homology Domain-Containing Gene Family in Solanaceae" International Journal of Molecular Sciences 24, no. 10: 8782. https://doi.org/10.3390/ijms24108782
APA StyleDai, Y., Ma, S., Guo, Y., Zhang, X., Liu, D., Gao, Y., Zhai, C., Chen, Q., Xiao, S., Zhang, Z., & Yu, L. (2023). Evolution and Expression of the Meprin and TRAF Homology Domain-Containing Gene Family in Solanaceae. International Journal of Molecular Sciences, 24(10), 8782. https://doi.org/10.3390/ijms24108782






