Abstract
Duchenne muscular dystrophy (DMD) is an X-linked progressive muscle degenerative disease caused by mutations in the dystrophin gene, resulting in death by the end of the third decade of life at the latest. A key aspect of the DMD clinical phenotype is dilated cardiomyopathy, affecting virtually all patients by the end of the second decade of life. Furthermore, despite respiratory complications still being the leading cause of death, with advancements in medical care in recent years, cardiac involvement has become an increasing cause of mortality. Over the years, extensive research has been conducted using different DMD animal models, including the mdx mouse. While these models present certain important similarities to human DMD patients, they also have some differences which pose a challenge to researchers. The development of somatic cell reprograming technology has enabled generation of human induced pluripotent stem cells (hiPSCs) which can be differentiated into different cell types. This technology provides a potentially endless pool of human cells for research. Furthermore, hiPSCs can be generated from patients, thus providing patient-specific cells and enabling research tailored to different mutations. DMD cardiac involvement has been shown in animal models to include changes in gene expression of different proteins, abnormal cellular Ca2+ handling, and other aberrations. To gain a better understanding of the disease mechanisms, it is imperative to validate these findings in human cells. Furthermore, with the recent advancements in gene-editing technology, hiPSCs provide a valuable platform for research and development of new therapies including the possibility of regenerative medicine. In this article, we review the DMD cardiac-related research performed so far using human hiPSCs-derived cardiomyocytes (hiPSC-CMs) carrying DMD mutations.
1. Duchenne Muscular Dystrophy
Duchenne muscular dystrophy (DMD) is an X-linked progressive muscle degenerative disease caused by mutations in the dystrophin gene with an estimated prevalence between 1.3 and 2.1 per 10,000 live male births [1,2,3]. Dystrophin is the longest gene in the human DNA spanning 2.4 Mbp, with its 14 kb transcript consisting of 79 exons; it encodes the 427 kDa dystrophin protein [4,5]. Dystrophin is a major structural protein which is also involved in important metabolic processes [6,7,8]. In skeletal and cardiac muscle, dystrophin provides mechanical stability essential for contracting myocytes, and anchors the cellular cytoskeleton to the extracellular matrix (ECM) via the transmembrane dystrophin–glycoprotein complex (DGC) which links directly to extracellular laminin-2 [9,10,11]. Dystrophin can be grossly divided into three major domains: (1) the N-terminus and actin-binding domain (ABD) (exons 1–8), (2) the central rod segment consisting of 24 spectrin repeats (exons 9–63), and (3) the DGC-binding domain and C-terminus (exons 64–79) [12,13]. The majority of DMD mutations are deletions of one or more exons (60–65%), while duplications make up to 5–10% of the cases. The remaining (25–35%) are single-nucleotide variants, small deletions or insertions in the coding sequence, or splice site variants [14,15].
DMD symptoms start at an early age, usually around 2–3 years, when proximal muscles of the lower extremities begin to weaken. Gradually, the weakness progresses to the distal muscles and the upper limbs. With age, symptoms become more prominent, and by the early teens, patients are usually wheelchair-dependent. The cardiac involvement of DMD includes dilated cardiomyopathy (DCM) which is present in virtually all patients by their late teen years, along with conduction abnormalities, various arrhythmias and extensive fibrosis. Eventually, patients die by their late 20s or early 30s due to respiratory and cardiac failure [16,17,18].
The current gold-standard treatment of DMD includes glucocorticoids (GCs), usually from the age of 4 years, aimed at improving motor and pulmonary function, while also potentially delaying the onset of DCM [16,19,20]. Beyond known side-effects including weight gain, hirsutism and other Cushing’s syndrome symptoms, GCs do not change the disease outcome, but rather can only slow its course [19,21]. Angiotensin-converting enzyme (ACE) inhibitors and angiotensin receptor blockers (ARBs) are administered from around the age of 10 years to reduce mechanical stress on the heart [16,17]. Novel therapies include Eteplirsen, an exon 51 skipping drug, and Ataluren (PTC124), which promotes ribosomal readthrough of nonsense mutations [22,23]. However, these treatments are not intended for all DMD mutations, and there is a need for additional research and therapies to be developed.
The primary animal model used for DMD research is the mdx mouse, which carries a nonsense point mutation in exon 23 of the dystrophin gene [24,25]. Although mdx mice exhibit chronic degeneration of myofibers, they do not manifest some prominent symptoms of DMD. mdx mice display a slower disease progression compared to human DMD patients, and their relative lifespan is significantly longer. The slow progression of muscle pathology does not lead to extensive fibrosis as in humans, and the mice retain their mobility. Cardiac involvement follows a different course than in humans, as mdx mice initially develop hypertrophic cardiomyopathy (HCM), while human DMD patients suffer from contractile dysfunction and DCM [26,27,28]. It has been previously found that mdx mice heart mitochondria display an increase in Ca2+ uptake rate via activation of Ca2+ transport, possibly compensating for a defective sarcoplasmic reticulum (SR) [29,30,31]. This adaptiveness of mdx mice may be a key feature differentiating this model from human patients [32]. Indeed, these challenges led to the development of the D2.mdx model which exhibits a significantly more prominent disease phenotype [33]. An additional limitation of the mouse model lies in gender differences between female and male mdx mice. These differences include a more prominent cardiac involvement and skeletal muscle degeneration in female compared to male mice [34,35], contrary to slower disease progression in human female carriers [36].
Another important animal model was developed in rats by means of TALENs targeting DMD exon 23 [37]. mdx rats manifest progressive muscle degeneration accompanied by a reduction in muscle force, as well as dilated cardiomyopathy. Importantly, mdx rats display significant fibrosis in skeletal and cardiac muscle, similar to human patients but contrary to mdx mice [37,38]. However, some differences remain between mdx rats and human patients, as well as cells derived from human patients, including the lack of muscle calcifications [37], normal L-type Ca2+ current (ICa,L) [39,40,41], and unimpaired β-adrenergic cascade [39,42,43,44] in mdx rats.
2. Human Induced Pluripotent Stem Cells
In 2006 Takahashi and Yamanaka published their successful attempt to reprogram differentiated rat somatic cells into induced pluripotent stem cells (iPSCs) by means of the induction of four factors: Oct3/4, SOX2, c-Myc, and KLF4 [45]; in 2007, these breakthroughs were repeated in human somatic cells [46]. Human iPSCs (hiPSCs) present classic embryonic stem-cell (ESC) characteristics including trilineage differentiation capability [47,48]. Thus, provided proper culture and media conditions, hiPSCs can be differentiated into various cell types. Like ESCs, hiPSCs and hiPSC-derived cells can be used for disease modeling and drug testing [49,50,51,52,53]. Furthermore, hiPSCs can generate a potentially endless pool of differentiated cells from a minute biopsy of a single living human donor, whereas ESC generation requires the sacrifices of embryos [54]. This enables previously unmatched research capabilities of various human diseases without the limitations of different animal models. Indeed, in the past years, numerous papers utilized the reprogramming technique for disease modeling and regenerative medicine. Patient-specific hiPSCs served as a means for many discoveries and advancements in research of different diseases [51,52,55,56,57].
Due to the multitude of different mutations causing DMD [58], patient-specific hiPSCs provide a valuable approach to investigate the precise disease mechanisms resulting from these mutations. Furthermore, hiPSC research enables the potential development of new drugs and therapeutic approaches targeting specific mutations with higher efficacy than previous generic treatments. Importantly, investigating human-derived cells is preferable to using animal models which display different disease course and characteristics, compared to human patients.
3. Gene Expression Changes in DMD hiPSC-CMs
DMD has been previously linked to changes in cellular gene expression in different models including mdx mice, as well as in human patients [59,60,61,62]. To support hiPSCs as a valid model for DMD modeling and gain better understanding of the disease mechanisms, it is also imperative to investigate gene expression changes in DMD hiPSC-CMs. Lin et al. [41] discovered that DMD hiPSC-CMs exhibit higher rates of cell death compared to healthy cells, demonstrated by increased staining for CASP3 and elevated levels of DNA fragmentation. Additionally, they found extensive changes in gene expression including important apoptosis regulators such as CASP3, CASP8, CASP9 and antiapoptotic XIAP, [63], genes involved in contractility such as MYL2, MYL3, ACTN1 and TPM1 [64], and genes associated with heart diseases such as MAPK11, COL3A1 and CALM1 [65,66,67]. Moreover, bio-functional enrichment analysis of the genes revealed that categories related to heart disease conditions were positively enriched, while others involved in muscle development and contractility were negatively enriched in DMD hiPSC-CMs. Importantly, these enrichment patterns are consistent with clinical observations in DMD patients [68,69,70,71]. By means of whole-transcriptome sequencing analysis, Lin and coworkers discovered that alongside elevated expression of CASP3 and DIABLO, XIAP was decreased in DMD hiPSC-CMs, indicating that possible mitochondrial involvement in DMD hiPSC-CMs increased apoptosis [63]. Accordingly, transmission electron microscopy (TEM) demonstrated swollen mitochondria in DMD hiPSC-CMs [41]. Furthermore, FACS analysis of DMD hiPSC-CMs using JC-1 mitochondrial membrane potential staining revealed an increase in damaged mitochondria in DMD hiPSC-CMs [41]. Overall, Lin et al.’s findings suggest that a common mitochondria-mediated signaling network is involved in elevated apoptosis in DMD hiPSC-CMs.
Chang and coauthors [72] investigated the involvement of telomere shortening in various cardiomyopathies including DMD. Quantitative fluorescence in situ hybridization (Q-FISH) and quantitative polymerase chain reaction (qPCR) demonstrated increased telomere shortening in DMD compared to healthy hiPSC-CMs. The telomere has been shown be involved in gene expression of endothelial cells, and overexpression of telomerase protein (TERT) in mouse cardiomyocytes was proven to protect from myocardial ischemia [73,74,75]. Interestingly, mdx mice, which do not express cardiac symptoms as extensive as human DMD patients, have substantially longer telomeres than human patients [76]. Indeed, genetically engineered mdx mice with shortened telomeres developed severe heart failure similar to humans. The same group later discovered [77] by means of qRT-PCR, decreased transcript levels of telomere repeat-binding proteins (TRF1 and TRF2) and shelterin complex proteins (RAP1, TIN2, and POT1) in DMD compared to healthy cardiomyocytes, indicating possible involvement of these absent proteins in telomere shortening. Additionally, this group also found upregulation of p53 and p53-binding protein 1 (53BP1), indicating activation of the DNA damage response, as also demonstrated by increased apoptotic markers caspase-3, cleaved PARP, and accumulation of β-galactosidase signal in DMD hiPSC-CMs compared to healthy cells. Lastly, blocking the contraction of cardiomyocytes with blebbistatin, which locks the myosin heads in a low-affinity state preventing actin binding [78], abolished telomere shortening. These findings highlight aberrant contraction as an important factor involved in telomere shortening and shelterin downregulation, eliciting a p53-dependent DNA damage response.
Farini et al. [79] investigated the involvement of immunoproteasome (IP) dysregulation in the cellular pathophysiology of DMD. The group reported an increase in the expression of IP subunits PSMB8 and PSMB9 in DMD compared to healthy cardiomyocytes. Accordingly, administration of ONX-0914, an IP inhibitor, decreased intracellular Ca2+ levels, as well as the release of cTnl, implying increased cell survival. Additionally, IP inhibition resulted in downregulation of TGF-β and type III collagen-α, suggesting reduced fibrosis [80]. Overall, these results demonstrate IP as a potential pharmacological target for DMD patients and its involvement in cardiac pathophysiology.
Our group [40] reported expression changes in genes encoding ion channels in DMD hiPSC-CMs, including HCN which encodes the channel responsible for the pacemaker funny current (If) during phase 4 of the cardiac action potential, as well as reduced If density compared to healthy cells. Additionally, we found in DMD cardiomyocytes, increased expression of the CACNA1C gene, which encodes the channel responsible for the ICa,L current, and we discovered that ICa,L density increased accordingly.
Kamdar and colleagues [44] reported increased expression of known fibrosis genes COL1A1, ADAMTS2, COL6A1 and THY1 in DMD cardiomyocytes compared to healthy cells, consistent with other reports [79]. Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of dysregulated genes found that DMD hiPSC-CMs demonstrated upregulation of genes associated with extracellular matrix (ECM) organization, cellular proliferation and fibrosis, as well as downregulation of genes associated with intracellular Ca2+ handing and contraction.
Jelinkova et al. [42] found an increase in K+ channel Kir2.1 expression, despite no difference in action potential parameters between DMD and healthy hiPSC-CMs, contrary to other studies [40]. Furthermore, immunostaining demonstrated increased levels of the ICa,L channel expression in DMD cardiomyocytes. However, contrary to our report [40], at the mRNA level there was no difference, indicating increased cellular localization rather than differences in transcription.
Our group also reported [43] changes in gene expression patterns in DMD hiPSC-CMs. RNA-seq analysis revealed 119 genes included in KEGG pathways for cardiac intracellular Ca2+ handling, contraction and adrenergic signaling. Western blot (WB) analysis of SERCA2 demonstrated overexpression in DMD compared to healthy cells, possibly a compensatory attempt for impaired intracellular Ca2+ handling. Furthermore, downregulation of the β1 adrenergic receptor (ADRβ1) and adenylate cyclase (AC) demonstrated by RNA-seq, combined with depleted SR Ca2+ stores, likely underlies the blunted β-adrenergic positive inotropic response of DMD cells.
Yasutake et al. [81] investigated the involvement of the yes-associated protein (YAP, an important transcriptional factor involved in cell proliferation, growth and regeneration) in DMD cardiomyopathy. Previous research found decreased YAP activity in dystrophic muscles, resulting in poor regeneration and damage [82]. Interestingly, in DMD cardiomyocytes, the YAP nuclear/cytoplasmic (N/C) ratio was decreased compared to healthy hiPSC-CMs, indicating a lower degree of nuclear localization and active transcription of YAP. Accordingly, DMD hiPSC-CMs exhibited lower proliferation rate (measured by Ki67 expression) compared to healthy cells, suggesting a novel target for further research and therapeutic development.
Bremner and coauthors [83] used hiPSC-CMs in the form of engineered heart tissues (EHTs) that provide a 3D physiological cell culture platform to faithfully mimic mature cardiac tissue structure [84,85,86]. Contrary to a 2D culture, comparison of DMD and healthy EHT cardiomyocytes demonstrated dysregulation of genes related to cardiac function, including many related to cardiac muscle development, action potential and contraction, as well as extracellular matrix organization in DMD cardiomyocytes. Gene Ontology (GO) analysis indicated a dysregulation of genes related to cardiac muscle development, contraction, membrane potential, intracellular Ca2+ handling, and extracellular matrix organization. In addition to supporting the notion of dysregulated genes in DMD cardiac pathophysiology, these results emphasize the important effect of culture structure on the maturity and gene expression patterns in hiPSC-CMs.
Marini and colleagues [87] utilized hiPSC-CMs to generate cardiac organoids (COs), 3D cellular structures possessing organotypic characteristics such as cytoarchitecture and tissue-specific physiological mechanisms [88,89,90]. Regarding changes in gene expression, Marini and coworkers found that DMD hiPSC-CMs expressed lower levels of α-actinin (ACTN2), pacemaker current channel HCN4 and troponin-related genes (TNNI1, TNNC1, and TNNI3) compared to healthy cells, consistent with other reports [40,41]. DMD cardiac organoids were larger in size than healthy organoids, consistent with cellular hypertrophy, a known hallmark of cardiomyopathy [91]. Contrary to healthy COs, DMD COs lost α, β, γ, and δ-sarcoglycan expression over time, likely due to a lack of connection to dystrophin. RT-qPCR demonstrated upregulation of genes related to cardiac contractility in DMD compared to healthy Cos, including ACTN1, IRX4, MYBPC3, MYL2, MYOM1, TNNC2 and TMP1. Marini et al. also found that ARCN1 and GORASP2, known endoplasmic reticulum (ER) stress markers, were increased in DMD cardiomyocytes compared to healthy cells, indicating a higher level of ER stress. Furthermore, immunofluorescence staining detected higher levels of NOX4, suggesting increased oxidative stress [92]. Histological examination revealed development of fibrotic-like structures in DMD COs, a finding also supported by upregulation of known fibrosis markers including COL1A2, COL3A1 and FN1. Additionally, this group discovered increased formation of adipose tissue in DMD COs by means of H&E staining, which was validated by BODIPY staining for lipid droplets and immunostaining of PDGFRα+, an adipocyte marker. These findings show that DMD COs exhibit fibrotic and adipogenic characteristics, resembling clinical features of DMD cardiomyopathy [93]. Importantly, the advantages of the 3D model over 2D hiPSC-CMs are further manifested by this group’s previous report [94], which demonstrated immaturity of dystrophin-associated protein complex in 2D structures lacking protein level expression of α-, γ-, and δ-sarcoglycan. In summary, this 3D CO model enables mimicking some of the pathological findings in DMD cardiomyopathy.
4. Abnormal Excitation–Contraction Coupling Machinery in DMD hiPSC-CMs
Due to the important structural role of dystrophin, it is not surprising that its absence has been linked to impaired intracellular Ca2+ homeostasis, resulting from extracellular influx [95,96]. Additionally, elevated cellular inflammatory mediators resulting from dystrophin absence promote an increase in inducible nitric oxide synthase (iNOS), which leads to destabilization of SR ryanodine receptors (RyRs), causing SR Ca2+ leak into the cytosol [97,98]. Barthélémy et al. [99] further validated the key role of defective RyRs in DMD pathophysiology by demonstrating that RyR stabilizers including dantrolene, as well as Rycals S107 and ARM210, improve exon skipping efficiency in hiPSC-derived myotubes. Another possible mechanism was reported by Uchimura and Sakurai [100] who found that inhibiting store-operated Ca2+ channel (SOC) components STIM1 and Orai1 prevented Ca2+ overload and accordingly improved contractility in DMD hiPSC-derived myotubes, thus pointing to the role of these channels in excess Ca2+ entry. Overall, excess cytosolic Ca2+ can activate various proapoptotic regulators leading to cell death [101,102,103,104]. Accordingly, several studies of mdx mice demonstrated altered Ca2+ handling [105,106,107,108,109]. Guan et al. [110] were the first to demonstrate abnormal Ca2+ handling in hiPSC-CMs generated from DMD urine-derived stem cells (USCs), which included a lower recovery rate of Ca2+ transients. Additionally, early mitochondrial permeability pore opening was demonstrated in DMD hiPSC-CMs, which may be attributed to Ca2+ overload [111].
Additional ion channel abnormalities in DMD cells include sodium and potassium channels. mdx cardiomyocytes demonstrated a reduction in NaV1.5 channel expression and subsequently lower inward Na+ current [112]. In addition, a reduction in the inward rectifier K+ current via the Kir2.1 channel was also observed in mdx mice, despite unchanged channel expression levels [113]. Interestingly, Jelinkova et al. [42] found increased Kir2.1 expression but no subsequent electrophysiological changes in DMD hiPSC-CMs. mdx mice cardiomyocytes also displayed a decrease in KATP current, IK,ATP [114,115] which is known to be associated with cardiomyopathy development [114,115]. Lastly, our group demonstrated reduced If current in DMD hiPSC-CMs despite normal HCN channel expression [40].
Contrary to enhanced ICa,L reported in mdx mice [106], Lin and et al. [41] found decreased ICa,L in DMD hiPSC-CMs. Using Ca2+ imaging, this group reported a high resting cytosolic Ca2+concentration. Moreover, treating DMD hiPSC-CMs with Poloxamer P188, a membrane sealant, decreased cytosolic Ca2+ and repressed CASP3 activation and apoptosis. These findings suggest that improving cell membrane integrity may be a beneficial strategy for prevention of cardiomyocyte loss in DMD patients.
Caluori and coauthors [116] used a system consisting of a microelectrode array (MEA) and simultaneous probing by a cantilever from an atomic force microscope (AFM) to couple electrical and mechanical recordings. In response to progressively increasing Ca2+ concentrations, DMD hiPSC-CMs demonstrated a lower proportional decrease in spontaneous beat rate compared to healthy CMs, which may be attributed to pre-existing Ca2+ stress impairing additional inward flux [117]. Introduction of verapamil, an ICa,L channel blocker, also yielded a lower proportional decrease in spontaneous beat rate compared to healthy cells.
Our group [40] reported in DMD hiPSC-CMs the presence of arrhythmias represented by delayed afterdepolarizations (DADs) and irregular spontaneous firing patterns (Figure 1). DADs are usually attributed to Ca2+ overload [118], and this finding supports this notion. Additionally, we found increased ICa,L density consistent with previous reports in mdx mice [106]. Accordingly, we discovered an elevated expression of the CACNA1C gene, which encodes the ion channel responsible for the ICa,L. Furthermore, we found a prolongation of action potential duration (APD) stemming mainly from phase 2 of the cardiac action potential, likely due to the increased ICa,L density [119]. Additionally, increased ICa,L density may be involved in the decreased response to verapamil previously reported [116].
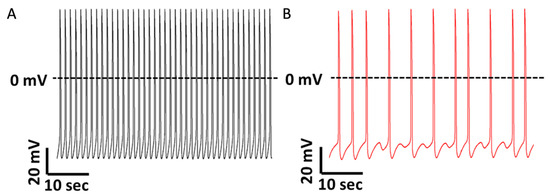
Figure 1.
Representative spontaneous action potential recordings from healthy and DMD iPSC-CMs. (A) iPSC-CMs generated from a healthy donor do not manifest arrhythmias during regular spontaneous activity. (B) iPSC-CMs generated from a DMD patient carrying a deletion of dystrophin exons 8–12 display delayed afterdepolarizations (DADs) and irregular firing pattern (modified with permission from Eisen et al., 2019 [40]).
Tsurumi et al. [120] found that diastolic and systolic Ca2+ concentrations measured by indo-1 fluorescence were elevated in DMD compared to healthy cardiomyocytes. Under mechanical stretching, diastolic and systolic Ca2+ concentrations were further increased in DMD cardiomyocytes but not in healthy cells. The elevation in intracellular Ca2+ concentrations, especially during diastole, correlates with diastolic dysfunction in DMD patients [121] and emphasizes the aberrant Ca2+ handling in DMD cardiomyocytes.
Pioner and colleagues [122] investigated mechanical and structural abnormalities in DMD hiPSC-CMs and found that they exhibited hypertrophy manifested by increased cellular length, diameter and cross-sectional area compared to healthy cardiomyocytes. Ultrastructurally, DMD hiPSC-CMs displayed sarcomeric changes including shortening of A- and Z-bands, increased percentage of identifiable I-bands, and overall greater variability in structural organization, suggesting underdeveloped sarcomere ultrastructure, consistent with previous reports of decreased transcription of sarcomeric genes [41] and reduced actin turnover [123]. These authors also demonstrated that in response to extracellular Ca2+ administration DMD hiPSC-CMs displayed reduced mechanical tension and prolonged relaxation. Furthermore, pCa50 was greater in DMD hiPSC-CMs, indicating increased Ca2+ sensitivity of tension generation, likely a compensatory mechanism aimed at increasing contractility in defective cells. Paced DMD hiPSC-CMs demonstrated a slower rate of contraction and relaxation compared to healthy cardiomyocytes. Post-resting period stimulation of DMD hiPSC-CMs yielded decreased twitch amplitude compared to healthy cells, suggesting a defective contractile reserve, likely due to impaired Ca2+ handling. Measurement of Ca2+ transients demonstrated low rates of rise and decay in DMD hiPSC-CMs, further supporting the notion of abnormal cellular Ca2+ handling. Lastly, similar to our published results [40], the spontaneous beat rate was lower in DMD compared to healthy hiPSC-CMs. Taken together, these results demonstrate the defective Ca2+ handling and contraction machinery in DMD hiPSC-CMs.
Kamdar and coauthors [44] tested β-adrenergic cascade characteristics in DMD hiPSC-CMs and reported an increased rate of baseline arrhythmogenic Ca2+ transients compared to healthy CMs. Under β-adrenergic stress introduced by isoproterenol, this group observed an increase in the frequency of arrhythmogenic Ca2+ traces, which were reduced by the β-adrenergic receptor blocker propranolol. Additionally, Kamdar et al. found a downregulation of genes associated with cardiac contraction and Ca2+ homeostasis. Overall, their results suggest an impaired β-adrenergic response in DMD cardiomyocytes, possibly correlated with autonomic dysfunction, known to affect DMD patients [124].
Jelinkova et al. [42] investigated molecular aspects of the excitation–contraction coupling (ECC) machinery and autonomic response in DMD hiPSC-CMs. Firstly, they found that compared to healthy cells, DMD hiPSCs presented lower differentiation efficiency to cardiomyocytes, manifested by a decreased fraction of spontaneously beating cells. Regarding Ca2+ transients, lower rates of rise and decay, as well as increased transient duration, were observed in DMD cardiomyocytes. These data support the notion that dystrophin deficiency results in ECC disturbances via Ca2+ handling abnormalities. They also found increased prevalence of delayed small-amplitude Ca2+ release events in DMD cells, which may correlate with the arrhythmogenic DADs we reported in DMD cardiomyocytes [40]. Other groups also found a weaker contraction force measured by AFM [125,126,127] in DMD cardiomyocytes. Contrary to our report, Jelinkova et al. found no difference in the spontaneous beat rate. However, similar to our findings, they found increased beat rate variability (BRV) measures in DMD cardiomyocytes compared to healthy cells [40]. DMD cardiomyocytes also displayed an abnormal response to isoproterenol and metoprolol, strengthening the notion of an abnormal β-adrenergic cascade in DMD cells. Importantly, these investigators found increased expression of β1- and β2-adrenergic receptors in DMD cardiomyocytes, likely a compensatory attempt of the defective cells.
Our group [43] investigated the β-adrenergic responsiveness and intracellular Ca2+ handling in DMD hiPSC-CMs. Under isoproterenol-induced β-adrenergic stimulation, DMD cardiomyocytes displayed a blunted positive inotropic response including decreased Ca2+ transient parameters such as amplitude, activation and relaxation rates, accompanied by corresponding changes in contraction parameters. Additionally, DMD cardiomyocytes did not exhibit a depressed chronotropic response to isoproterenol, suggesting the mechanism underlying the blunted inotropic response is not an impaired β-adrenergic cascade. In response to increasing extracellular Ca2+ concentration, known to induce augmented SR Ca2+ release and positive inotropy, DMD cells displayed smaller Ca2+ and contraction parameters compared to healthy cardiomyocytes. Lastly, we tested the effect of caffeine, which induces SR Ca2+ release, the common pathway of β-adrenergic stimulation and Ca2+ administration inducing positive inotropy. DMD cardiomyocytes displayed a blunted response to caffeine, including smaller Ca2+ amplitude and a shorter recovery time compared to healthy cells. In support of the notion of depleted SR Ca2+ stores in DMD cardiomyocytes, administration of ryanodine and cyclopiazonic acid (CPA), a SERCA inhibitor, resulted in a smaller inhibitory effect compared to healthy cells represented by a smaller relative decrease in Ca2+ amplitude. These results, combined with the reported changes in expression of genes related to Ca2+ handling, SERCA2, ADRβ1 and AC, point to abnormal Ca2+ handling as a key factor underlying abnormal β-adrenergic response in DMD.
YAP activity is known to increase due to mechano-transduction converting physical to electrochemical stimulus, regulated by actin dynamics [128,129]. Therefore, Yasutake et al. [81] assessed the actin state to determine its possible involvement in altered YAP activity in DMD hiPSC-CMs. Indeed, DMD cardiomyocytes were found to be initially smaller and rounder than healthy CMs and had a lower number of nuclei, in addition to failing to morphologically change with maturation as healthy cardiomyocytes, supporting the notion that DMD cardiomyocytes fail to progress morphologically in a proper manner. Immunofluorescence staining demonstrated a disrupted actin structure in DMD hiPSC-CMs as well, indicating a possible link between abnormal actin and YAP activity in DMD cardiomyocytes.
Chang and colleagues [77] devised a unique bioengineered traction force microscopy platform which enables mimicking stiffness of fibrotic heart tissue, a key feature of DMD cardiomyopathy, as well as controlling cardiomyocyte orientation. DMD hiPSC-CMs displayed increased resting [Ca2+], decreased Ca2+ transient amplitude, increased transient duration and increased transient decay rate, measured by Fura 2 fluorescence. These results demonstrate that impaired Ca2+ handling is involved in DMD pathophysiology at the tissue level.
Bremner and coauthors [83] measured contractile force in electrically stimulated hiPSC-CMs 3D EHT to mimic mature cardiac tissue structure [84,85,86]. DMD cardiomyocytes generated decreased twitch force and contraction kinetic parameters compared to healthy cardiomyocytes, implying lower contractile performance. Staining of Z-disc and α-actinin followed by confocal imaging revealed that DMD cardiomyocytes had shorter sarcomere length compared to healthy cardiomyocytes, likely contributing to the lower contractile force. Using Fura-2 fluorescence, it was found that DMD cardiomyocytes displayed a higher baseline of cytosolic [Ca2+] and increased transient peak, but overall decreased amplitude compared to healthy cells. DMD cells also exhibited slower rates of rise and decay of Ca2+ transients. These results emphasize once more abnormal Ca2+ handling as a key pathophysiological feature of DMD. Consistent with our previous report [40], DMD EHTs displayed increased BRV compared to healthy cells, strengthening the 3D structure as a valid important model for investigating DMD cardiomyopathy.
Pioner et al. [130] investigated in DMD hiPSC-CMs the maturation of the Ca2+ handling machinery and adaptation to changes in substrate stiffness. Analysis of Ca2+ transients revealed that DMD cardiomyocytes exhibited an initial smaller Ca2+ transient amplitude and a smaller increase in the transient amplitude with maturation compared to healthy cardiomyocytes. Additionally, post-rest contraction potentiation was lower in DMD compared to healthy cardiomyocytes, indicating lower SR calcium storage and release. Ca2+/calmodulin-dependent protein kinase II (CaMKII) levels were higher in DMD compared to healthy cardiomyocytes; consequently, the RyR2 CaMKII phosphorylation site S2814, was more phosphorylated. This increased phosphorylation may have accounted for the SR Ca2+ leak, resulting in the reduced Ca2+ transient amplitude in DMD cardiomyocytes. Compared to healthy cells, DMD hiPSC-CMs exhibited reduced Ca2+ transient amplitude on both soft and stiff substrates. Furthermore, while healthy cardiomyocytes displayed an accelerated decay of Ca2+ transient amplitude when cultured on a stiffer substrate, DMD cells did not differ in their kinetics between the substrates, indicating failure to adapt normally to differences in tissue stiffness. These findings demonstrate the abnormal Ca2+ handling of DMD cardiomyocytes and specifically the inability to adapt their Ca2+ homeostasis in response to changes in ECM stiffness, which may be a key factor of the DMD cardiac pathophysiology.
5. Alterations in Cellular Energy and Metabolism in DMD hiPSC-CMs
Emerging evidence from recent years suggests that metabolic and mitochondrial dysfunction play prominent roles in DMD pathophysiology [131,132,133,134,135,136,137]. Afzal et al. [138] tested the effects of nicorandil, a known NO donor, KATP channel opener and an antioxidant [139,140,141] on DMD hiPSC-CMs. The group found that in DMD hiPSC-CMs, nicorandil reduced H2O2-induced stress related LDH release and cell death to levels similar to healthy cells, through a mechanism involving the NO–cGMP pathway and KATP channel opening. Nicorandil also decreased ROS levels and maintained mitochondrial membrane integrity following oxidative stress, an effect mediated via increase of SOD2 expression which converts superoxide to hydrogen peroxide. Lastly, nicorandil was also shown to decrease cytosolic hydrogen peroxide production in DMD hiPSC-CMs. Although further research is required, these findings provide evidence that nicorandil may have a potential to protect against stress-induced cardiac involvement in DMD.
Gartz and colleagues [142] investigated cardioprotective effects of exosomes on DMD cardiomyocytes. They found that exosomes collected from conditioned media of both DMD and healthy hiPSC-CMs decreased injury-induced ROS levels in DMD cells. Additionally, exosomes led to inhibition of Bax expression and mitochondrial translocation, which are known to trigger opening of the mitochondrial permeability transition pore (mPTP), resulting in caspase activation and cell death [143]. Furthermore, exosomes were found to reduce stress-induced loss of mitochondrial membrane integrity and early mPTP opening time in DMD cardiomyocytes, as well as decreasing caspase 3/7 levels and subsequent apoptosis. By pretreating exosomes with trypsin, the researchers discovered that surface proteins are required to exert the protective effect. Treated exosomes failed to initiate ERK1/2 phosphorylation, an important component of the mitogen-activated protein kinase (MAPK)-mediated response which is responsible for dictating cell death or survival [144]. Additionally, inhibition of p38 MAPK reversed exosome-protective effects, indicating its specific involvement in cardio-protection. These results demonstrate the protective potential of cardiomyocyte-secreted exosomes in DMD cardiomyocytes, specifically the involvement of surface proteins ERK1/2 and p38 MAPK. Gartz and coauthors [145] further investigated the involvement of secreted exosomes in the pathological response of DMD hiPSC-CMs to metabolic stress. Exosomes, known to play an important role in intercellular communication and signaling [142,146,147,148], were studied in DMD [146,147,148,149,150] and found to be involved in modulation of inflammatory processes, oxidative injury, mitochondrial respiration, myocyte differentiation and cell death. In this study, the researchers identified several exosomal micro RNAs (exomiRs) dysregulated in DMD. Specifically, mir-339-5p was found to be upregulated in DMD exosomes and DMD hiPSC-CMs. The mir-339-5p level was also found to increase with cardiac injury and age in mdx mice, rendering it a potential cardiomyopathy-sensitive target in DMD. Interestingly, in DMD hiPSC-CMs, miR-339-5p-containing exosomes were found to be involved in stress-related mitochondrial-dependent cell death. Specifically, miR-339-5p, known to directly bind to MDM2, a p53 regulator leading to its downregulation [151], was also found to decrease MDM2 levels in DMD hiPSC-CMs. MDM2 was then reported to be downregulated in stressed DMD hiPSC-CMs. Lastly, miR-339-5p knockdown in DMD cardiomyocytes led to the preservation of mitochondrial membrane potential and reduction in stress-induced cell death, indicating its important role in modulating the cellular response to stress in DMD hiPSC-CMs. Collectively, these findings emphasize the importance of exosomal miR-339-5p involvement in cellular stress response, which may also serve as a basis for future research as a therapeutic target.
Duelen and coauthors [152] investigated the involvement of NOX4 in oxidative stress in DMD hiPSC-CMs. NOX enzymes, known to generate reactive oxygen species (ROS) in various cellular processes [153], are also involved in cardiovascular diseases [154,155]. On the basis of this knowledge, these researchers targeted NOX4 (mainly expressed in cardiomyocytes) in DMD hiPSC-CMs. Contrary to our report and that in mdx mice [40,106], DMD cardiomyocytes displayed a reduction in ICa,L density, although, similarly to our findings, they observed arrhythmogenic firing patterns including DADs and oscillatory prepotentials (OPPs). Another finding in line with ours was APD prolongation in DMD cells. Cell death, measured by flow cytometric analyses using annexin V and 7-amino-actinomycin D (7AAD), revealed that DMD cardiomyocytes underwent accelerated cell death compared to healthy cells. Furthermore, DMD cells had a higher ROS content compared to healthy cardiomyocytes. Treatment with N-acetyl-L-cysteine (NAC), ataluren (PTC124) and idebenone increased DMD cell survival and reduced ROS levels. By means of JC-1, a fluorescent voltage-sensitive dye with membrane-permeant fluorescent lipophilic cationic properties, Lin and coworkers measured mitochondrial membrane potential. Similar to Lin et al. [41], DMD hiPSC-CMs displayed increased levels of mitochondrial depolarization, which was improved by treatment with NAC, ataluren, and idebenone. Consistent with previous reports of increased NOX4 expression in heart failure [154,155,156,157], NOX4 was also upregulated in DMD hiPSC-CMs. Treatment with PTC124, NAC and idebenone reduced NOX4 expression. To assess the role of NOX4 in increased ROS levels, antisense locked nucleic acid (LNA) GapmeRs was used to degrade NOX4 mRNA. Indeed, NOX4 degradation led to reduced NADPH-dependent ROS production. DMD cardiomyocytes displayed increased NOX4 levels, and following addition of idebenone, ROS production was reduced. Furthermore, addition of ATP, a NOX4 regulator, and idebenone treatment also reduced ROS production. Lastly, contractile function assessed in 3D EHT constructs showed that DMD cardiomyocytes displayed lower contraction amplitude compared to healthy cells; this decrease was rescued by PTC124, NAC, and idebenone.
Our group investigated various metabolic and bioenergetic aberrations in DMD cardiomyocytes [158]. We first focused on polar metabolites (such as phospho-sugars, organic acids, nucleotides, and fatty acids) by means of liquid chromatography/mass spectrometry (LC–MS)-based metabolomics to study the central carbon metabolism of DMD cardiomyocytes. Indeed, nine metabolites differed in DMD cardiomyocytes compared to healthy cells, pointing primarily toward impairment in fatty acid oxidation. Next, we used labeled glucose tracing to study cellular glucose-derived metabolites. Indeed, DMD cardiomyocytes exhibited lower levels of glucose-derived carbon into the TCA cycle, indicating decreased mitochondrial glucose oxidation. Seahorse XFe96 flux analyzer was used to assess mitochondrial respiration and mitochondrial oxidative phosphorylation by measuring the oxygen consumption rate (OCR). DMD cardiomyocytes displayed decreased basal respiration and ATP production coupled to oxygen consumption, compared to healthy CMs. Interestingly, uncoupling ATP synthesis from the mitochondrial respiratory chain restored respiration rate, suggesting defective mitochondrial ATP synthase (Complex V). To examine mitochondrial number and morphology, electron microscopy (EM) analysis was performed. DMD cells contained a higher number of mitochondria compared to healthy CMs. Additionally, a higher frequency of aberrations, including increased size, disrupted cristae and multiple focal swelling areas, were seen in DMD cardiomyocytes. To further investigate the association between morphological abnormalities and impaired mitochondrial function, we measured mitochondrial activity by means of live confocal 3D imaging. Florescent staining revealed that mitochondrial activity was attenuated compared to healthy cells. Taken together, these findings suggest that bioenergetic and metabolic impairments are involved in DMD cardiac pathophysiology and should be further investigated in search of novel therapeutic targets.
6. Gene Therapy and Gene Editing in DMD hiPSC-CMs
Current treatments for DMD can at best slow the disease course and alleviate some symptoms [19,159,160]. However, these treatments do not fundamentally change the outcome, and patients rarely live beyond their late 20 s [18,161]. Furthermore, since around one-third of DMD cases originate from de novo mutations with no familial background [162,163], preconception genetic testing is expected to miss a significant proportion of cases. Hence, to truly cure DMD, genetic correction is needed.
Different methods can be used to attempt genetic repair (Figure 2). Early efforts to genetically correct DMD hiPSCs were conducted using gene therapy. Kazuki et al. [164] demonstrated the successful delivery of a human artificial chromosome (HAC) containing the complete dystrophin gene. Zatti and coauthors [165] successfully delivered an HAC containing WT dystrophin into DMD hiPSC-CMs; these authors found that differentiated cardiomyocytes expressed all dystrophin isoforms and displayed corrected protein localization, in addition to normal Ca2+ transients. Choi and colleagues [166] also reported correction of abnormal dystrophin gene expression profiles upon HAC delivery into DMD cardiomyocytes. Dick and coauthors [167] chose minigene delivery and exon-skipping to restore dystrophin expression in DMD cardiomyocytes. Additionally, Howard et al. [168] found that micro-dystrophin transgene containing the main functional domains was sufficient to preserve cardiac function and prevent fibrosis and inflammation in a DMD mouse model. While these techniques are useful tools for research, clinical application is not easily feasible as delivered HACs and minigenes are only transiently expressed in host cells [169]. In addition, delivery of such large complexes is a difficult challenge and carries a risk of nonspecific integration to the host DNA [170,171].
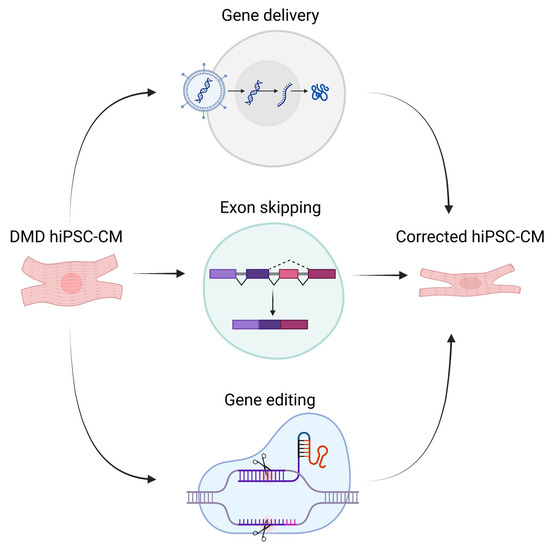
Figure 2.
Illustration of different genetic therapeutic methods for DMD (created with BioRender.com).
Another approach to correct DMD mutations is by editing the mutated gene directly. Gene editing requires a break in the DNA sequence at a specific position, followed by repair, which includes replacement of excised DNA with a new sequence. Cellular DNA repair mechanisms include two options [172,173,174]. The first is homology-directed repair (HDR), which is based on a new sequence introduced to repair double-strand breaks (DSBs). This enables precise correction of mutations in the DNA sequence. The DNA template used for repair can be coupled and delivered together with an endonuclease which performs the DSBs. Delivering a template to be utilized by HDR is the preferred mechanism for correction of small genetic mutations which require an insertion of a relatively small new sequence [175]. The second option is nonhomologous end-joining (NHEJ), which joins two DNA break ends, but at the cost of frequent small insertions/deletions (indels), potentially disrupting the open reading frame (ORF) and subsequently leading to RNA degradation through the nonsense-mediated decay (NMD) mechanism or production of a nonfunctional protein. NHEJ is useful to correct large frame-shifting deletions or duplications, by means of deleting additional base pairs to achieve ORF restoration or excising the duplicated segment to accomplish complete correction, respectively [175,176].
To achieve safeness and effectiveness, the process of introducing an exogenous endonuclease alongside a DNA template must be highly precise with minimal off-target effects. To date, three main systems are in use for preforming DNA DSBs and editing:
- Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) endonuclease comprises an anti-bacteriophage bacterial system [177]. CRISPR are bacterial DNA segments used to store antibacterial genetic information. Upon infection with a virus containing RNA homologous to RNA transcribed from the CRISPR, effector proteins can be guided by the latter to cleave the virus. Cleavage is performed by endonucleases such as Cas9, Cpf1, and Cas12. The guiding RNA sequence (gRNA) can be edited and, together with the endonuclease, delivered into cells using different types of vectors [178,179].
- Zinc finger nucleases (ZFNs) are a group of enzymes capable of cleaving specific sites in DNA. ZFNs consist of DNA-binding zinc finger arrays and a FokI-based endonuclease [180]. Despite high specificity of ZFN binding, the process of producing precise zinc fingers for specific DNA segments is considered relatively difficult and expensive [181].
- Transcription activator-like effector nucleases (TALENs) are based on proteins produced by Xanthomonas bacteria which are capable of plant gene expression alteration [182,183]. Like ZFNs, TALENs production is difficult and expensive compared to CRISPR and its associated endonucleases [184,185].
Off-target effects are a major concern when considering the clinical application of gene editing. These include possible disruption of oncogenes, tumor suppression genes and DNA repair genes [176,186,187]. An additional issue is the possibility of an immune response against elements of the delivered DNA-binding and endonuclease complex, or even against corrected gene products [176,188]. Despite these concerns, it is widely agreed among researchers and clinicians that genome editing carries the potential to cure genetic diseases previously considered incurable.
To develop an approach which potentially benefits a larger target population, Young et al. [189] utilized the CRISPR/Cas9 system to delete exons 45–55 and most of their adjacent introns. The rationale is that, since most DMD patients suffer from out-of-frame mutations located within the “hotspot” of exons 45–55, deletion of these exons and restoration of the ORF will potentially benefit most of the patients [190,191]. Moreover, in-frame mutations in the same region are associated with the milder dystropathy, Becker muscular dystrophy (BMD) [192]. The successful restoration of the reading frame was confirmed via sequencing, and the corresponding protein expression was validated by immunohistochemistry and Western blot (WB). As a functional test, hiPSC-CMs and skeletal myocytes were exposed to osmotic stress. Indeed, in hypotonic solution, creatine kinase (CK) release levels were reduced in corrected cells to levels similar to healthy cardiomyocytes, indicating that the restored dystrophin provided sufficient membrane stability. Furthermore, miR-31 levels, which were reportedly high in DMD cells [193,194], were also reduced to levels comparable to BMD cells. Lastly, immunostaining and WB demonstrated successful restoration of β-dystroglycan in corrected skeletal myocytes. Additionally, hiPSC-derived skeletal myocytes were injected into NOD-scid IL2Rgamma (NSG) mdx mice, and dystrophin and β-dystroglycan were found only in the corrected and healthy cardiomyocytes. In conclusion, ORF restoration reinstated key functional cellular aspects of dystrophin, and this approach was proven as an important therapeutic strategy.
Zhang and colleagues [195] applied a similar principle, as skipping exon 51 can potentially restore the ORF in ~13% of DMD exon deletion mutations [196]. This group used two Cpf1-mediated CRISPR approaches to restore the ORF in hiPSCs carrying an out-of-frame deletion of exons 48–50. The first approach involved a single gRNA aimed at introducing indels in exon 51 by nonhomologous end joining (NHEJ). Indeed, corrected hiPSCs were reframed successfully and displayed an in-frame connection of exons 47 and 51. Another approach involved two gRNAs to achieve ORF restoration by skipping exon 51 and connecting exon 47 and 52. Restoration of the truncated protein was confirmed by WB analysis and immunocytochemistry. As a functional test, the mitochondrial DNA (mtDNA) and cellular respiration rate of hiPSC-CMs were measured. Corrected cells displayed increased mitochondrial numbers and oxygen consumption rate (OCR) compared to DMD cells and comparable to WT. Overall, these results demonstrate another promising approach in which individual DMD patients with different mutations may benefit from a “generic” ORF restoration.
Kyrychenko and coauthors [197] compared three approaches to restore the ORF in hiPSCs carrying a DMD mutation (deletion of exons 8–9) involving the actin-binding domain-1 (ABD-1) [12]. The first approach was deleting exons 3–7, leaving out actin-binding sites 1 and 2 (ABS-1 and ABS-2). Another strategy involved deleting exons 6–7 and excising ABS-3. The third method was deleting exons 7–11, leaving all three ABS regions intact. The respective dystrophin expression was validated by WB analysis and immunocytochemistry. As a functional assessment, spontaneous Ca2+ transients were analyzed in hiPSC-CMs. Indeed, release and reuptake parameters, including time to peak, Ca2+ decay rate, and transient duration, were higher in the uncorrected DMD hiPSC-CMs compared to WT and isogenic cells. Interestingly, while corrected cells with exons 3–9 deleted displayed normalized Ca2+ transient properties, corrected hiPSC-CMs carrying a deletion of exons 6–9 or 7–11 displayed only partial improvement of the Ca2+ transient parameters. These results indicate that, among the three approaches of ORF restoration, the one leading to a deletion of exons 3–9 yields the most functioning protein. Additional functional assessment was performed by means of engineered heart muscle (EHM). Consistent with the Ca2+ transient parameters results, enhanced contractile performance was observed in all corrected cell lines, with the most prominent effect in hiPSC-CMs carrying the deletion of exons 3–9. Overall, these results suggest that, of the three approaches, the correction leading to a deletion of exons 3–9 was the most effective in restoring functionality, implying the importance of ABS-3 over ABS-1 and ABS-2 of the ABD-1 domain. This also demonstrates the importance of functional testing, as they serve as a measure to evaluate the different approaches, specifically because there is no conclusive explanation as to why the correction resulting in the three ABS regions being intact (and a deletion of exons 7–11) was less effective than the approach leaving out ABS-1 and ABS-2.
Long et al. [198] identified single gRNAs capable of skipping and restoring the ORF in ~60% of DMD mutations together with the CRISPR/Cas9 system [199]. As a demonstration, they treated three different types of mutations. DMD hiPSCs carrying a deletion of exons 48–50 were corrected by destruction of the splice acceptor in exon 51, thereby allowing splicing of exon 47 to exon 52 and restoration of the ORF. DMD cells carrying a nonsense point mutation in intron 47 were corrected directly leading to permanent skipping of the pseudo-exon and restoring full-length dystrophin. Lastly, DMD hiPSCs carrying a duplication mutation of exons 55–59 were corrected by targeting the junction of intron 54 and exon 55, thus leaving out the duplicated region. Correction was confirmed via RT-PCR, and dystrophin levels were measured by immunocytochemistry and WB. Three-dimensional engineered heart muscle (3D-EHM) was used to assess functionality of WT, DMD, and corrected hiPSC-CMs; the force of contraction (FOC) was improved in corrected cells. Furthermore, this group found that 30–50% of cardiomyocytes need to be repaired for partial (30%) or maximal (50%) rescue of the contractile phenotype. In summary, these results support the notion of developing treatments benefiting groups of patients rather than individual mutations. Furthermore, their insistence on using single gRNAs simplifies the process and potentially increases efficacy.
Yuan and coauthors [200] utilized the previously published method of fusing nuclease-inactivated Cas9 with an activation-induced cytidine deaminase (AID) to convert targeted C to T or G to A, and subsequently restore the ORF in DMD hiPSCs lacking exon 51 [201,202]. The group successfully skipped exon 50 of dystrophin, thus restoring the ORF in 99.9% of dystrophin transcripts, as demonstrated by high-throughput sequencing (HTS). Corrected hiPSC-CMs released normal levels of creatine kinase (CK). Additionally, expression of miR-31 was suppressed in the corrected hiPSC-CMs, and β-dystroglycan levels were increased to levels comparable with healthy hiPSC-CMs. Importantly, contrary to CRISPR/Cas9 approaches which involve DSBs, targeted AID-mediated mutagenesis (TAM) does not require NHEJ or HDR; therefore, it is significantly less prone to indels, with higher correction efficiency [203]. These benefits strongly suggest that TAM is superior to other CRISPR/Cas9 correction methods for DMD and should be favored when applicable.
Sato and colleagues [204] investigated the effects of exon skipping on Ca2+ handling in DMD hiPSC-CMs. They used phosphorodiamidate morpholino oligomers (PMOs) to skip exon 45 and restore the ORF in cells carrying a deletion of dystrophin exons 46–55. By means of Fluo-4 they measured Ca2+ transient parameters following treatment and detected increased amplitude, as well as a decrease in the rates of rise and decay, all improvements resembling healthy cells. Additionally, the number of fluorescent-detected irregular pattern events suggestive of arrhythmogenicity decreased following exon 45 skipping. Taken together, these results demonstrate the effectiveness of the exon-skipping strategy to improve functional abnormalities in DMD cardiomyocytes.
Chemello et al. [205] investigated correction of exon deletion mutations using base and prime editing. For base editing, they used an adenine base editor approach with ABEmax-SpCas9 to induce exon 50 skipping by means of swapping the GT to GC sequence and restoring the ORF in DMD cells carrying an exon 51 deletion. The prime-editing strategy was used to introduce a swap of the TT to GT sequence in exon 52, resulting in ORF restoration. RT-PCR, sequencing, WB analysis, and immunocytochemistry confirmed correction and the resulting truncated protein. Next, the researchers tested the effect of correction on cardiomyocytes treated with isoproterenol. Indeed, corrected DMD cardiomyocytes displayed a reduction in arrhythmogenic Ca2+ transients compared to DMD cardiomyocytes, similar to healthy cardiomyocytes. Overall, these results demonstrate the feasibility of both genetic correction approaches in alleviating DMD cardiac pathophysiology.
Zhang and coauthors [206] devised a new approach for delivering Cas9 as part of CRISPR/Cas9 genome editing. Instead of using Streptococcus pyogenes Cas9 (SpCas9), which requires a dedicated adeno-associated virus (AAV) vector aside from the sgRNA vector, they used a Cas9 ortholog from Staphylococcus aureus (SaCas9), which is small enough to be packed together with sgRNA into a single AAV vector. This strategy was then utilized to deliver SaCas9-mediated single-cut gene editing to correct an exon 48–50 DMD deletion, avoiding possible unwanted genomic modifications due to the “double-cut” strategy with dual sgRNAs [207,208]. Their aim was restoration of the ORF of exon 51 via a two-nucleotide deletion. Indeed, the reading frame of hiPSCs carrying a DMD deletion of exons 48–50 was corrected accordingly. Furthermore, following correction, Ca2+ transient parameters in hiPSC-CMs, such as time to peak and rate of decay, were comparable to healthy cells, contrary to their abnormal elevation prior to correction. Additionally, genotoxicity analysis demonstrated no significant off-target editing, suggesting that this method is safe and effective at restoring the ORF and achieving functional restoration in DMD hiPSC-CMs.
Atmanli and colleagues [209] investigated various effects of CRISPR/Cas9-mediated corrections in DMD hiPSC-CMs. They generated two different hiPSC lines to restore the ORF in cells carrying a deletion of DMD exon 44: one using a single-nucleotide insertion to achieve reframing and the other by skipping exon 45 to restore the ORF. The correction of DMD hiPSC-CMs restored cellular area, volume and sarcomere length back to normal. DMD cardiomyocytes showed Ca2+ handling abnormalities including increased release and reuptake times, in addition to impaired localization of RYR2s, all of which were corrected with the truncated dystrophin lines. To test the effects of the corrections on contractility, the researchers used a polydimethylsiloxane matrix as a substrate for seeding cardiomyocytes, allowing auxotonic contraction. By means of video detection they demonstrated in DMD cells decreased systolic force and prolonged relaxation time, which were corrected in the truncated DMD lines. Furthermore, they found that ORF restoration reversed changes in gene expression, including two hallmark DCM transcripts, NPPA and NPPB [210], which were downregulated in DMD compared to corrected hiPSC-CMs. The researchers also tested changes in membrane tension by means of fluorescent membrane tension probe Flipper-TR [211]. These researchers found that DMD cardiomyocytes displayed higher membrane tension compared to healthy and corrected cells, demonstrating the important structural role of dystrophin, which is also preserved with the truncated protein. Lastly, the investigators corrected DMD cardiomyocytes directly, after the appearance of cellular abnormalities. Interestingly, after loading cardiomyocytes with the voltage-sensitive probe FluoVolt to measure arrhythmogenicity, they found a reduction in arrhythmogenicity in corrected and healthy cells compared to DMD cardiomyocytes. In summary, this robust work demonstrated the important functional, transcriptional and structural abnormalities in DMD cardiomyocytes, which were all rescued with CRISPR/Cas9-mediated ORF restoration.
7. Summary
This article reviewed the DMD cardiomyopathy-related research conducted in recent years in hiPSCs-CMs, as summarized in Table 1. The importance of investigating DMD pathophysiology in an in vitro human model is emphasized by the limitations of the current popular animal model, the mdx mouse [26,27,28,212]. Furthermore, the breakthrough reprograming technique enables the generation of patient- and mutation-specific hiPSC lines, providing an endless pool of cells carrying specific mutations for research [49,50,51,55,56]. Indeed, research on DMD hiPSC-CMs has yielded novel findings thus far, as well as validation of results previously reported in other models. Furthermore, with the introduction of gene-editing techniques, promising therapeutic possibilities are becoming more likely, including complete genetic cure. These need to be tested first in vitro, and hiPSCs are the ultimate model for such research. Furthermore, gene editing enables the generation of isogenic controls which provide significantly stronger validation of results obtained using the hiPSC model. Lastly, although extensive research is needed before clinical application, hiPSCs and hiPSC-derived cells may be used in the future in regenerative medicine to repair damaged tissue which may otherwise not be treatable.

Table 1.
Summary of DMD hiPSC-CM research papers in recent years.
Author Contributions
Writing—original draft preparation, review and editing, B.E.; writing—review and editing, O.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Israel Science Foundation (ISF) (#824/19 to O.B.), the Niedersachsen Foundation (#ZN3452 to O.B.), the US–Israel Binational Foundation (BSF) (#2019039 to O.B.), the Association Duchenne Israel (A.D.I, #580545317, to O.B), and the Duchenne Parent Project Netherlands (DPP NL) (#2029771 to O.B).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moat, S.J.; Bradley, D.M.; Salmon, R.; Clarke, A.; Hartley, L. Newborn Bloodspot Screening for Duchenne Muscular Dystrophy: 21 Years Experience in Wales (UK). Eur. J. Hum. Genet. 2013, 21, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Romitti, P.A.; Zhu, Y.; Puzhankara, S.; James, K.A.; Nabukera, S.K.; Zamba, G.K.D.; Ciafaloni, E.; Cunniff, C.; Druschel, C.M.; Mathews, K.D.; et al. Prevalence of Duchenne and Becker Muscular Dystrophies in the United States. Pediatrics 2015, 135, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Ryder, S.; Leadley, R.M.; Armstrong, N.; Westwood, M.; de Kock, S.; Butt, T.; Jain, M.; Kleijnen, J. The Burden, Epidemiology, Costs and Treatment for Duchenne Muscular Dystrophy: An Evidence Review. Orphanet J. Rare Dis. 2017, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Ahn, A.H.; Kunkel, L.M. The Structural and Functional Diversity of Dystrophin. Nat. Genet. 1993, 3, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Tennyson, C.N.; Klamut, H.J.; Worton, R.G. The Human Dystrophin Gene Requires 16 Hours to Be Transcribed and Is Cotranscriptionally Spliced. Nat. Genet. 1995, 9, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Brenman, J.E.; Chao, D.S.; Gee, S.H.; McGee, A.W.; Craven, S.E.; Santillano, D.R.; Wu, Z.; Huang, F.; Xia, H.; Peters, M.F.; et al. Interaction of Nitric Oxide Synthase with the Postsynaptic Density Protein PSD-95 and Al-pha1-Syntrophin Mediated by PDZ Domains. Cell 1996, 84, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Desguerre, I.; Mayer, M.; Leturcq, F.; Barbet, J.-P.; Gherardi, R.K.; Christov, C. Endomysial Fibrosis in Duchenne Muscular Dystrophy: A Marker of Poor Outcome Associated with Macrophage Alternative Activation. J. Neuropathol. Exp. Neurol. 2009, 68, 762–773. [Google Scholar] [CrossRef]
- Millay, D.P.; Goonasekera, S.A.; Sargent, M.A.; Maillet, M.; Aronow, B.J.; Molkentin, J.D. Calcium Influx Is Sufficient to Induce Muscular Dystrophy through a TRPC-Dependent Mechanism. Proc. Natl. Acad. Sci. USA 2009, 106, 19023–19028. [Google Scholar] [CrossRef]
- Xi, H.; Shin, W.S.; Suzuki, J.-I.; Nakajima, T.; Kawada, T.; Uehara, Y.; Nakazawa, M.; Toyo-oka, T. Dystrophin Disruption Might Be Related to Myocardial Cell Apoptosis Caused by Isoproterenol. J. Cardiovasc. Pharmacol. 2000, 36, S25-9. [Google Scholar] [CrossRef]
- Brown, S.C.; Fassati, A.; Popplewell, L.; Page, A.M.; Henry, M.D.; Campbell, K.P.; Dickson, G. Dystrophic Phenotype Induced in Vitro by Antibody Blockade of Muscle Alpha-Dystroglycan-Laminin Interaction. J. Cell Sci. 1999, 112, 209–216. [Google Scholar] [CrossRef]
- Sciandra, F.; Bozzi, M.; Bianchi, M.; Pavoni, E.; Giardina, B.; Brancaccio, A. Dystroglycan and Muscular Dystrophies Related to the Dystrophin-Glycoprotein Complex. Ann. Ist. Super. Sanita 2003, 39, 173–181. [Google Scholar] [PubMed]
- Kamdar, F.; Garry, D.J. Dystrophin-Deficient Cardiomyopathy. J. Am. Coll. Cardiol. 2016, 67, 2533–2546. [Google Scholar] [CrossRef]
- Rybakova, I.N.; Patel, J.R.; Ervasti, J.M. The Dystrophin Complex Forms a Mechanically Strong Link between the Sarcolemma and Costameric Actin. J. Cell Biol. 2000, 150, 1209–1214. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, Y.; Yagi, M.; Okizuka, Y.; Awano, H.; Zhang, Z.; Yamauchi, Y.; Nishio, H.; Matsuo, M. Mutation Spectrum of the Dystrophin Gene in 442 Duchenne/Becker Muscular Dystrophy Cases from One Japanese Referral Center. J. Hum. Genet. 2010, 55, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; Van Deutekom, J.C.T.; Fokkema, I.F.; Van Ommen, G.-J.B.; Den Dunnen, J.T. Entries in the Lei-den Duchenne Muscular Dystrophy Mutation Database: An Overview of Mutation Types and Paradoxical Cases That Confirm the Reading-Frame Rule. Muscle Nerve 2006, 34, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Birnkrant, D.J.; Bushby, K.; Bann, C.M.; Apkon, S.D.; Blackwell, A.; Brumbaugh, D.; Case, L.E.; Clemens, P.R.; Hadjiyannakis, S.; Pandya, S.; et al. Diagnosis and Management of Duchenne Muscular Dystrophy, Part 1: Diagnosis, and Neuromuscular, Rehabilitation, Endocrine, and Gastrointestinal and Nutritional Management. Lancet Neurol. 2018, 17, 251–267. [Google Scholar] [CrossRef]
- McNally, E.M.; Kaltman, J.R.; Benson, D.W.; Canter, C.E.; Cripe, L.H.; Duan, D.; Finder, J.D.; Groh, W.J.; Hoffman, E.P.; Judge, D.P.; et al. Contemporary Cardiac Issues in Duchenne Muscular Dystrophy. Working Group of the National Heart, Lung, and Blood Institute in Collaboration with Parent Project Muscular Dystrophy. Circulation 2015, 131, 1590–1598. [Google Scholar] [CrossRef]
- Passamano, L.; Taglia, A.; Palladino, A.; Viggiano, E.; D’Ambrosio, P.; Scutifero, M.; Rosaria Cecio, M.; Torre, V.; DE Luca, F.; Picillo, E.; et al. Improvement of Survival in Duchenne Muscular Dystrophy: Retrospective Analysis of 835 Patients. Acta Myol. 2012, 31, 121–125. [Google Scholar]
- Gloss, D.; Moxley, R.T.; Ashwal, S.; Oskoui, M. Practice Guideline Update Summary: Corticosteroid Treatment of Duchenne Muscular Dystrophy: Report of the Guideline Development Subcommittee of the American Academy of Neurology. Neurology 2016, 86, 465–472. [Google Scholar] [CrossRef]
- Matthews, E.; Brassington, R.; Kuntzer, T.; Jichi, F.; Manzur, A.Y. Corticosteroids for the Treatment of Duchenne Muscular Dystrophy. Cochrane Database Syst. Rev. 2016, 5, CD003725. [Google Scholar] [CrossRef]
- Griggs, R.C.; Moxley, R.T.; Mendell, J.R.; Fenichel, G.M.; Brooke, M.H.; Pestronk, A.; Miller, J.P. Prednisone in Duchenne Dystrophy. A Randomized, Controlled Trial Defining the Time Course and Dose Response. Clinical Investigation of Duchenne Dystrophy Group. Arch. Neurol. 1991, 48, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Goemans, N.; Lowes, L.P.; Alfano, L.N.; Berry, K.; Shao, J.; Kaye, E.M.; Mercuri, E.; Eteplirsen Study Group and Telethon Foundation DMD Italian Network. Longitudinal Effect of Eteplirsen versus Historical Control on Ambulation in Duchenne Muscular Dystrophy. Ann. Neurol. 2016, 79, 257–271. [Google Scholar] [CrossRef]
- McDonald, C.M.; Campbell, C.; Torricelli, R.E.; Finkel, R.S.; Flanigan, K.M.; Goemans, N.; Heydemann, P.; Kaminska, A.; Kirschner, J.; Muntoni, F.; et al. Ataluren in Patients with Nonsense Mutation Duchenne Muscular Dystrophy (ACT DMD): A Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 390, 1489–1498. [Google Scholar] [CrossRef]
- Bulfield, G.; Siller, W.G.; Wight, P.A.; Moore, K.J. X Chromosome-Linked Muscular Dystrophy (Mdx) in the Mouse. Proc. Natl. Acad. Sci. USA 1984, 81, 1189–1192. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.; Banks, G.B.; Hall, J.K.; Muir, L.A.; Ramos, J.N.; Wicki, J.; Odom, G.L.; Konieczny, P.; Seto, J.; Chamberlain, J.R.; et al. Animal Models of Muscular Dystrophy. Prog. Mol. Biol. Transl. Sci. 2012, 105, 83–111. [Google Scholar] [CrossRef]
- Carnwath, J.W.; Shotton, D.M. Muscular Dystrophy in the Mdx Mouse: Histopathology of the Soleus and Extensor Digitorum Longus Muscles. J. Neurol. Sci. 1987, 80, 39–54. [Google Scholar] [CrossRef] [PubMed]
- DiMario, J.X.; Uzman, A.; Strohman, R.C. Fiber Regeneration Is Not Persistent in Dystrophic (MDX) Mouse Skeletal Muscle. Dev. Biol. 1991, 148, 314–321. [Google Scholar] [CrossRef]
- Quinlan, J.G.; Hahn, H.S.; Wong, B.L.; Lorenz, J.N.; Wenisch, A.S.; Levin, L.S. Evolution of the Mdx Mouse Cardiomyopathy: Physiological and Morphological Findings. Neuromuscul. Disord. 2004, 14, 491–496. [Google Scholar] [CrossRef]
- Ascah, A.; Khairallah, M.; Daussin, F.; Bourcier-Lucas, C.; Godin, R.; Allen, B.G.; Petrof, B.J.; Des Rosiers, C.; Burelle, Y. Stress-Induced Opening of the Permeability Transition Pore in the Dystrophin-Deficient Heart Is Attenuated by Acute Treatment with Sildenafil. Am. J. Physiol. Heart Circ. Physiol. 2011, 300, 144–153. [Google Scholar] [CrossRef]
- Jung, C.; Martins, A.S.; Niggli, E.; Shirokova, N. Dystrophic Cardiomyopathy: Amplification of Cellular Damage by Ca2+ Signalling and Reactive Oxygen Species-Generating Pathways. Cardiovasc. Res. 2008, 77, 766–773. [Google Scholar] [CrossRef]
- Kyrychenko, V.; Poláková, E.; Janíček, R.; Shirokova, N. Mitochondrial Dysfunctions during Progression of Dystrophic Cardiomyopathy. Cell Calcium 2015, 58, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Garbincius, J.F.; Luongo, T.S.; Elrod, J.W. The Debate Continues—What Is the Role of MCU and Mitochondrial Calcium Uptake in the Heart? J. Mol. Cell Cardiol. 2020, 143, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Coley, W.D.; Bogdanik, L.; Vila, M.C.; Yu, Q.; Van Der Meulen, J.H.; Rayavarapu, S.; Novak, J.S.; Nearing, M.; Quinn, J.L.; Saunders, A.; et al. Effect of Genetic Background on the Dystrophic Phenotype in Mdx Mice. Hum. Mol. Genet. 2015, 25, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Bostick, B.; Yue, Y.; Duan, D. Gender Influences Cardiac Function in the Mdx Model of Duchenne Cardiomyopathy. Muscle Nerve 2010, 42, 600–603. [Google Scholar] [CrossRef]
- Guéniot, L.; Latroche, C.; Thépenier, C.; Chatre, L.; Mazeraud, A.; Fiole, D.; Goossens, P.L.; Chrétien, F.; Jouvion, G. The Female Mdx Mouse: An Unexpected Vascular Story. J. Neurol. Neuromedicine 2016, 1, 41–53. [Google Scholar] [CrossRef]
- Ishizaki, M.; Kobayashi, M.; Adachi, K.; Matsumura, T.; Kimura, E. Female Dystrophinopathy: Review of Current Literature. Neuromuscul. Disord. 2018, 28, 572–581. [Google Scholar] [CrossRef]
- Larcher, T.; Lafoux, A.; Tesson, L.; Remy, S.V.; Thepenier, V.; François, V.; Guiner, C.L.; Goubin, H.; Dutilleul, M.V.; Guigand, L.; et al. Characterization of Dystrophin Deficient Rats: A New Model for Duchenne Muscular Dystrophy. PLoS ONE 2014, 9, e110371. [Google Scholar] [CrossRef]
- Klingler, W.; Jurkat-Rott, K.; Lehmann-Horn, F.; Schleip, R. The Role of fibrosis in Duchenne Muscular Dystrophy. Acta Myologica 2012, 31, 184–195. [Google Scholar]
- Szabó, P.L.; Ebner, J.; Koenig, X.; Hamza, O.; Watzinger, S.; Trojanek, S.; Abraham, D.; Todt, H.; Kubista, H.; Schicker, K.; et al. Cardiovascular Phenotype of the Dmdmdx Rat—A Suitable Animal Model for Duchenne Muscular Dystrophy. Dis. Model. Mech. 2021, 14, 047704. [Google Scholar] [CrossRef]
- Eisen, B.; Ben Jehuda, R.; Cuttitta, A.J.; Mekies, L.N.; Shemer, Y.; Baskin, P.; Reiter, I.; Willi, L.; Freimark, D.; Gherghiceanu, M.; et al. Electrophysiological Abnormalities in Induced Pluripotent Stem Cell-Derived Cardiomyocytes Generated from Duchenne Muscular Dystrophy Patients. J. Cell. Mol. Med. 2019, 23, 2125–2135. [Google Scholar] [CrossRef]
- Lin, B.; Li, Y.; Han, L.; Kaplan, A.D.; Ao, Y.; Kalra, S.; Bett, G.C.L.; Rasmusson, R.L.; Denning, C.; Yang, L. Model-ing and Study of the Mechanism of Dilated Cardiomyopathy Using Induced Pluripotent Stem Cells Derived from Individuals with Duchenne Muscular Dystrophy. Dis. Model. Mech. 2015, 8, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Jelinkova, S.; Vilotic, A.; Pribyl, J.; Aimond, F.; Salykin, A.; Acimovic, I.; Pesl, M.; Caluori, G.; Klimovic, S.; Urban, T.; et al. DMD Pluripotent Stem Cell Derived Cardiac Cells Recapitulate in Vitro Human Cardiac Pathophysiology. Front. Bioeng. Biotechnol. 2020, 8, 535. [Google Scholar] [CrossRef] [PubMed]
- Mekies, L.N.; Regev, D.; Eisen, B.; Fernandez-Gracia, J.; Baskin, P.; Ben Jehuda, R.; Shulman, R.; Reiter, I.; Palty, R.; Arad, M.; et al. Depressed β-Adrenergic Inotropic Responsiveness and Intracellular Calcium Handling Abnormalities in Duchenne Muscular Dystrophy Patients’ Induced Pluripotent Stem Cell–Derived Cardiomyocytes. J. Cell. Mol. Med. 2021, 25, 3922–3934. [Google Scholar] [CrossRef] [PubMed]
- Kamdar, F.; Das, S.; Gong, W.; Klaassen Kamdar, A.; Meyers, T.A.; Shah, P.; Ervasti, J.M.; Townsend, D.W.; Kamp, T.J.; Wu, J.C.; et al. Stem Cell-Derived Cardiomyocytes and Beta-Adrenergic Receptor Blockade in Duchenne Muscular Dystrophy Cardiomyopathy. J. Am. Coll. Cardiol. 2020, 75, 1159–1174. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Guenther, M.G.; Frampton, G.M.; Soldner, F.; Hockemeyer, D.; Mitalipova, M.; Jaenisch, R.; Young, R.A. Chroma-tin Structure and Gene Expression Programs of Human Embryonic and Induced Pluripotent Stem Cells. Cell Stem Cell 2010, 7, 249–257. [Google Scholar] [CrossRef]
- Zhao, M.-T.; Chen, H.; Liu, Q.; Shao, N.-Y.; Sayed, N.; Wo, H.-T.; Zhang, J.Z.; Ong, S.-G.; Liu, C.; Kim, Y.; et al. Molecular and Functional Resemblance of Differentiated Cells Derived from Isogenic Human IPSCs and SCNT-Derived ESCs. Proc. Natl. Acad. Sci. USA 2017, 114, E11111–E11120. [Google Scholar] [CrossRef]
- Soldner, F.; Hockemeyer, D.; Beard, C.; Gao, Q.; Bell, G.W.; Cook, E.G.; Hargus, G.; Blak, A.; Cooper, O.; Mitalipova, M.; et al. Parkinson’s Disease Patient-Derived Induced Pluripotent Stem Cells Free of Viral Reprogramming Fac-tors. Cell 2009, 136, 964–977. [Google Scholar] [CrossRef]
- Maehr, R.; Chen, S.; Snitow, M.; Ludwig, T.; Yagasaki, L.; Goland, R.; Leibel, R.L.; Melton, D.A. Generation of Pluripotent Stem Cells from Patients with Type 1 Diabetes. Proc. Natl. Acad. Sci. USA 2009, 106, 15768–15773. [Google Scholar] [CrossRef]
- Ben Jehuda, R.; Eisen, B.; Shemer, Y.; Mekies, L.N.; Szantai, A.; Reiter, I.; Cui, H.; Guan, K.; Haron-Khun, S.; Freimark, D.; et al. CRISPR Correction of the PRKAG2 Gene Mutation in the Patient’s Induced Pluripotent Stem Cell-Derived Cardiomyocytes Eliminates Electrophysiological and Structural Abnormalities. Heart Rhythm. 2018, 15, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Hallas, T.; Eisen, B.; Shemer, Y.; Ben Jehuda, R.; Mekies, L.N.; Naor, S.; Schick, R.; Eliyahu, S.; Reiter, I.; Vlodavsky, E.; et al. Investigating the Cardiac Pathology of SCO2-Mediated Hypertrophic Cardiomyopathy Using Patients Induced Pluripotent Stem Cell-Derived Cardiomyocytes. J. Cell. Mol. Med. 2017, 22, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Eisen, B.; Ben Jehuda, R.; Cuttitta, A.J.; Mekies, L.N.; Reiter, I.; Ramchandren, S.; Arad, M.; Michele, D.E.; Binah, O. Generation of Duchenne Muscular Dystrophy Patient-Specific Induced Pluripotent Stem Cell Line Lacking Exons 45-50 of the Dystrophin Gene (IITi001-A). Stem Cell Res. 2018, 29, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R.; et al. Induced Pluripotent Stem Cells Generated from Patients with ALS Can Be Differentiated into Motor Neurons. Science 2008, 321, 1218–1221. [Google Scholar] [CrossRef]
- Carlson, C.; Koonce, C.; Aoyama, N.; Einhorn, S.; Fiene, S.; Thompson, A.; Swanson, B.; Anson, B.; Kattman, S. Phenotypic Screening with Human IPS Cell-Derived Cardiomyocytes: HTS-Compatible Assays for Interrogating Cardiac Hypertrophy. J. Biomol. Screen. 2013, 18, 1203–1211. [Google Scholar] [CrossRef]
- Novak, A.; Barad, L.; Lorber, A.; Gherghiceanu, M.; Reiter, I.; Eisen, B.; Eldor, L.; Itskovitz-Eldor, J.; Eldar, M.; Arad, M.; et al. Functional Abnormalities in IPSC-Derived Cardiomyocytes Generated from CPVT1 and CPVT2 Patients Carrying Ryanodine or Calsequestrin Mutations. J. Cell. Mol. Med. 2015, 19, 2006–2018. [Google Scholar] [CrossRef]
- Schick, R.; Mekies, L.N.; Shemer, Y.; Eisen, B.; Hallas, T.; Ben Jehuda, R.; Ben-Ari, M.; Szantai, A.; Willi, L.; Shul-man, R.; et al. Functional Abnormalities in Induced Pluripotent Stem Cell-Derived Cardiomyocytes Generated from Titin-Mutated Patients with Dilated Cardiomyopathy. PLoS ONE 2018, 13, e0205719. [Google Scholar] [CrossRef]
- Flanigan, K.M.; Dunn, D.M.; von Niederhausern, A.; Soltanzadeh, P.; Gappmaier, E.; Howard, M.T.; Sampson, J.B.; Mendell, J.R.; Wall, C.; King, W.M.; et al. Mutational Spectrum of DMD Mutations in Dystrophinopathy Patients: Application of Modern Diagnostic Techniques to a Large Cohort. Hum. Mutat. 2009, 30, 1657–1666. [Google Scholar] [CrossRef]
- Porter, J.D.; Merriam, A.P.; Leahy, P.; Gong, B.; Feuerman, J.; Cheng, G.; Khanna, S. Temporal Gene Expression Profiling of Dystrophin-Deficient (Mdx) Mouse Diaphragm Identifies Conserved and Muscle Group-Specific Mechanisms in the Pathogenesis of Muscular Dystrophy. Hum. Mol. Genet. 2004, 13, 257–269. [Google Scholar] [CrossRef]
- Camerino, G.M.; Cannone, M.; Giustino, A.; Massari, A.M.; Capogrosso, R.F.; Cozzoli, A.; De Luca, A. Gene Ex-pression in Mdx Mouse Muscle in Relation to Age and Exercise: Aberrant Mechanical-Metabolic Coupling and Implications for Pre-Clinical Studies in Duchenne Muscular Dystrophy. Hum. Mol. Genet. 2014, 23, 5720–5732. [Google Scholar] [CrossRef]
- Haslett, J.N.; Sanoudou, D.; Kho, A.T.; Bennett, R.R.; Greenberg, S.A.; Kohane, I.S.; Beggs, A.H.; Kunkel, L.M. Gene Expression Comparison of Biopsies from Duchenne Muscular Dystrophy (DMD) and Normal Skeletal Muscle. Proc. Natl. Acad. Sci. USA 2002, 99, 15000–15005. [Google Scholar] [CrossRef] [PubMed]
- Haslett, J.N.; Sanoudou, D.; Kho, A.T.; Han, M.; Bennett, R.R.; Kohane, I.S.; Beggs, A.H.; Kunkel, L.M. Gene Ex-pression Profiling of Duchenne Muscular Dystrophy Skeletal Muscle. Neurogenetics 2003, 4, 163–171. [Google Scholar] [CrossRef]
- Verhagen, A.M.; Ekert, P.G.; Pakusch, M.; Silke, J.; Connolly, L.M.; Reid, G.E.; Moritz, R.L.; Simpson, R.J.; Vaux, D.L. Identification of DIABLO, a Mammalian Protein That Promotes Apoptosis by Binding to and Antagonizing IAP Proteins. Cell 2000, 102, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Marston, S. The Molecular Mechanisms of Mutations in Actin and Myosin That Cause Inherited Myopathy. Int. J. Mol. Sci. 2018, 19, 2020. [Google Scholar] [CrossRef] [PubMed]
- Chun Yang, X.; Hui Zhao, D.; Bond Lau, W.; Qiang Liu, K.; Yu Tian, J.; Chao Cheng, Z.; Liang Ma, X.; Hua Liu, J.; Fan, Q. LncRNA ENSMUST00000134285 Increases MAPK11 Activity, Regulating Aging-Related Myocardial Apoptosis. J. Gerontol. A Biol. Sci. Med. Sci. 2018, 73, 1010–1017. [Google Scholar] [CrossRef]
- Deng, Y.; Wei, S.; Hu, S.; Chen, J.; Tan, Z.; Yang, Y. Ehlers-Danlos Syndrome Type IV Is Associated with a Novel G984R COL3A1 Mutation. Mol. Med. Rep. 2015, 12, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, X.; Yu, H.; Pei, J.; Zhang, Y.; Gong, J.; Pu, J. Common Variants in TRDN and CALM1 Are Associated with Risk of Sudden Cardiac Death in Chronic Heart Failure Patients in Chinese Han Population. PLoS ONE 2015, 10, e0132459. [Google Scholar] [CrossRef]
- Finsterer, J.; Stöllberger, C. Cardiac Involvement in Becker Muscular Dystrophy. Can. J. Cardiol. 2008, 24, 786–792. [Google Scholar] [CrossRef]
- Yilmaz, A.; Gdynia, H.-J.; Baccouche, H.; Mahrholdt, H.; Meinhardt, G.; Basso, C.; Thiene, G.; Sperfeld, A.-D.; Ludolph, A.C.; Sechtem, U. Cardiac Involvement in Patients with Becker Muscular Dystrophy: New Diagnostic and Pathophysiological Insights by a CMR Approach. J. Cardiovasc. Magn. Reson. 2008, 10, 50. [Google Scholar] [CrossRef]
- Fayssoil, A.; Nardi, O.; Orlikowski, D.; Annane, D. Cardiomyopathy in Duchenne Muscular Dystrophy: Patho-genesis and Therapeutics. Heart Fail. Rev. 2010, 15, 103–107. [Google Scholar] [CrossRef]
- Romfh, A.; McNally, E.M. Cardiac Assessment in Duchenne and Becker Muscular Dystrophies. Curr. Heart Fail. Rep. 2010, 7, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.C.Y.; Chang, A.C.H.; Kirillova, A.; Sasagawa, K.; Su, W.; Weber, G.; Lin, J.; Termglinchan, V.; Karakikes, I.; Seeger, T.; et al. Telomere Shortening Is a Hallmark of Genetic Cardiomyopathies. Proc. Natl. Acad. Sci. USA 2018, 115, 9276–9281. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Magdinier, F.; Stadler, G.; Wagner, K.R.; Shay, J.W.; Wright, W.E. Telomere Position Effect: Regulation of Gene Expression with Progressive Telomere Shortening over Long Distances. Genes. Dev. 2014, 28, 2464–2476. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Taffet, G.E.; Youker, K.A.; Entman, M.L.; Overbeek, P.A.; Michael, L.H.; Schneider, M.D. Telomerase Re-verse Transcriptase Promotes Cardiac Muscle Cell Proliferation, Hypertrophy, and Survival. Proc. Natl. Acad. Sci. USA 2001, 98, 10308–10313. [Google Scholar] [CrossRef]
- Bär, C.; Bernardes de Jesus, B.; Serrano, R.; Tejera, A.; Ayuso, E.; Jimenez, V.; Formentini, I.; Bobadilla, M.; Mizrahi, J.; de Martino, A.; et al. Telomerase Expression Confers Cardioprotection in the Adult Mouse Heart after Acute Myocardial Infarction. Nat. Commun. 2014, 5, 5863. [Google Scholar] [CrossRef]
- Sacco, A.; Mourkioti, F.; Tran, R.; Choi, J.; Llewellyn, M.; Kraft, P.; Shkreli, M.; Delp, S.; Pomerantz, J.H.; Artandi, S.E.; et al. Short Telomeres and Stem Cell Exhaustion Model Duchenne Muscular Dystrophy in Mdx/MTR Mice. Cell 2010, 143, 1059–1071. [Google Scholar] [CrossRef]
- Chang, A.C.Y.; Pardon, G.; Chang, A.C.H.; Wu, H.; Ong, S.G.; Eguchi, A.; Ancel, S.; Holbrook, C.; Ramunas, J.; Ribeiro, A.J.S.; et al. Increased Tissue Stiffness Triggers Contractile Dysfunction and Telomere Shortening in Dys-trophic Cardiomyocytes. Stem Cell Rep. 2021, 16, 2169–2181. [Google Scholar] [CrossRef]
- Skwarek-Maruszewska, A.; Hotulainen, P.; Mattila, P.K.; Lappalainen, P. Contractility-Dependent Actin Dynam-ics in Cardiomyocyte Sarcomeres. J. Cell Sci. 2009, 122, 2119–2126. [Google Scholar] [CrossRef]
- Farini, A.; Gowran, A.; Bella, P.; Sitzia, C.; Scopece, A.; Castiglioni, E.; Rovina, D.; Nigro, P.; Villa, C.; Fortunato, F.; et al. Fibrosis Rescue Improves Cardiac Function in Dystrophin-Deficient Mice and Duchenne Patient-Specific Cardiomyocytes by Immunoproteasome Modulation. Am. J. Pathol. 2019, 189, 339–353. [Google Scholar] [CrossRef]
- Meng, X.M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-β: The Master Regulator of Fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Yasutake, H.; Lee, J.K.; Hashimoto, A.; Masuyama, K.; Li, J.; Kuramoto, Y.; Higo, S.; Hikoso, S.; Hidaka, K.; Naito, A.T.; et al. Decreased YAP Activity Reduces Proliferative Ability in Human Induced Pluripotent Stem Cell of Duchenne Muscular Dystrophy Derived Cardiomyocytes. Sci. Rep. 2021, 11, 10351. [Google Scholar] [CrossRef] [PubMed]
- Vita, G.L.; Polito, F.; Oteri, R.; Arrigo, R.; Ciranni, A.M.; Musumeci, O.; Messina, S.; Rodolico, C.; Di Giorgio, R.M.; Vita, G.; et al. Hippo Signaling Pathway Is Altered in Duchenne Muscular Dystrophy. PLoS ONE 2018, 13, e0205514. [Google Scholar] [CrossRef] [PubMed]
- Bremner, S.B.; Mandrycky, C.J.; Leonard, A.; Padgett, R.M.; Levinson, A.R.; Rehn, E.S.; Pioner, J.M.; Sniadecki, N.J.; Mack, D.L. Full-Length Dystrophin Deficiency Leads to Contractile and Calcium Transient Defects in Human Engineered Heart Tissues. J. Biophys. J. 2022, 121, 419a. [Google Scholar] [CrossRef]
- Bielawski, K.S.; Leonard, A.; Bhandari, S.; Murry, C.E.; Sniadecki, N.J. Real-Time Force and Frequency Analysis of Engineered Human Heart Tissue Derived from Induced Pluripotent Stem Cells Using Magnetic Sensing. Tissue Eng. Part C Methods 2016, 22, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.; Bertero, A.; Powers, J.D.; Beussman, K.M.; Bhandari, S.; Regnier, M.; Murry, C.E.; Sniadecki, N.J. Afterload Promotes Maturation of Human Induced Pluripotent Stem Cell Derived Cardiomyocytes in Engineered Heart Tissues. J. Mol. Cell. Cardiol. 2018, 118, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Bremner, S.; Goldstein, A.J.; Higashi, T.; Sniadecki, N.J. Engineered Heart Tissues for Contractile, Structural, and Transcriptional Assessment of Human Pluripotent Stem Cell-Derived Cardiomyocytes in a Three-Dimensional, Auxotonic Environment. Methods Mol. Biol. 2022, 2485, 87–97. [Google Scholar] [CrossRef]
- Marini, V.; Marino, F.; Aliberti, F.; Giarratana, N.; Pozzo, E.; Duelen, R.; Cortés Calabuig, Á.; La Rovere, R.; Vervliet, T.; Torella, D.; et al. Long-Term Culture of Patient-Derived Cardiac Organoids Recapitulated Duchenne Muscular Dystrophy Cardiomyopathy and Disease Progression. Front. Cell Dev. Biol. 2022, 10, 878311. [Google Scholar] [CrossRef]
- Velasco, V.; Shariati, S.A.; Esfandyarpour, R. Microtechnology-Based Methods for Organoid Models. Microsyst. Nanoeng. 2020, 6, 76. [Google Scholar] [CrossRef]
- Heydari, Z.; Moeinvaziri, F.; Agarwal, T.; Pooyan, P.; Shpichka, A.; Maiti, T.K.; Timashev, P.; Baharvand, H.; Vosough, M. Organoids: A Novel Modality in Disease Modeling. Bio-Design Manuf. 2021, 4, 689–716. [Google Scholar] [CrossRef]
- Scalise, M.; Marino, F.; Salerno, L.; Cianflone, E.; Molinaro, C.; Salerno, N.; De Angelis, A.; Viglietto, G.; Urbanek, K.; Torella, D. From Spheroids to Organoids: The Next Generation of Model Systems of Human Cardiac Regeneration in a Dish. Int. J. Mol. Sci. 2021, 22, 13180. [Google Scholar] [CrossRef]
- Vainzof, M.; Souza, L.S.; Gurgel-Giannetti, J.; Zatz, M. Sarcoglycanopathies: An Update. Neuromuscul. Disord. 2021, 31, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, C.D.; Wu, R.F.; Terada, L.S. ROS Signaling and ER Stress in Cardiovascular Disease. Mol. Aspects Med. 2018, 63, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Simón, E.; Suárez-Calvet, X.; Carrasco-Rozas, A.; Piñol-Jurado, P.; López-Fernández, S.; Pons, G.; Bech Serra, J.J.; de la Torre, C.; de Luna, N.; Gallardo, E.; et al. RhoA/ROCK2 Signalling Is Enhanced by PDGF-AA in Fibro-Adipogenic Progenitor Cells: Implications for Duchenne Muscular Dystrophy. J. Cachexia Sarcopenia Muscle 2022, 13, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, G.; Kadur Nagaraju, C.; Duelen, R.; Amoni, M.; Bobin, P.; Eschenhagen, T.; Roderick, H.L.; Sampaolesi, M.; Sipido, K.R. Incomplete Assembly of the Dystrophin-Associated Protein Complex in 2D and 3D-Cultured Hu-man Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Front. Cell Dev. Biol. 2021, 9, 2869. [Google Scholar] [CrossRef]
- Fong, P.Y.; Turner, P.R.; Denetclaw, W.F.; Steinhardt, R.A. Increased Activity of Calcium Leak Channels in Myo-tubes of Duchenne Human and Mdx Mouse Origin. Science 1990, 250, 673–676. [Google Scholar] [CrossRef]
- Blake, D.J.; Weir, A.; Newey, S.E.; Davies, K.E.; Spaulding, H.R.; Quindry, T.; Hammer, K.; Quindry, J.C.; Selsby, J.T.; Amirouche, A.; et al. Function and Genetics of Dystrophin and Dystrophin-Related Proteins in Muscle. Physiol. Rev. 2002, 82, 291–329. [Google Scholar] [CrossRef]
- Tidball, J.G.; Villalta, S.A. NO May Prompt Calcium Leakage in Dystrophic Muscle. Nat. Med. 2009, 15, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Bellinger, A.M.; Reiken, S.; Carlson, C.; Mongillo, M.; Liu, X.; Rothman, L.; Matecki, S.; Lacampagne, A.; Marks, A.R. Hypernitrosylated Ryanodine Receptor Calcium Release Channels Are Leaky in Dystrophic Muscle. Nat. Med. 2009, 15, 325–330. [Google Scholar] [CrossRef]
- Barthélémy, F.; Wang, R.T.; Hsu, C.; Douine, E.D.; Marcantonio, E.E.; Nelson, S.F.; Miceli, M.C. Targeting RyR Activity Boosts Antisense Exon 44 and 45 Skipping in Human DMD Skeletal or Cardiac Muscle Culture Models. Mol. Ther. Nucleic Acids 2019, 18, 580–589. [Google Scholar] [CrossRef]
- Uchimura, T.; Sakurai, H. Orai1–Stim1 Regulates Increased Ca2+ Mobilization, Leading to Contractile Duchenne Muscular Dystrophy Phenotypes in Patient-Derived Induced Pluripotent Stem Cells. Biomedicines 2021, 9, 1589. [Google Scholar] [CrossRef]
- Bodensteiner, J.B.; Engel, A.G. Intracellular Calcium Accumulation in Duchenne Dystrophy and Other Myopathies: A Study of 567,000 Muscle Fibers in 114 Biopsies. Neurology 1978, 28, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.R.; Westwood, T.; Regen, C.M.; Steinhardt, R.A. Increased Protein Degradation Results from Elevated Free Calcium Levels Found in Muscle from Mdx Mice. Nature 1988, 335, 735–738. [Google Scholar] [CrossRef]
- Spencer, M.J.; Croall, D.E.; Tidball, J.G. Calpains Are Activated in Necrotic Fibers from Mdx Dystrophic Mice. J. Biol. Chem. 1995, 270, 10909–10914. [Google Scholar] [CrossRef]
- Robert, V.; Massimino, M.L.; Tosello, V.; Marsault, R.; Cantini, M.; Sorrentino, V.; Pozzan, T. Alteration in Calci-um Handling at the Subcellular Level in Mdx Myotubes. J. Biol. Chem. 2001, 276, 4647–4651. [Google Scholar] [CrossRef] [PubMed]
- Fauconnier, J.; Thireau, J.; Reiken, S.; Cassan, C.; Richard, S.; Matecki, S.; Marks, A.R.; Lacampagne, A. Leaky RyR2 Trigger Ventricular Arrhythmias in Duchenne Muscular Dystrophy. Proc. Natl. Acad. Sci. USA 2010, 107, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Koenig, X.; Rubi, L.; Obermair, G.J.; Cervenka, R.; Dang, X.B.; Lukacs, P.; Kummer, S.; Bittner, R.E.; Kubista, H.; Todt, H.; et al. Enhanced Currents through L-Type Calcium Channels in Cardiomyocytes Disturb the Electro-physiology of the Dystrophic Heart. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H564–H573. [Google Scholar] [CrossRef]
- Vandebrouck, C.; Martin, D.; Schoor, M.C.-V.; Debaix, H.; Gailly, P. Involvement of TRPC in the Abnormal Calcium Influx Observed in Dystrophic (Mdx) Mouse Skeletal Muscle Fibers. J. Cell Biol. 2002, 158, 1089–1096. [Google Scholar] [CrossRef]
- Mallouk, N.; Allard, B. Ca2+ Influx and Opening of Ca2+-Activated K+ Channels in Muscle Fibers from Control and Mdx Mice. Biophys. J. 2002, 82, 3012–3021. [Google Scholar] [CrossRef]
- Alderton, J.M.; Steinhardt, R.A. Calcium Influx through Calcium Leak Channels Is Responsible for the Elevated Levels of Calcium-Dependent Proteolysis in Dystrophic Myotubes. J. Biol. Chem. 2000, 275, 9452–9460. [Google Scholar] [CrossRef]
- Guan, X.; Mack, D.L.; Moreno, C.M.; Strande, J.L.; Mathieu, J.; Shi, Y.; Markert, C.D.; Wang, Z.; Liu, G.; Lawlor, M.W.; et al. Dystrophin-Deficient Cardiomyocytes Derived from Human Urine: New Biologic Reagents for Drug Discovery. Stem Cell Res. 2014, 12, 467–480. [Google Scholar] [CrossRef]
- Bauer, T.M.; Murphy, E. Role of Mitochondrial Calcium and the Permeability Transition Pore in Regulating Cell Death. Circ. Res. 2020, 126, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Gavillet, B.; Rougier, J.S.; Domenighetti, A.A.; Behar, R.; Boixel, C.; Ruchat, P.; Lehr, H.A.; Pedrazzini, T.; Abriel, H. Cardiac Sodium Channel Nav1.5 Is Regulated by a Multiprotein Complex Composed of Syntrophins and Dystrophin. Circ. Res. 2006, 99, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Rubi, L.; Koenig, X.; Kubista, H.; Todt, H.; Hilber, K. Decreased Inward Rectifier Potassium Current IK1 in Dystrophin-Deficient Ventricular Cardiomyocytes. Channels 2017, 11, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Bienengraeber, M.; Olson, T.M.; Selivanov, V.A.; Kathmann, E.C.; O’Cochlain, F.; Gao, F.; Karger, A.B.; Ballew, J.D.; Hodgson, D.M.; Zingman, L.V.; et al. ABCC9 Mutations Identified in Human Dilated Cardiomyopathy Disrupt Catalytic KATP Channel Gating. Nat. Genet. 2004, 36, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Farid, T.A.; Nair, K.; Massé, S.; Azam, M.A.; Maguy, A.; Lai, P.F.H.; Umapathy, K.; Dorian, P.; Chauhan, V.; Varró, A.; et al. Role of KATP Channels in the Maintenance of Ventricular Fibrillation in Cardiomyopathic Human Hearts. Circ. Res. 2011, 109, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Caluori, G.; Pribyl, J.; Pesl, M.; Jelinkova, S.; Rotrekl, V.; Skladal, P.; Raiteri, R. Non-Invasive Electromechanical Cell-Based Biosensors for Improved Investigation of 3D Cardiac Models. Biosens. Bioelectron. 2019, 124–125, 129–135. [Google Scholar] [CrossRef]
- Fanchaouy, M.; Polakova, E.; Jung, C.; Ogrodnik, J.; Shirokova, N.; Niggli, E. Pathways of Abnormal Stress-Induced Ca2+ Influx into Dystrophic Mdx Cardiomyocytes. Cell. Calcium 2009, 46, 114–121. [Google Scholar] [CrossRef]
- Katra, R.P.; Laurita, K.R. Cellular Mechanism of Calcium-Mediated Triggered Activity in the Heart. Circ. Res. 2005, 96, 535–542. [Google Scholar] [CrossRef]
- Grant, A.O. Cardiac Ion Channels. Circ. Arrhythm. Electrophysiol. 2009, 2, 185–194. [Google Scholar] [CrossRef]
- Tsurumi, F.; Baba, S.; Yoshinaga, D.; Umeda, K.; Hirata, T.; Takita, J.; Heike, T. The Intracellular Ca2+ Concentration Is Elevated in Cardiomyocytes Differentiated from HiPSCs Derived from a Duchenne Muscular Dystrophy Patient. PLoS ONE 2019, 14, e0213768. [Google Scholar] [CrossRef]
- Judge, D.P.; Kass, D.A.; Thompson, W.R.; Wagner, K.R. Pathophysiology and Therapy of Cardiac Dysfunction in Duchenne Muscular Dystrophy. Am. J. Cardiovasc. Drugs 2011, 11, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Pioner, J.M.; Guan, X.; Klaiman, J.M.; Racca, A.W.; Pabon, L.; Muskheli, V.; Macadangdang, J.; Ferrantini, C.; Hoopmann, M.R.; Moritz, R.L.; et al. Absence of Full-Length Dystrophin Impairs Normal Maturation and Con-traction of Cardiomyocytes Derived from Human-Induced Pluripotent Stem Cells. Cardiovasc. Res. 2020, 116, 368–382. [Google Scholar] [CrossRef]
- Macadangdang, J.; Guan, X.; Smith, A.S.T.; Lucero, R.; Czerniecki, S.; Childers, M.K.; Mack, D.L.; Kim, D.H. Nanopatterned Human IPSC-Based Model of a Dystrophin-Null Cardiomyopathic Phenotype. Cell. Mol. Bioeng. 2015, 8, 320–332. [Google Scholar] [CrossRef]
- Thomas, T.O.; Jefferies, J.L.; Lorts, A.; Anderson, J.B.; Gao, Z.; Woodrow Benson, D.; Hor, K.N.; Cripe, L.H.; Urbina, E.M. Autonomic Dysfunction: A Driving Force for Myocardial Fibrosis in Young Duchenne Muscular Dystrophy Patients? Pediatr. Cardiol. 2015, 36, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Pesl, M.; Acimovic, I.; Pribyl, J.; Hezova, R.; Vilotic, A.; Fauconnier, J.; Vrbsky, J.; Kruzliak, P.; Skladal, P.; Kara, T.; et al. Forced Aggregation and Defined Factors Allow Highly Uniform-Sized Embryoid Bodies and Functional Cardiomyocytes from Human Embryonic and Induced Pluripotent Stem Cells. Heart Vessels 2014, 29, 834–846. [Google Scholar] [CrossRef]
- Pesl, M.; Pribyl, J.; Acimovic, I.; Vilotic, A.; Jelinkova, S.; Salykin, A.; Lacampagne, A.; Dvorak, P.; Meli, A.C.; Skladal, P.; et al. Atomic Force Microscopy Combined with Human Pluripotent Stem Cell Derived Cardiomyo-cytes for Biomechanical Sensing. Biosens. Bioelectron. 2016, 85, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Acimovic, I.; Refaat, M.M.; Moreau, A.; Salykin, A.; Reiken, S.; Sleiman, Y.; Souidi, M.; Přibyl, J.; Kajava, A.V.; Richard, S.; et al. Post-Translational Modifications and Diastolic Calcium Leak Associated to the Novel RyR2-D3638A Mutation Lead to CPVT in Patient-Specific HiPSC-Derived Cardiomyocytes. J. Clin. Med. 2018, 7, 423. [Google Scholar] [CrossRef]
- Greiner, A.M.; Chen, H.; Spatz, J.P.; Kemkemer, R. Cyclic Tensile Strain Controls Cell Shape and Directs Actin Stress Fiber Formation and Focal Adhesion Alignment in Spreading Cells. PLoS ONE 2013, 8, e77328. [Google Scholar] [CrossRef]
- Wada, K.I.; Itoga, K.; Okano, T.; Yonemura, S.; Sasaki, H. Hippo Pathway Regulation by Cell Morphology and Stress Fibers. Development 2011, 138, 3907–3914. [Google Scholar] [CrossRef]
- Pioner, J.M.; Santini, L.; Palandri, C.; Langione, M.; Grandinetti, B.; Querceto, S.; Martella, D.; Mazzantini, C.; Scellini, B.; Giammarino, L.; et al. Calcium Handling Maturation and Adaptation to Increased Substrate Stiffness in Human IPSC-Derived Cardiomyocytes: The Impact of Full-Length Dystrophin Deficiency. Front. Physiol. 2022, 13, 1030920. [Google Scholar] [CrossRef]
- Rybalka, E.; Timpani, C.A.; Cooke, M.B.; Williams, A.D.; Hayes, A. Defects in Mitochondrial ATP Synthesis in Dystrophin-Deficient Mdx Skeletal Muscles May Be Caused by Complex I Insufficiency. PLoS ONE 2014, 9, e115763. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, J.; Trimarco, B.; Iaccarino, G.; Santulli, G. New Insights in Cardiac Calcium Handling and Excita-tion-Contraction Coupling. Adv. Exp. Med. Biol. 2018, 1067, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, J.; Sorriento, D.; Ciccarelli, M.; Del Giudice, C.; Fiordelisi, A.; Napolitano, L.; Trimarco, B.; Iaccarino, G.; Santulli, G. Functional Role of Mitochondria in Arrhythmogenesis. Adv. Exp. Med. Biol. 2017, 982, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, A.; Chamberlain, C.M.; Witthuhn, B.A.; Lowe, D.A.; Ervasti, J.M. Dystrophinopathy-Associated Dysfunction of Krebs Cycle Metabolism. Hum. Mol. Genet. 2019, 28, 942–951. [Google Scholar] [CrossRef]
- Ramos, S.V.; Hughes, M.C.; Delfinis, L.J.; Bellissimo, C.A.; Perry, C.G.R. Mitochondrial Bioenergetic Dysfunction in the D2.Mdx Model of Duchenne Muscular Dystrophy Is Associated with Microtubule Disorganization in Skeletal Muscle. PLoS ONE 2020, 15, e0237138. [Google Scholar] [CrossRef]
- Stapleton, D.I.; Lau, X.; Flores, M.; Trieu, J.; Gehrig, S.M.; Chee, A.; Naim, T.; Lynch, G.S.; Koopman, R. Dysfunctional Muscle and Liver Glycogen Metabolism in Mdx Dystrophic Mice. PLoS ONE 2014, 9, e91514. [Google Scholar] [CrossRef]
- Strakova, J.; Kamdar, F.; Kulhanek, D.; Razzoli, M.; Garry, D.J.; Ervasti, J.M.; Bartolomucci, A.; Townsend, D.W. Integrative Effects of Dystrophin Loss on Metabolic Function of the Mdx Mouse. Sci. Rep. 2018, 8, 13624. [Google Scholar] [CrossRef]
- Afzal, M.Z.; Reiter, M.; Gastonguay, C.; McGivern, J.V.; Guan, X.; Ge, Z.D.; Mack, D.L.; Childers, M.K.; Ebert, A.D.; Strande, J.L. Nicorandil, a Nitric Oxide Donor and ATP-Sensitive Potassium Channel Opener, Protects Against Dystrophin-Deficient Cardiomyopathy. J. Cardiovasc. Pharmacol. Ther. 2016, 21, 549–562. [Google Scholar] [CrossRef]
- Taira, N. Nicorandil as a Hybrid between Nitrates and Potassium Channel Activators. Am. J. Cardiol. 1989, 63, J18–J24. [Google Scholar] [CrossRef]
- Mano, T.; Shinohara, R.; Nagasaka, A.; Nakagawa, H.; Uchimura, K.; Hayashi, R.; Nakano, I.; Tsugawa, T.; Watanabe, F.; Kobayashi, T.; et al. Scavenging Effect of Nicorandil on Free Radicals and Lipid Peroxide in Streptozotocin-Induced Diabetic Rats. Metabolism 2000, 49, 427–431. [Google Scholar] [CrossRef]
- Serizawa, K.I.; Yogo, K.; Aizawa, K.; Tashiro, Y.; Ishizuka, N. Nicorandil Prevents Endothelial Dysfunction Due to Antioxidative Effects via Normalisation of NADPH Oxidase and Nitric Oxide Synthase in Streptozotocin Diabetic Rats. Cardiovasc. Diabetol. 2011, 10, 105. [Google Scholar] [CrossRef]
- Gartz, M.; Darlington, A.; Afzal, M.Z.; Strande, J.L. Exosomes Exert Cardioprotection in Dystrophin-Deficient Cardiomyocytes via ERK1/2-P38/MAPK Signaling. Sci. Rep. 2018, 8, 16519. [Google Scholar] [CrossRef]
- Smaili, S.S.; Hsu, Y.T.; Sanders, K.M.; Russell, J.T.; Youle, R.J. Bax Translocation to Mitochondria Subsequent to a Rapid Loss of Mitochondrial Membrane Potential. Cell Death Differ. 2001, 8, 909–920. [Google Scholar] [CrossRef]
- Wada, T.; Penninger, J.M. Mitogen-Activated Protein Kinases in Apoptosis Regulation. Oncogene 2004, 23, 2838–2849. [Google Scholar] [CrossRef]
- Gartz, M.; Beatka, M.; Prom, M.J.; Strande, J.L.; Lawlor, M.W. Cardiomyocyte-Produced MiR-339-5p Mediates Pa-thology in Duchenne Muscular Dystrophy Cardiomyopathy. Hum. Mol. Genet. 2021, 30, 2347–2361. [Google Scholar] [CrossRef]
- Ibrahim, A.G.E.; Cheng, K.; Marbán, E. Exosomes as Critical Agents of Cardiac Regeneration Triggered by Cell Therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef]
- Stahl, P.D.; Raposo, G. Exosomes and Extracellular Vesicles: The Path Forward. Essays Biochem. 2018, 62, 119–124. [Google Scholar] [CrossRef]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as Intercellular Signaling Organelles Involved in Health and Disease: Basic Science and Clinical Applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef]
- Bier, A.; Berenstein, P.; Kronfeld, N.; Morgoulis, D.; Ziv-Av, A.; Goldstein, H.; Kazimirsky, G.; Cazacu, S.; Meir, R.; Popovtzer, R.; et al. Placenta-Derived Mesenchymal Stromal Cells and Their Exosomes Exert Therapeutic Effects in Duchenne Muscular Dystrophy. Biomaterials 2018, 174, 67–78. [Google Scholar] [CrossRef]
- Rogers, R.G.; Fournier, M.; Sanchez, L.; Ibrahim, A.G.; Aminzadeh, M.A.; Lewis, M.I.; Marbán, E. Disease-Modifying Bioactivity of Intravenous Cardiosphere-Derived Cells and Exosomes in Mdx Mice. J. Clin. Investig. 2019, 4, 130202. [Google Scholar] [CrossRef]
- Jansson, M.D.; Damas, N.D.; Lees, M.; Jacobsen, A.; Lund, A.H. MiR-339-5p Regulates the P53 Tumor-Suppressor Pathway by Targeting MDM2. Oncogene 2015, 34, 1908–1918. [Google Scholar] [CrossRef]
- Duelen, R.; Costamagna, D.; Gilbert, G.; De Waele, L.; Goemans, N.; Desloovere, K.; Verfaillie, C.M.; Sipido, K.R.; Buyse, G.M.; Sampaolesi, M. Human IPSC Model Reveals a Central Role for NOX4 and Oxidative Stress in Duchenne Cardiomyopathy. Stem Cell Rep. 2022, 17, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Vermot, A.; Petit-Härtlein, I.; Smith, S.M.E.; Fieschi, F. NADPH Oxidases (NOX): An Overview from Discovery, Molecular Mechanisms to Physiology and Pathology. Antioxidants 2021, 10, 890. [Google Scholar] [CrossRef]
- Ago, T.; Kuroda, J.; Pain, J.; Fu, C.; Li, H.; Sadoshima, J. Upregulation of Nox4 by Hypertrophic Stimuli Promotes Apoptosis and Mitochondrial Dysfunction in Cardiac Myocytes. Circ. Res. 2010, 106, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Perino, A.; Ghigo, A.; Hirsch, E.; Shah, A.M. NADPH Oxidases in Heart Failure: Poachers or Game-keepers? Antioxid. Redox Signal. 2013, 18, 1024–1041. [Google Scholar] [CrossRef]
- Spurney, C.F.; Knoblach, S.; Pistilli, E.E.; Nagaraju, K.; Martin, G.R.; Hoffman, E.P. Dystrophin-Deficient Cardio-myopathy in Mouse: Expression of Nox4 and Lox Are Associated with Fibrosis and Altered Functional Parameters in the Heart. Neuromuscul. Disord. 2008, 18, 371–381. [Google Scholar] [CrossRef]
- Varga, Z.V.; Pipicz, M.; Baán, J.A.; Baranyai, T.; Koncsos, G.; Leszek, P.; Kusmierczyk, M.; Sánchez-Cabo, F.; García-Pavía, P.; Brenner, G.J.; et al. Alternative Splicing of NOX4 in the Failing Human Heart. Front. Physiol. 2017, 8, 935. [Google Scholar] [CrossRef]
- Willi, L.; Abramovich, I.; Fernandez-Garcia, J.; Agranovich, B.; Shulman, M.; Milman, H.; Baskin, P.; Eisen, B.; Michele, D.E.; Arad, M.; et al. Bioenergetic and Metabolic Impairments in Induced Pluripotent Stem Cell-Derived Cardiomyocytes Generated from Duchenne Muscular Dystrophy Patients. Int. J. Mol. Sci. 2022, 23, 9808. [Google Scholar] [CrossRef]
- Schram, G.; Fournier, A.; Leduc, H.; Dahdah, N.; Therien, J.; Vanasse, M.; Khairy, P. All-Cause Mortality and Cardiovascular Outcomes with Prophylactic Steroid Therapy in Duchenne Muscular Dystrophy. J. Am. Coll. Cardiol. 2013, 61, 948–954. [Google Scholar] [CrossRef]
- Bach, J.R.; Martinez, D.; Saulat, B. Duchenne Muscular Dystrophy: The Effect of Glucocorticoids on Ventilator Use and Ambulation. Am. J. Phys. Med. Rehabil. 2010, 89, 620–624. [Google Scholar] [CrossRef]
- Eagle, M.; Bourke, J.; Bullock, R.; Gibson, M.; Mehta, J.; Giddings, D.; Straub, V.; Bushby, K. Managing Duchenne Muscular Dystrophy--the Additive Effect of Spinal Surgery and Home Nocturnal Ventilation in Improving Survival. Neuromuscul. Disord. 2007, 17, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi-Ikeda, M.; Takeshima, Y.; Lee, T.; Nishiyama, M.; Awano, H.; Yagi, M.; Unzaki, A.; Nozu, K.; Nishio, H.; Matsuo, M.; et al. Next-Generation Sequencing Discloses a Nonsense Mutation in the Dystrophin Gene from Long Preserved Dried Umbilical Cord and Low-Level Somatic Mosaicism in the Proband Mother. J. Hum. Genet. 2016, 61, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Voit, T.; Neuen-Jacob, E.; Mahler, V.; Jauch, A.; Cremer, M. Somatic Mosaicism for a Deletion of the Dystrophin Gene in a Carrier of Becker Muscular Dystrophy. Eur. J. Pediatr. 1992, 151, 112–116. [Google Scholar] [CrossRef]
- Kazuki, Y.; Hiratsuka, M.; Takiguchi, M.; Osaki, M.; Kajitani, N.; Hoshiya, H.; Hiramatsu, K.; Yoshino, T.; Kazuki, K.; Ishihara, C.; et al. Complete Genetic Correction of IPS Cells from Duchenne Muscular Dystrophy. Mol. Ther. 2010, 18, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Zatti, S.; Martewicz, S.; Serena, E.; Uno, N.; Giobbe, G.; Kazuki, Y.; Oshimura, M.; Elvassore, N. Complete Restoration of Multiple Dystrophin Isoforms in Genetically Corrected Duchenne Muscular Dystrophy Patient–Derived Cardiomyocytes. Mol. Ther. Methods Clin. Dev. 2014, 1, 1. [Google Scholar] [CrossRef]
- Choi, I.Y.; Lim, H.; Estrellas, K.; Mula, J.; Cohen, T.V.; Zhang, Y.; Donnelly, C.J.; Richard, J.-P.; Kim, Y.J.; Kim, H.; et al. Concordant but Varied Phenotypes among Duchenne Muscular Dystrophy Patient-Specific Myoblasts De-rived Using a Human IPSC-Based Model. Cell Rep. 2016, 15, 2301–2312. [Google Scholar] [CrossRef]
- Dick, E.; Kalra, S.; Anderson, D.; George, V.; Ritso, M.; Laval, S.H.; Barresi, R.; Aartsma-Rus, A.; Lochmüller, H.; Denning, C. Exon Skipping and Gene Transfer Restore Dystrophin Expression in Human Induced Pluripotent Stem Cells-Cardiomyocytes Harboring DMD Mutations. Stem Cells Dev. 2013, 22, 2714–2724. [Google Scholar] [CrossRef]
- Howard, Z.M.; Dorn, L.E.; Lowe, J.; Gertzen, M.D.; Ciccone, P.; Rastogi, N.; Odom, G.L.; Accornero, F.; Chamber-lain, J.S.; Rafael-Fortney, J.A. Micro-Dystrophin Gene Therapy Prevents Heart Failure in an Improved Duchenne Muscular Dystrophy Cardiomyopathy Mouse Model. J. Clin. Investig. 2021, 6, 146511. [Google Scholar] [CrossRef]
- Oshimura, M.; Uno, N.; Kazuki, Y.; Katoh, M.; Inoue, T. A Pathway from Chromosome Transfer to Engineering Resulting in Human and Mouse Artificial Chromosomes for a Variety of Applications to Bio-Medical Challenges. Chromosome Res. 2015, 23, 111–133. [Google Scholar] [CrossRef]
- Colella, P.; Ronzitti, G.; Mingozzi, F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018, 8, 87–104. [Google Scholar] [CrossRef]
- Kouprina, N.; Larionov, V. Recent Advances in Chromosome Engineering. Chromosome Res. 2015, 23, 1–5. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Ben Jehuda, R.; Shemer, Y.; Binah, O. Genome Editing in Induced Pluripotent Stem Cells Using CRISPR/Cas9. Stem Cell Rev. Rep. 2018, 14, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Paix, A.; Folkmann, A.; Goldman, D.H.; Kulaga, H.; Grzelak, M.J.; Rasoloson, D.; Paidemarry, S.; Green, R.; Reed, R.R.; Seydoux, G. Precision Genome Editing Using Synthesis-Dependent Repair of Cas9-Induced DNA Breaks. Proc. Natl. Acad. Sci. USA 2017, 114, E10745–E10754. [Google Scholar] [CrossRef]
- Yin, H.; Kauffman, K.J.; Anderson, D.G. Delivery Technologies for Genome Editing. Nat. Rev. Drug. Discov. 2017, 16, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Murugan, K.; Babu, K.; Sundaresan, R.; Rajan, R.; Sashital, D.G. The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol. Cell 2017, 68, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Badran, A.H.; Liu, D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017, 168, 20–36. [Google Scholar] [CrossRef]
- Ando, D.; Meyer, K. Gene Editing: Regulatory and Translation to Clinic. Hematol. Oncol. Clin. N. Am. 2017, 31, 797–808. [Google Scholar] [CrossRef]
- Eid, A.; Mahfouz, M.M. Genome Editing: The Road of CRISPR/Cas9 from Bench to Clinic. Exp. Mol. Med. 2016, 48, e265. [Google Scholar] [CrossRef]
- Smith, A.J.; Carter, S.P.; Kennedy, B.N. Genome Editing: The Breakthrough Technology for Inherited Retinal Disease? Expert Opin. Biol. Ther. 2017, 17, 1245–1254. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE Nuclease Architecture for Efficient Genome Editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Hockemeyer, D.; Wang, H.; Kiani, S.; Lai, C.S.; Gao, Q.; Cassady, J.P.; Cost, G.J.; Zhang, L.; Santiago, Y.; Miller, J.C.; et al. Genetic Engineering of Human Pluripotent Cells Using TALE Nucleases. Nat. Biotechnol. 2011, 29, 731–734. [Google Scholar] [CrossRef]
- Li, T.; Huang, S.; Zhao, X.; Wright, D.A.; Carpenter, S.; Spalding, M.H.; Weeks, D.P.; Yang, B. Modularly Assembled Designer TAL Effector Nucleases for Targeted Gene Knockout and Gene Replacement in Eukaryotes. Nucleic Acids Res. 2011, 39, 6315–6325. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-Fidelity CRISPR-Cas9 Nucleases with No Detectable Genome-Wide off-Target Effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A High-Fidelity Cas9 Mutant Delivered as a Ribonucleoprotein Complex Enables Ef-ficient Gene Editing in Human Hematopoietic Stem and Progenitor Cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Repessé, Y.; Peyron, I.; Dimitrov, J.D.; Dasgupta, S.; Moshai, E.F.; Costa, C.; Borel-Derlon, A.; Guillet, B.; D’Oiron, R.; Aouba, A.; et al. Development of Inhibitory Antibodies to Therapeutic Factor VIII in Severe Hemophilia A Is Associated with Microsatellite Polymorphisms in the HMOX1 Promoter. Haematologica 2013, 98, 1650–1655. [Google Scholar] [CrossRef]
- Young, C.S.; Hicks, M.R.; Ermolova, N.V.; Nakano, H.; Jan, M.; Younesi, S.; Karumbayaram, S.; Kumagai-Cresse, C.; Wang, D.; Zack, J.A.; et al. A Single CRISPR-Cas9 Deletion Strategy That Targets the Majority of DMD Patients Restores Dystrophin Function in HiPSC-Derived Muscle Cells. Cell Stem Cell 2016, 18, 533–540. [Google Scholar] [CrossRef]
- Ma, P.; Zhang, S.; Zhang, H.; Fang, S.; Dong, Y.; Zhang, Y.; Hao, W.; Wu, S.; Zhao, Y. Comprehensive Genetic Characteristics of Dystrophinopathies in China. Orphanet J. Rare Dis. 2018, 13, 109. [Google Scholar] [CrossRef]
- Chen, C.; Ma, H.; Zhang, F.; Chen, L.; Xing, X.; Wang, S.; Zhang, X.; Luo, Y. Screening of Duchenne Muscular Dystrophy (DMD) Mutations and Investigating Its Mutational Mechanism in Chinese Patients. PLoS ONE 2014, 9, e108038. [Google Scholar] [CrossRef] [PubMed]
- Monaco, A.P.; Bertelson, C.J.; Liechti-Gallati, S.; Moser, H.; Kunkel, L.M. An Explanation for the Phenotypic Differences between Patients Bearing Partial Deletions of the DMD Locus. Genomics 1988, 2, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Cacchiarelli, D.; Incitti, T.; Martone, J.; Cesana, M.; Cazzella, V.; Santini, T.; Sthandier, O.; Bozzoni, I. MiR-31 Modulates Dystrophin Expression: New Implications for Duchenne Muscular Dystrophy Therapy. EMBO Rep. 2011, 12, 136–141. [Google Scholar] [CrossRef]
- Guilbaud, M.; Gentil, C.; Peccate, C.; Gargaun, E.; Holtzmann, I.; Gruszczynski, C.; Falcone, S.; Mamchaoui, K.; Ben Yaou, R.; Leturcq, F.; et al. MiR-708-5p and MiR-34c-5p Are Involved in NNOS Regulation in Dystrophic Context. Skelet. Muscle 2018, 8, 15. [Google Scholar] [CrossRef]
- Zhang, Y.; Long, C.; Li, H.; McAnally, J.R.; Baskin, K.K.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. CRISPR-Cpf1 Correction of Muscular Dystrophy Mutations in Human Cardiomyocytes and Mice. Sci. Adv. 2017, 3, e1602814. [Google Scholar] [CrossRef] [PubMed]
- Cirak, S.; Arechavala-Gomeza, V.; Guglieri, M.; Feng, L.; Torelli, S.; Anthony, K.; Abbs, S.; Garralda, M.E.; Bourke, J.; Wells, D.J.; et al. Exon Skipping and Dystrophin Restoration in Patients with Duchenne Muscular Dystrophy after Systemic Phosphorodiamidate Morpholino Oligomer Treatment: An Open-Label, Phase 2, Dose-Escalation Study. Lancet 2011, 378, 595–605. [Google Scholar] [CrossRef]
- Kyrychenko, V.; Kyrychenko, S.; Tiburcy, M.; Shelton, J.M.; Long, C.; Schneider, J.W.; Zimmermann, W.-H.; Bassel-Duby, R.; Olson, E.N. Functional Correction of Dystrophin Actin Binding Domain Mutations by Genome Editing. J. Clin. Investig. 2017, 2, e95918. [Google Scholar] [CrossRef]
- Long, C.; Li, H.; Tiburcy, M.; Rodriguez-Caycedo, C.; Kyrychenko, V.; Zhou, H.; Zhang, Y.; Min, Y.-L.; Shelton, J.M.; Mammen, P.P.A.; et al. Correction of Diverse Muscular Dystrophy Mutations in Human Engineered Heart Muscle by Single-Site Genome Editing. Sci. Adv. 2018, 4, eaap9004. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Fokkema, I.; Verschuuren, J.; Ginjaar, I.; van Deutekom, J.; van Ommen, G.-J.; den Dunnen, J.T. Theoretic Applicability of Antisense-Mediated Exon Skipping for Duchenne Muscular Dystrophy Mutations. Hum. Mutat. 2009, 30, 293–299. [Google Scholar] [CrossRef]
- Yuan, J.; Ma, Y.; Huang, T.; Chen, Y.; Peng, Y.; Li, B.; Li, J.; Zhang, Y.; Song, B.; Sun, X.; et al. Genetic Modulation of RNA Splicing with a CRISPR-Guided Cytidine Deaminase. Mol. Cell 2018, 72, 380–394.e7. [Google Scholar] [CrossRef]
- Hess, G.T.; Frésard, L.; Han, K.; Lee, C.H.; Li, A.; Cimprich, K.A.; Montgomery, S.B.; Bassik, M.C. Directed Evolution Using DCas9-Targeted Somatic Hypermutation in Mammalian Cells. Nat. Methods 2016, 13, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Park, K.H.; Zhao, L.; Xu, J.; El Refaey, M.; Gao, Y.; Zhu, H.; Ma, J.; Han, R. CRISPR-Mediated Genome Editing Restores Dystrophin Expression and Function in Mdx Mice. Mol. Ther. 2016, 24, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Shiba, N.; Miyazaki, D.; Shiba, Y.; Echigoya, Y.; Yokota, T.; Takizawa, H.; Aoki, Y.; Takeda, S.; Nakamura, A. Amelioration of Intracellular Ca2+ Regulation by Exon-45 Skipping in Duchenne Muscular Dystrophy-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Biochem. Biophys. Res. Commun. 2019, 520, 179–185. [Google Scholar] [CrossRef]
- Chemello, F.; Chai, A.C.; Li, H.; Rodriguez-Caycedo, C.; Sanchez-Ortiz, E.; Atmanli, A.; Mireault, A.A.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Precise Correction of Duchenne Muscular Dystrophy Exon Deletion Mutations by Base and Prime Editing. Sci. Adv. 2021, 7, eabg4910. [Google Scholar] [CrossRef]
- Zhang, Y.; Nishiyama, T.; Li, H.; Huang, J.; Atmanli, A.; Sanchez-Ortiz, E.; Wang, Z.; Mireault, A.A.; Mammen, P.P.A.; Bassel-Duby, R.; et al. A Consolidated AAV System for Single-Cut CRISPR Correction of a Common Duchenne Muscular Dystrophy Mutation. Mol. Ther. Methods Clin. Dev. 2021, 22, 122–132. [Google Scholar] [CrossRef]
- Nelson, C.E.; Wu, Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Castellanos Rivera, R.M.; Collier, J.H.; et al. Long-Term Evaluation of AAV-CRISPR Genome Editing for Duchenne Muscular Dystrophy. Nat. Med. 2019, 25, 427–432. [Google Scholar] [CrossRef]
- Kwon, J.B.; Ettyreddy, A.R.; Vankara, A.; Bohning, J.D.; Devlin, G.; Hauschka, S.D.; Asokan, A.; Gersbach, C.A. In Vivo Gene Editing of Muscle Stem Cells with Adeno-Associated Viral Vectors in a Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther. Methods Clin. Dev. 2020, 19, 320–329. [Google Scholar] [CrossRef]
- Atmanli, A.; Chai, A.C.; Cui, M.; Wang, Z.; Nishiyama, T.; Bassel-Duby, R.; Olson, E.N. Cardiac Myoediting Attenuates Cardiac Abnormalities in Human and Mouse Models of Duchenne Muscular Dystrophy. Circ. Res. 2021, 129, 602–616. [Google Scholar] [CrossRef]
- Pettinato, A.M.; Ladha, F.A.; Mellert, D.J.; Legere, N.; Cohn, R.; Romano, R.; Thakar, K.; Chen, Y.S.; Hinson, J.T. Development of a Cardiac Sarcomere Functional Genomics Platform to Enable Scalable Interrogation of Human TNNT2 Variants. Circulation 2020, 142, 2262–2275. [Google Scholar] [CrossRef]
- Colom, A.; Derivery, E.; Soleimanpour, S.; Tomba, C.; Molin, M.D.; Sakai, N.; González-Gaitán, M.; Matile, S.; Roux, A. A Fluorescent Membrane Tension Probe. Nat. Chem. 2018, 10, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Dangain, J.; Vrbova, G. Muscle Development in Mdx Mutant Mice. Muscle Nerve 1984, 7, 700–704. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).