Structural-Activity Relationship of Rare Ginsenosides from Red Ginseng in the Treatment of Alzheimer’s Disease
Abstract
1. Introduction
2. Results
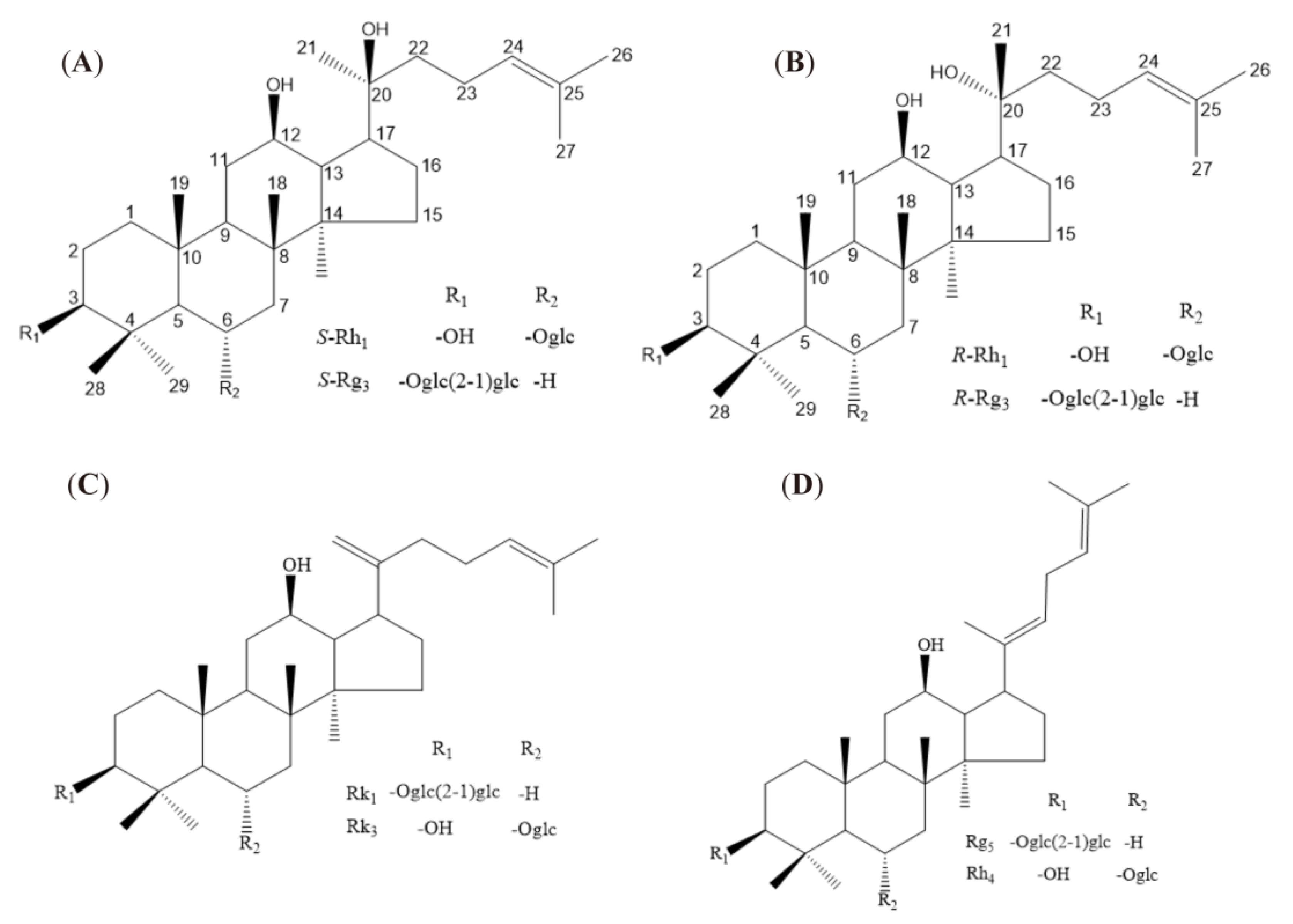
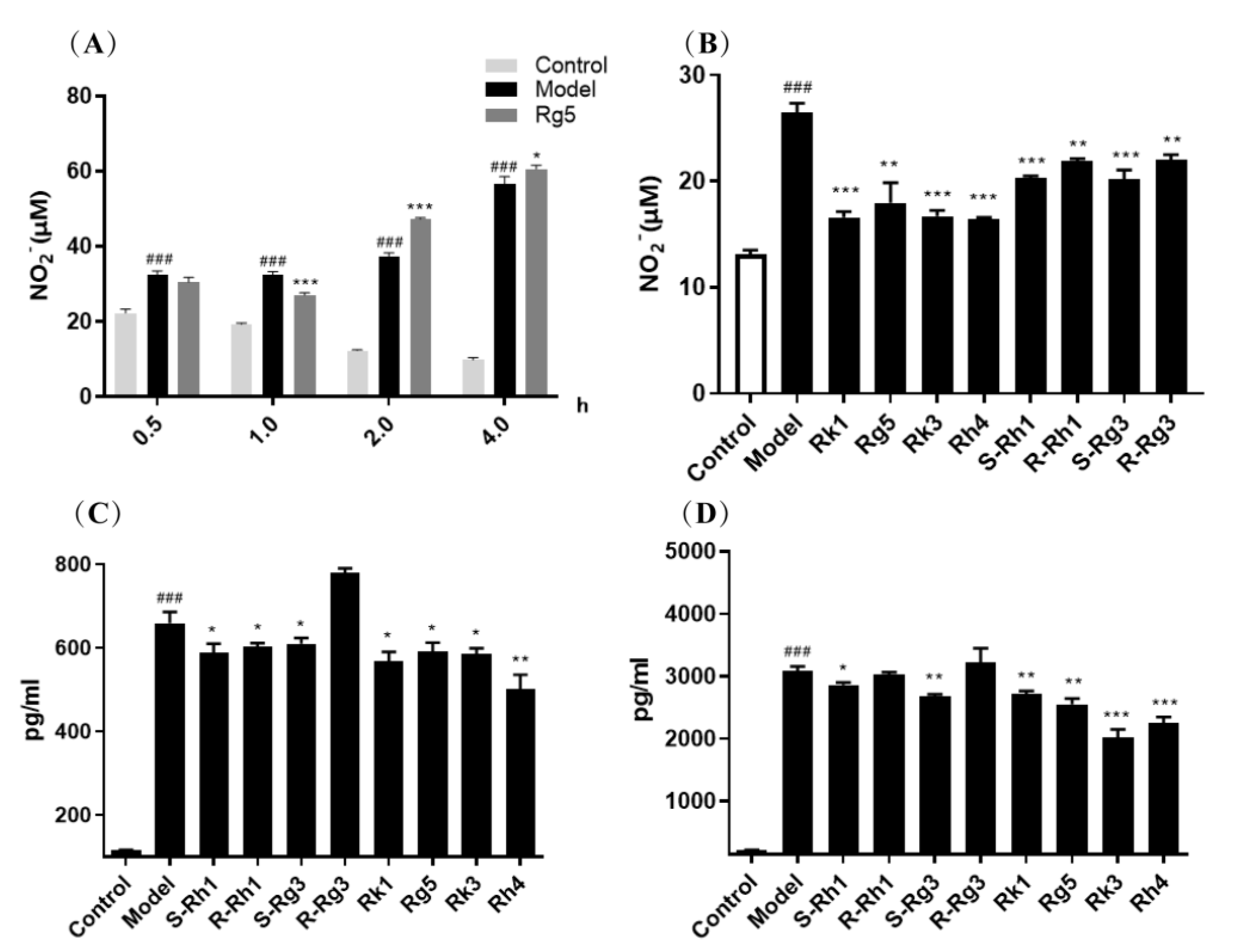
2.1. Protective Effect of Ginsenosides on LPS-Induced Microglial Inflammation
Ginsenosides for Inhibiting Inflammatory Factors Released
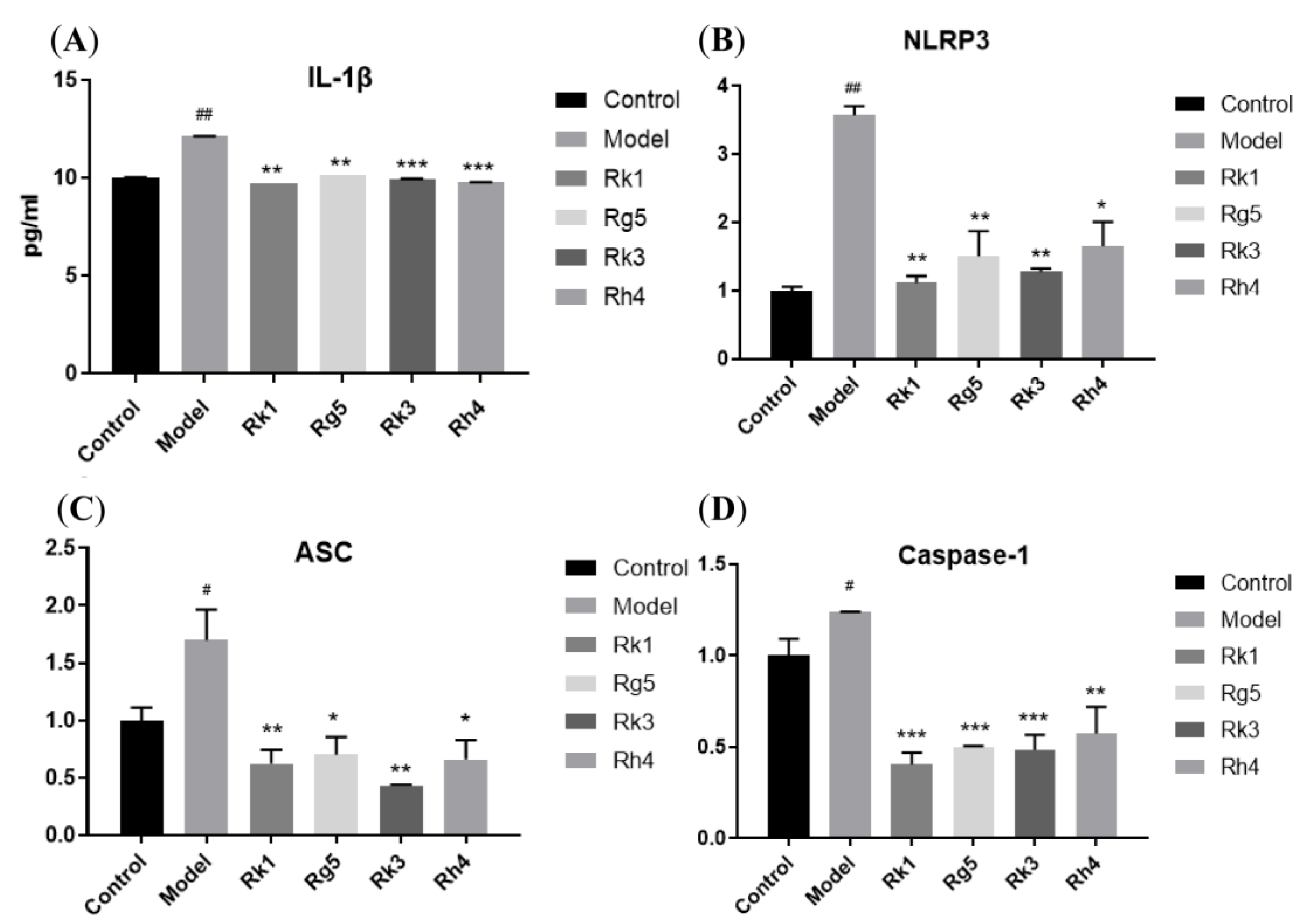
2.2. qRT-PCR Analysis of the Gene Expression of Inflammatory Factors
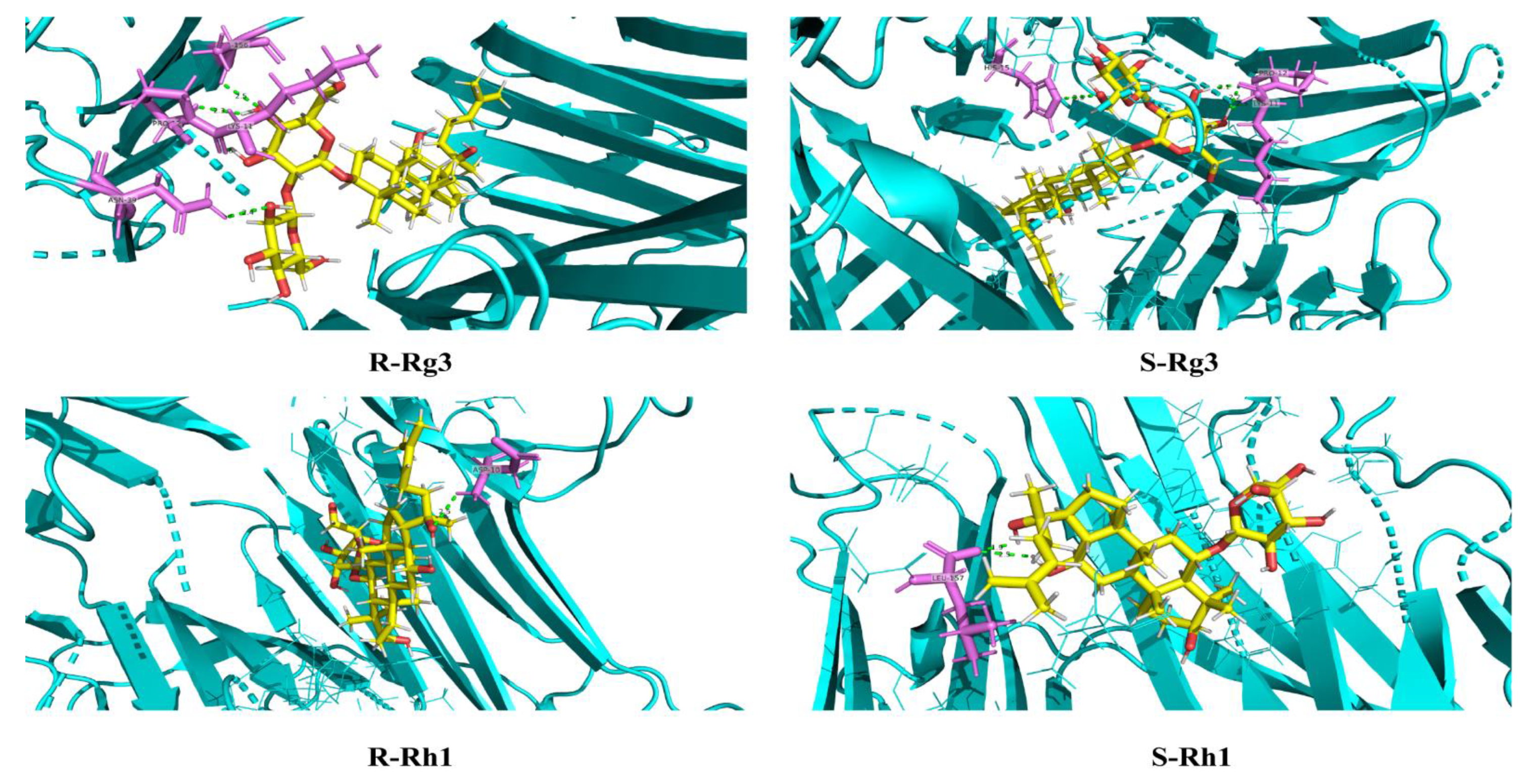
2.3. Molecular Docking Analysis for S and R Epimers of Ginsenosides
2.4. qRT-PCR Analysis of the Protein Expression of AD
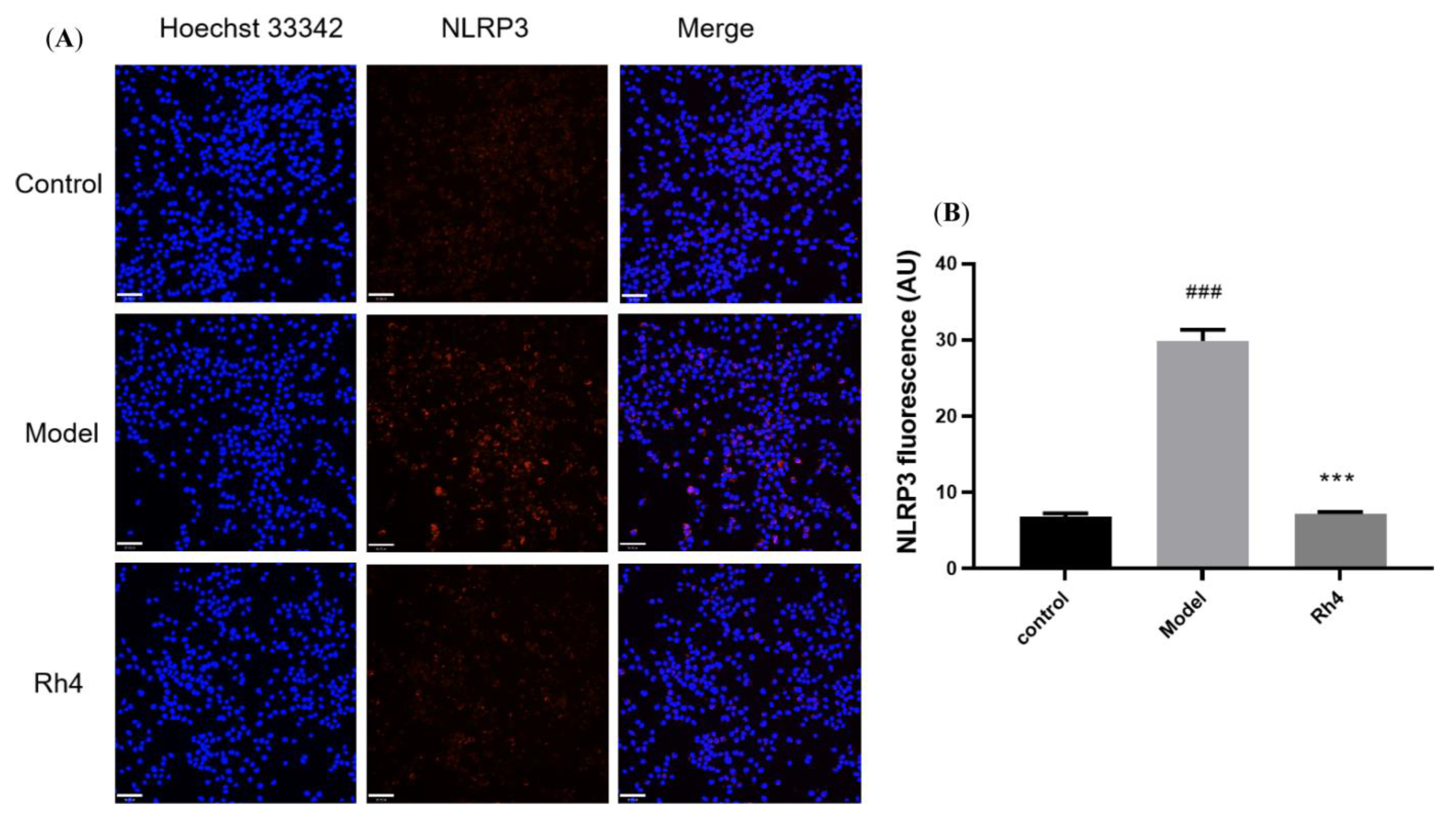
2.5. Laser Confocal Microscope Analysis of the Protein Expression of AD
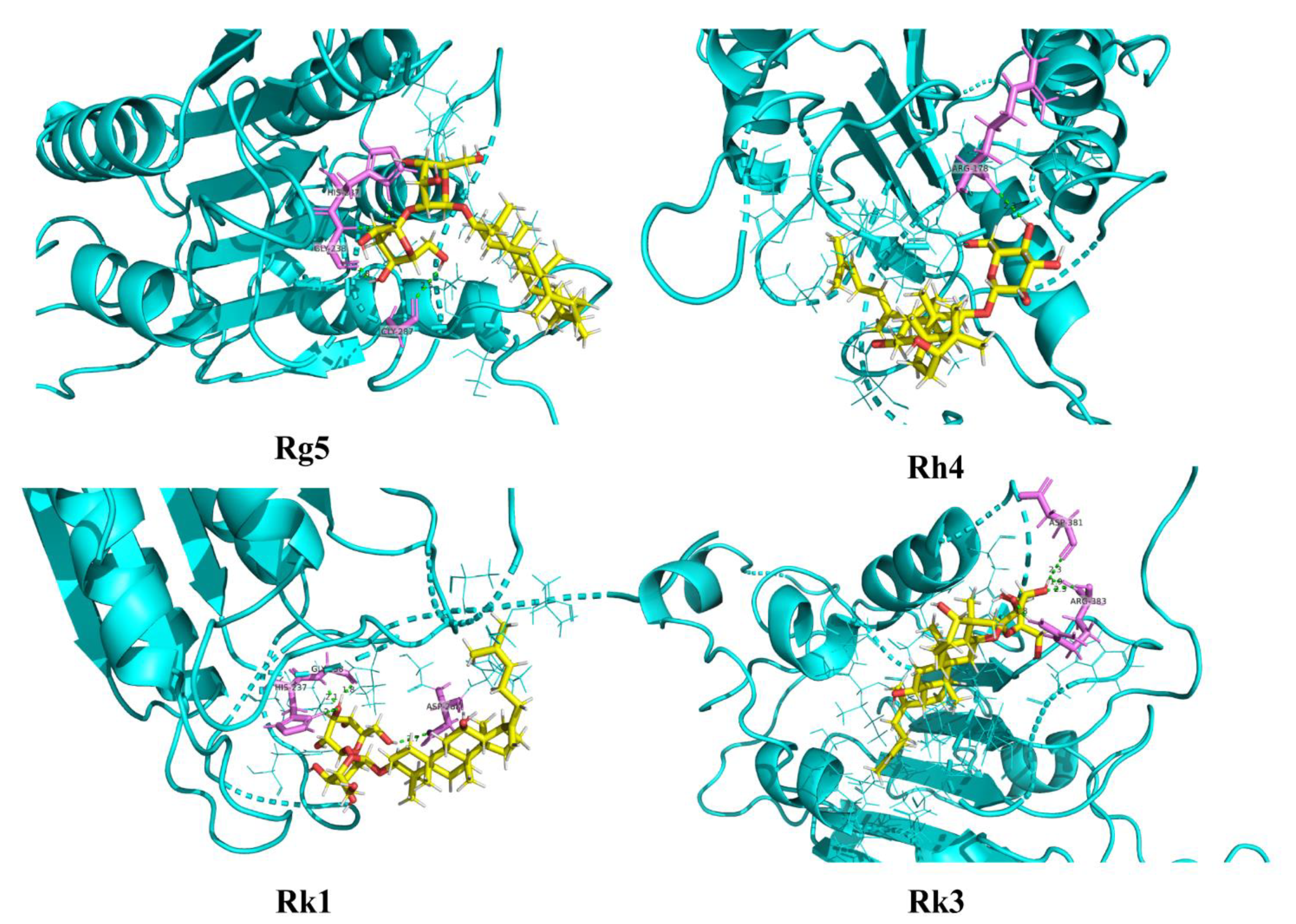
2.6. Molecular Docking Analysis for Ginsenosides to the Potential Proteins of AD
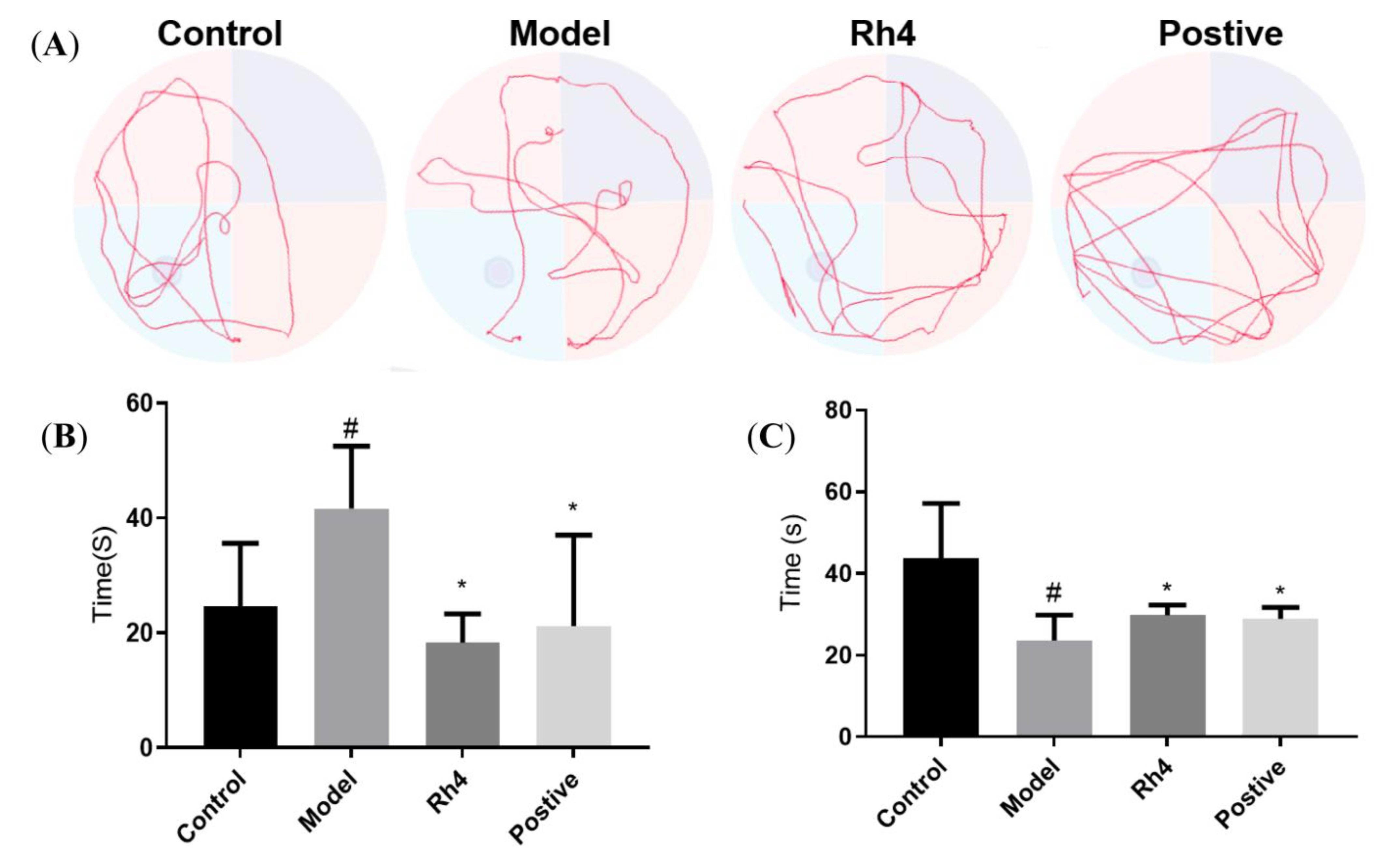
2.7. Rh4 Ameliorates Dementia Conditions in APP/PS1 Mice
Effect of Rh4 on APP/PS1 Mouse Behavior
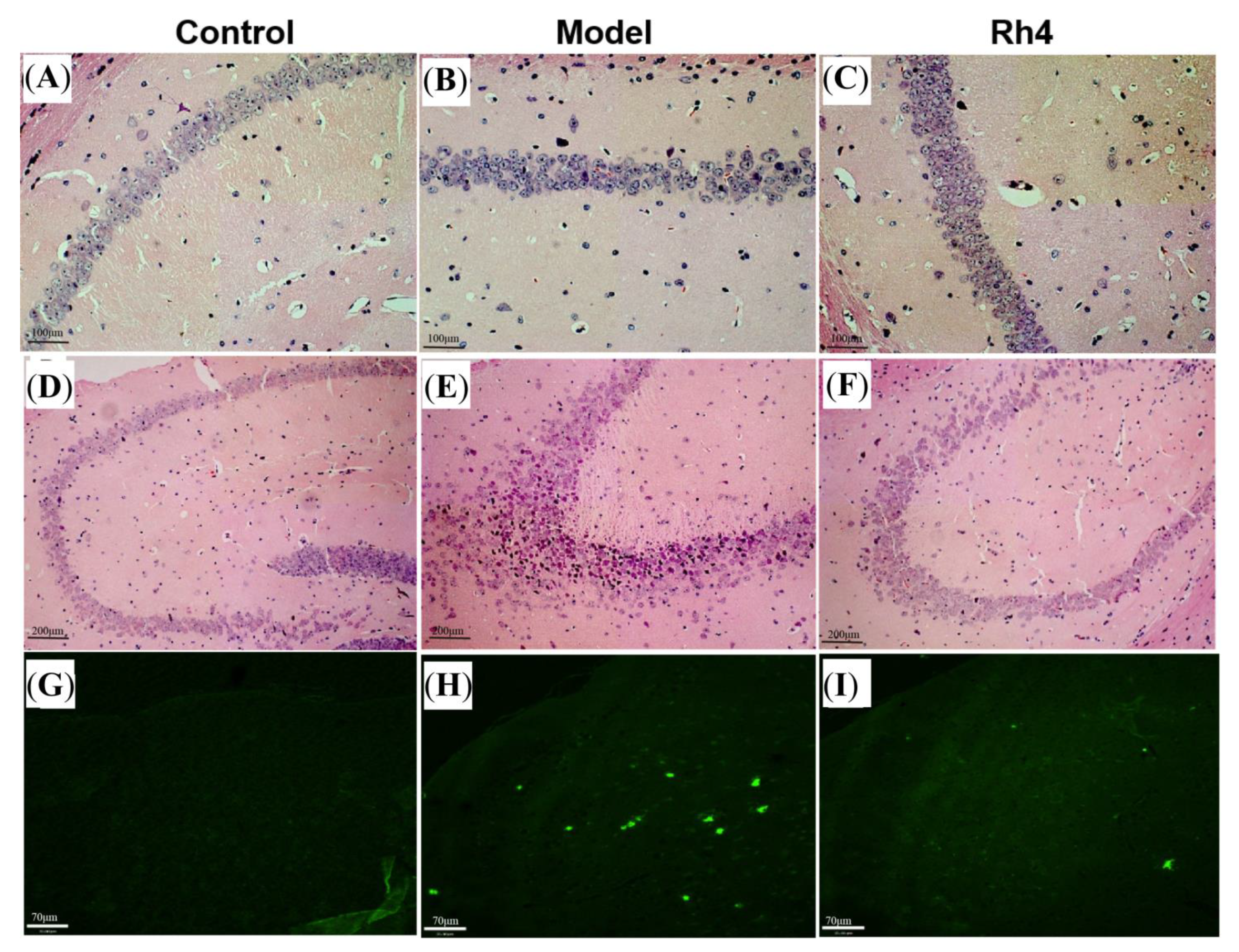
2.8. HE Observed Pathological Changes in the Mouse Brain
2.9. Th-S Staining to Observe the Changes of Aβ in the Mouse Brain
2.10. Rh4 Regulates Multiple Metabolic Pathways in AD Mice and Plays a Therapeutic Role
2.11. Analysis of Metabolic Pathway in Urine Samples
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell Experiment
4.2.1. Cell Culture and Treatment
4.2.2. Biochemical and Elisa Analysis
4.3. Quantitative PCR and Immunocytochemistry (ASC, NLRP3) in BV-2 Cell
4.4. Molecular Docking Analysis
4.5. Animal Experiment
Animal Grouping and Drug Administration
4.6. Morris Water Maze Test, HE Staining, and Thioflavin S Staining
4.7. Metabolomics Study of Urine
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Lee, H.; Kim, J.; Park, J.Y.; Kang, K.S.; Park, J.H.; Hwang, G.S. Processed Panax ginseng, sun ginseng, inhibits the differentiation and proliferation of 3T3-L1 preadipocytes and fat accumulation in Caenorhabditis elegans. J. Ginseng Res. 2017, 41, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.F.; Xu, S.Y.; Zhang, Y.; Zhang, H.; Liu, M.N.; Liu, H.; Gao, Y.; Xue, X.; Xiong, H.; Lin, R.C.; et al. Chemical Comparison of Two Drying Methods of Mountain Cultivated Ginseng by UPLC-QTOF-MS/MS and Multivariate Statistical Analysis. Molecules 2017, 22, 717. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.I.; Kim, J.K.; Kim, J.Y.; Han, M.J.; Kim, D.H. Fermented red ginseng and ginsenoside Rd alleviate ovalbumin-induced allergic rhinitis in mice by suppressing IgE, interleukin-4, and interleukin-5 expression. J. Ginseng Res. 2019, 43, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Oh, S.; Kim, M.J.; Sim, G.S.; Moon, T.W.; Lee, J. Oxidative stability of extracts from red ginseng and puffed red ginseng in bulk oil or oil-in-water emulsion matrix. J. Ginseng Res. 2018, 42, 320–326. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.Q.; Zhou, Y.D.; Hou, J.G.; Liu, Y.; Wang, Y.P.; Gong, X.J.; Lin, X.H.; Jiang, S.; Wang, Z. Rare Ginsenoside 20(R)-Rg3 Inhibits D-Galactose-Induced Liver and Kidney Injury by Regulating Oxidative Stress-Induced Apoptosis. Am. J. Chin. Med. 2020, 48, 1141–1157. [Google Scholar] [CrossRef]
- Qu, L.L.; Zhu, Y.Y.; Liu, Y.N.; Yang, H.X.; Zhu, C.H.; Ma, P.; Deng, J.J.; Fan, D.D. Protective effects of ginsenoside Rk3 against chronic alcohol-induced liver injury in mice through inhibition of inflammation, oxidative stress, and apoptosis. Food Chem. Toxicol. 2019, 126, 277–284. [Google Scholar] [CrossRef]
- Hong, Y.N.; Fan, D.D. Ginsenoside Rk1 induces cell cycle arrest and apoptosis in MDA-MB-231 triple negative breast cancer cells. Toxicology 2019, 418, 22–31. [Google Scholar] [CrossRef]
- Jaeschke, H. Comments on Caspase-Mediated Anti-Apoptotic Effect of Ginsenoside Rg5, a Main Rare Ginsenoside, on Acetaminophen Induced Hepatotoxicity in Mice. J. Agric. Food Chem. 2018, 66, 1732–1733. [Google Scholar] [CrossRef]
- Xu, X.F.; Gao, Y.; Xu, S.Y.; Liu, H.; Xue, X.; Zhang, Y.; Zhang, H.; Liu, M.N.; Xiong, H.; Lin, R.C.; et al. Remarkable impact of steam temperature on ginsenosides transformation from fresh ginseng to red ginseng. J. Ginseng Res. 2018, 42, 277–287. [Google Scholar] [CrossRef]
- Ye, X.-W.; Li, C.-S.; Zhang, H.-X.; Li, Q.; Cheng, S.-Q.; Wen, J.; Wang, X.; Ren, H.-M.; Xia, L.-J.; Wang, X.-X.; et al. Saponins of ginseng products: A review of their transformation in processing. Front. Pharmacol. 2023, 14, 1137. [Google Scholar] [CrossRef]
- Vina, J.; Sanz-Ros, J. Alzheimer’s disease: Only prevention makes sense. Eur. J. Clin. Investig. 2018, 48, e13005. [Google Scholar] [CrossRef]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; Jun, G.; DeStefano, A.L.; Bis, J.C.; Beecham, G.W.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef]
- Yokoyama, J.S.; Wang, Y.; Schork, A.J.; Thompson, W.K.; Karch, C.M.; Cruchaga, C.; McEvoy, L.K.; Witoelar, A.; Chen, C.-H.; Holland, D.; et al. Association Between Genetic Traits for Immune-Mediated Diseases and Alzheimer Disease. JAMA Neurol. 2016, 73, 691–697. [Google Scholar] [CrossRef]
- Gjoneska, E.; Pfenning, A.R.; Mathys, H.; Quon, G.; Kundaje, A.; Tsai, L.H.; Kellis, M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 2015, 518, 365–369. [Google Scholar] [CrossRef]
- Zhang, B.; Gaiteri, C.; Bodea, L.G.; Wang, Z.; McElwee, J.; Podtelezhnikov, A.A.; Zhang, C.S.; Xie, T.; Tran, L.; Dobrin, R.; et al. Integrated Systems Approach Identifies Genetic Nodes and Networks in Late-Onset Alzheimer’s Disease. Cell 2013, 153, 707–720. [Google Scholar] [CrossRef]
- Raj, T.; Rothamel, K.; Mostafavi, S.; Ye, C.; Lee, M.N.; Replogle, J.M.; Feng, T.; Lee, M.; Asinovski, N.; Frohlich, I.; et al. Polarization of the Effects of Autoimmune and Neurodegenerative Risk Alleles in Leukocytes. Science 2014, 344, 519–523. [Google Scholar] [CrossRef]
- Hickman, S.; Izzy, S.; Sen, P.; Morsett, L.; El Khoury, J. Microglia in neurodegeneration. Nat. Neurosci. 2018, 21, 1359–1369. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, X.; Ying, W.H.; Wu, D.H.; Zhong, P. Cryptotanshinone Attenuates Inflammatory Response of Microglia Cells via the Nrf2/HO-1 Pathway. Front. Neurosci. 2019, 13, 852. [Google Scholar] [CrossRef]
- Hansen, D.V.; Hanson, J.E.; Sheng, M. Microglia in Alzheimer’s disease. J. Cell Biol. 2018, 217, 459–472. [Google Scholar] [CrossRef]
- Sarlus, H.; Heneka, M.T. Microglia in Alzheimer’s disease. J. Clin. Investig. 2017, 127, 3240–3249. [Google Scholar] [CrossRef]
- Venegas, C.; Kumar, S.; Franklin, B.S.; Dierkes, T.; Brinkschulte, R.; Tejera, D.; Vieira-Saecker, A.; Schwartz, S.; Santarelli, F.; Kummer, M.P. Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature 2017, 552, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, C.; Li, Y.; Dong, Y.; Wu, C.Y.; Lee, R.H.; Brann, D.W.; Lin, H.W.; Zhang, Q. Long-term exercise pre-training attenuates Alzheimer’s disease-related pathology in a transgenic rat model of Alzheimer’s disease. Geroscience 2022, 44, 1457–1477. [Google Scholar] [CrossRef] [PubMed]
- Filip, T.; Mairinger, S.; Neddens, J.; Sauberer, M.; Flunkert, S.; Stanek, J.; Wanek, T.; Okamura, N.; Langer, O.; Hutter-Paier, B.; et al. Characterization of an APP/tau rat model of Alzheimer’s disease by positron emission tomography and immunofluorescent labeling. Alzheimers Res. 2021, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Joo, I.L.; Lam, W.W.; Oakden, W.; Hill, M.E.; Koletar, M.M.; Morrone, C.D.; Stanisz, G.J.; McLaurin, J.; Stefanovic, B. Early alterations in brain glucose metabolism and vascular function in a transgenic rat model of Alzheimer’s disease. Prog. Neurobiol. 2022, 217, 102327. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Fujita, M.; Itokawa, H.; Tanaka, O.; Ishii, T. Studies on the Constituents of Japanese and Chinese Crude Drugs. Xi. Panaxadiol, a Sapogenin of Ginseng Roots. Chem. Pharm. Bull. 1963, 11, 759–761. [Google Scholar] [CrossRef]
- Popovich, D.G.; Yeo, C.R.; Zhang, W. Ginsenosides derived from Asian (Panax ginseng), American ginseng (Panax quinquefolius) and potential cytoactivity. Int. J. Biomed. Pharm. Sci. 2012, 6, 56–62. [Google Scholar]
- Quan, K.; Liu, Q.; Wan, J.Y.; Zhao, Y.J.; Guo, R.Z.; Alolga, R.N.; Li, P.; Qi, L.W. Rapid preparation of rare ginsenosides by acid transformation and their structure-activity relationships against cancer cells. Sci. Rep. 2015, 5, 8598. [Google Scholar] [CrossRef]
- Wei, X.; Chen, J.; Su, F.; Su, X.; Hu, T.; Hu, S. Stereospecificity of ginsenoside Rg3 in promotion of the immune response to ovalbumin in mice. Int. Immunol. 2012, 24, 465–471. [Google Scholar] [CrossRef]
- Park, M.W.; Ha, J.; Chung, S.H. 20(S)-ginsenoside Rg3 enhances glucose-stimulated insulin secretion and activates AMPK. Biol. Pharm. Bull. 2008, 31, 748–751. [Google Scholar] [CrossRef]
- Wang, Y.-S.; Li, H.; Li, Y.; Zhu, H.; Jin, Y.-H. Identification of natural compounds targeting Annexin A2 with an anti-cancer effect. Protein Cell 2018, 9, 568–579. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Park, J.S.; Jung, J.S.; Kim, D.H.; Kim, H.S. Anti-Inflammatory Effect of Ginsenoside Rg5 in Lipopolysaccharide-Stimulated BV2 Microglial Cells. Int. J. Mol. Sci. 2013, 14, 9820–9833. [Google Scholar] [CrossRef]
- Ying, A.; Yu, Q.T.; Guo, L.; Zhang, W.S.; Liu, J.F.; Li, Y.; Song, H.; Li, P.; Qi, L.W.; Ge, Y.Z.; et al. Structural-Activity Relationship of Ginsenosides from Steamed Ginseng in the Treatment of Erectile Dysfunction. Am. J. Chin. Med. 2018, 46, 137–155. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, Z.; Song, W. NLRP3 inflammasome as a novel therapeutic target for Alzheimer’s disease. Signal Transduct. Target. Ther. 2020, 5, 37. [Google Scholar] [CrossRef]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef]
- Subhramanyam, C.S.; Wang, C.; Hu, Q.D.; Dheen, S.T. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin. Cell Dev. Biol. 2019, 94, 112–120. [Google Scholar] [CrossRef]
- White, C.S.; Lawrence, C.B.; Brough, D.; Rivers-Auty, J. Inflammasomes as therapeutic targets for Alzheimer’s disease. Brain Pathol. 2017, 27, 223–234. [Google Scholar] [CrossRef]
- Ising, C.; Venegas, C.; Zhang, S.S.; Scheiblich, H.; Schmidt, S.V.; Vieira-Saecker, A.; Schwartz, S.; Albasset, S.; McManus, R.M.; Tejera, D.; et al. NLRP3 inflammasome activation drives tau pathology. Nature 2019, 575, 669–673. [Google Scholar] [CrossRef]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef]
- Masumoto, J.; Taniguchi, S.; Ayukawa, K.; Sarvotham, H.; Kishino, T.; Niikawa, N.; Hidaka, E.; Katsuyama, T.; Higuchi, T.; Sagara, J. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 1999, 274, 33835–33838. [Google Scholar] [CrossRef]
- Chung, H.; Vilaysane, A.; Lau, A.; Stahl, M.; Morampudi, V.; Bondzi-Simpson, A.; Platnich, J.M.; Bracey, N.A.; French, M.C.; Beck, P.L.; et al. NLRP3 regulates a non-canonical platform for caspase-8 activation during epithelial cell apoptosis. Cell Death Differ. 2016, 23, 1331–1346. [Google Scholar] [CrossRef]
- Xie, K.; Qin, Q.; Long, Z.; Yang, Y.; Peng, C.; Xi, C.; Li, L.; Wu, Z.; Daria, V.; Zhao, Y.; et al. High-Throughput Metabolomics for Discovering Potential Biomarkers and Identifying Metabolic Mechanisms in Aging and Alzheimer’s Disease. Front. Cell Dev. Biol. 2021, 9, 602887. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Liu, J.; Huang, C.; Liu, J.; Cui, H.; Zhao, B. A Urinary Metabolomics Analysis Based on UPLC-MS and Effects of Moxibustion in APP/PS1 Mice. Curr. Alzheimer Res. 2020, 17, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Akyol, S.; Ugur, Z.; Yilmaz, A.; Ustun, I.; Gorti, S.K.K.; Oh, K.; McGuinness, B.; Passmore, P.; Kehoe, P.G.; Maddens, M.E.; et al. Lipid Profiling of Alzheimer’s Disease Brain Highlights Enrichment in Glycerol(phospho)lipid, and Sphingolipid Metabolism. Cells 2021, 10, 2591. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Yang, Q.; Yu, N.; Cao, Y.; Wang, X.; Wang, Z.; Qiu, W.Y.; Ma, C. Phenylalanine Metabolism Is Dysregulated in Human Hippocampus with Alzheimer’s Disease Related Pathological Changes. J. Alzheimers Dis. 2021, 83, 609–622. [Google Scholar] [CrossRef]
- Sharma, V.K.; Singh, T.G.; Prabhakar, N.K.; Mannan, A. Kynurenine Metabolism and Alzheimer’s Disease: The Potential Targets and Approaches. Neurochem. Res. 2022, 47, 1459–1476. [Google Scholar] [CrossRef]
- Marongiu, R. Accelerated Ovarian Failure as a Unique Model to Study Peri-Menopause Influence on Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 242. [Google Scholar] [CrossRef]
- Xiong, J.; Kang, S.S.; Wang, Z.; Liu, X.; Kuo, T.C.; Korkmaz, F.; Padilla, A.; Miyashita, S.; Chan, P.; Zhang, Z.; et al. FSH blockade improves cognition in mice with Alzheimer’s disease. Nature 2022, 603, 470–476. [Google Scholar] [CrossRef]



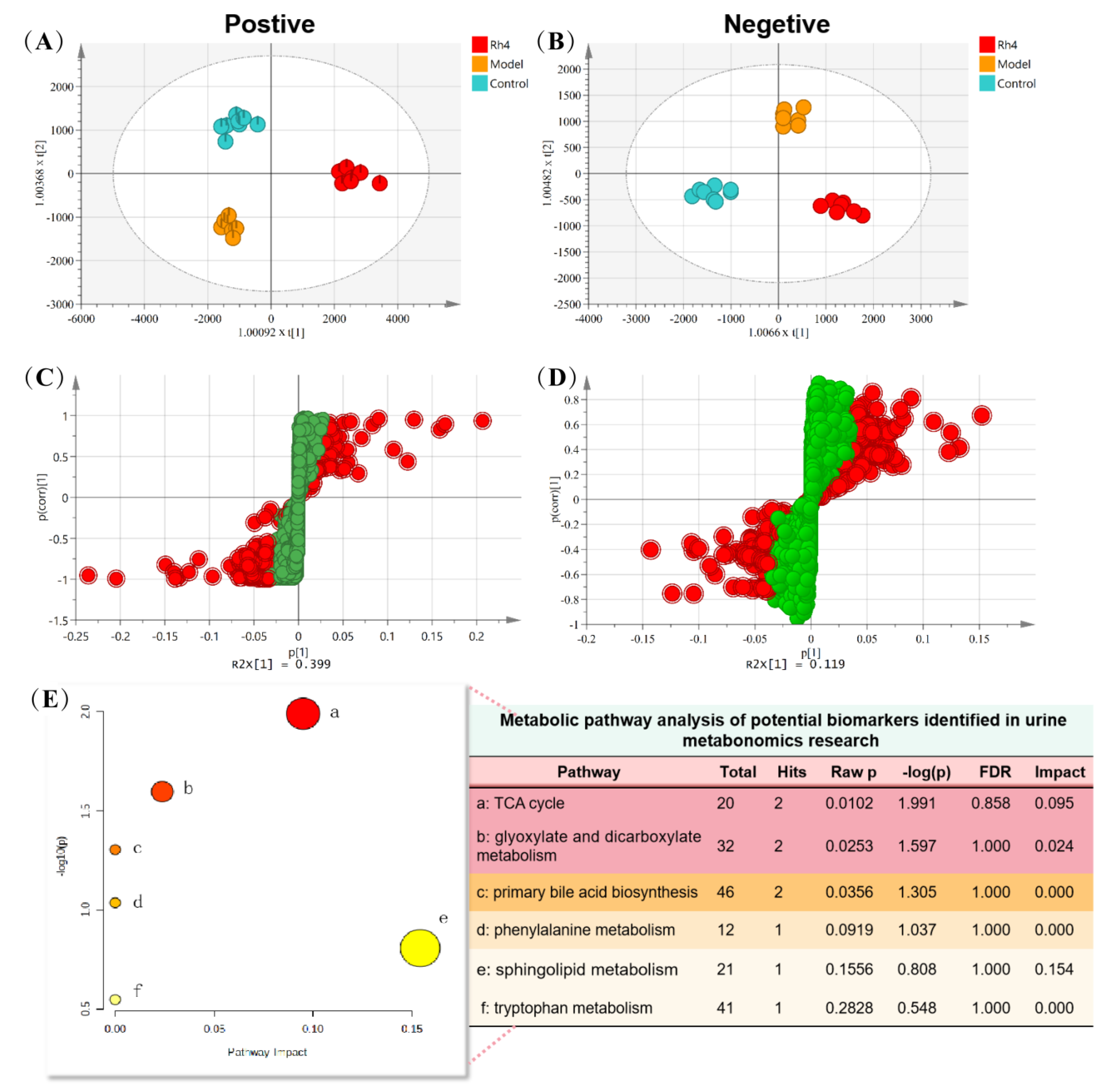
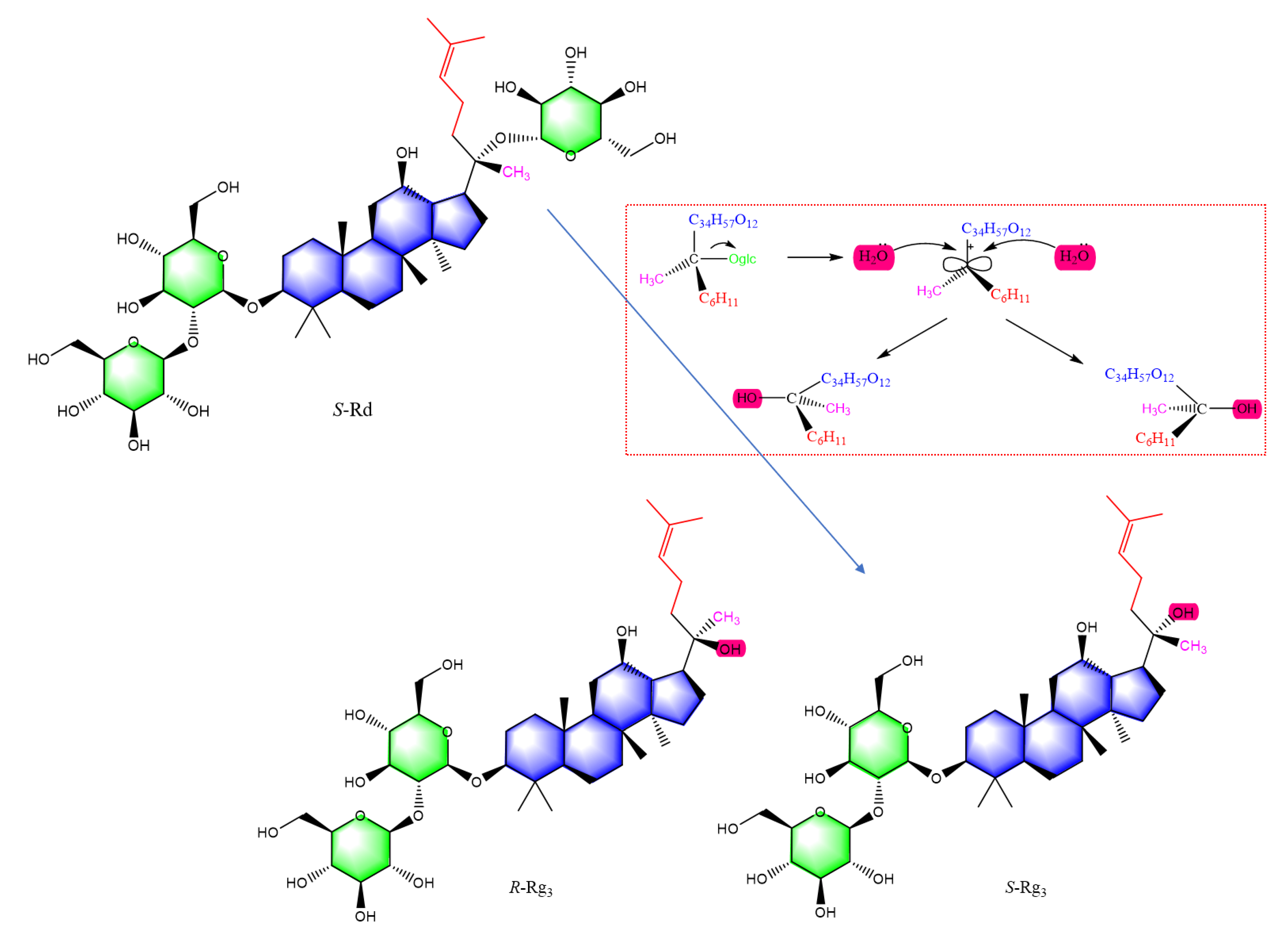
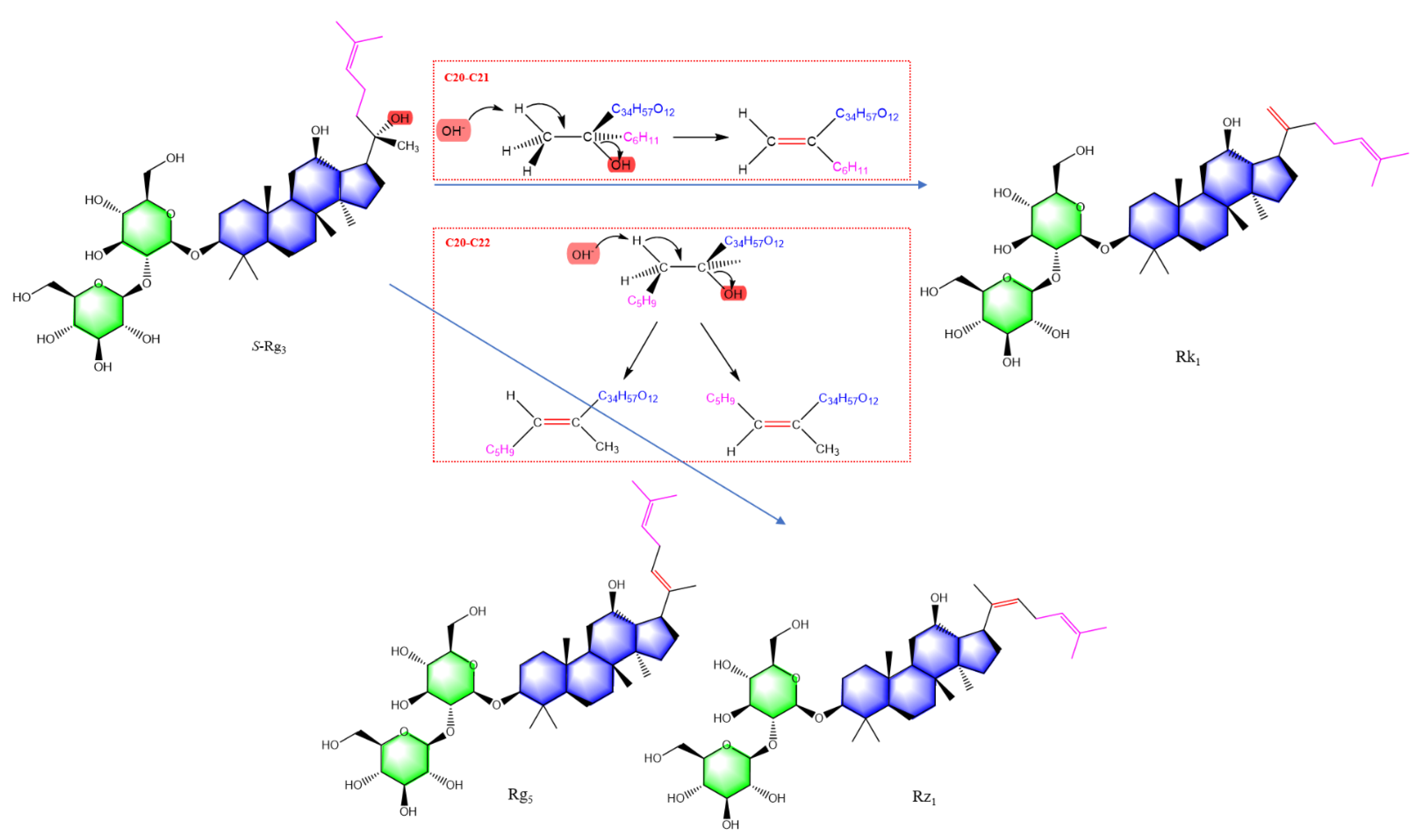
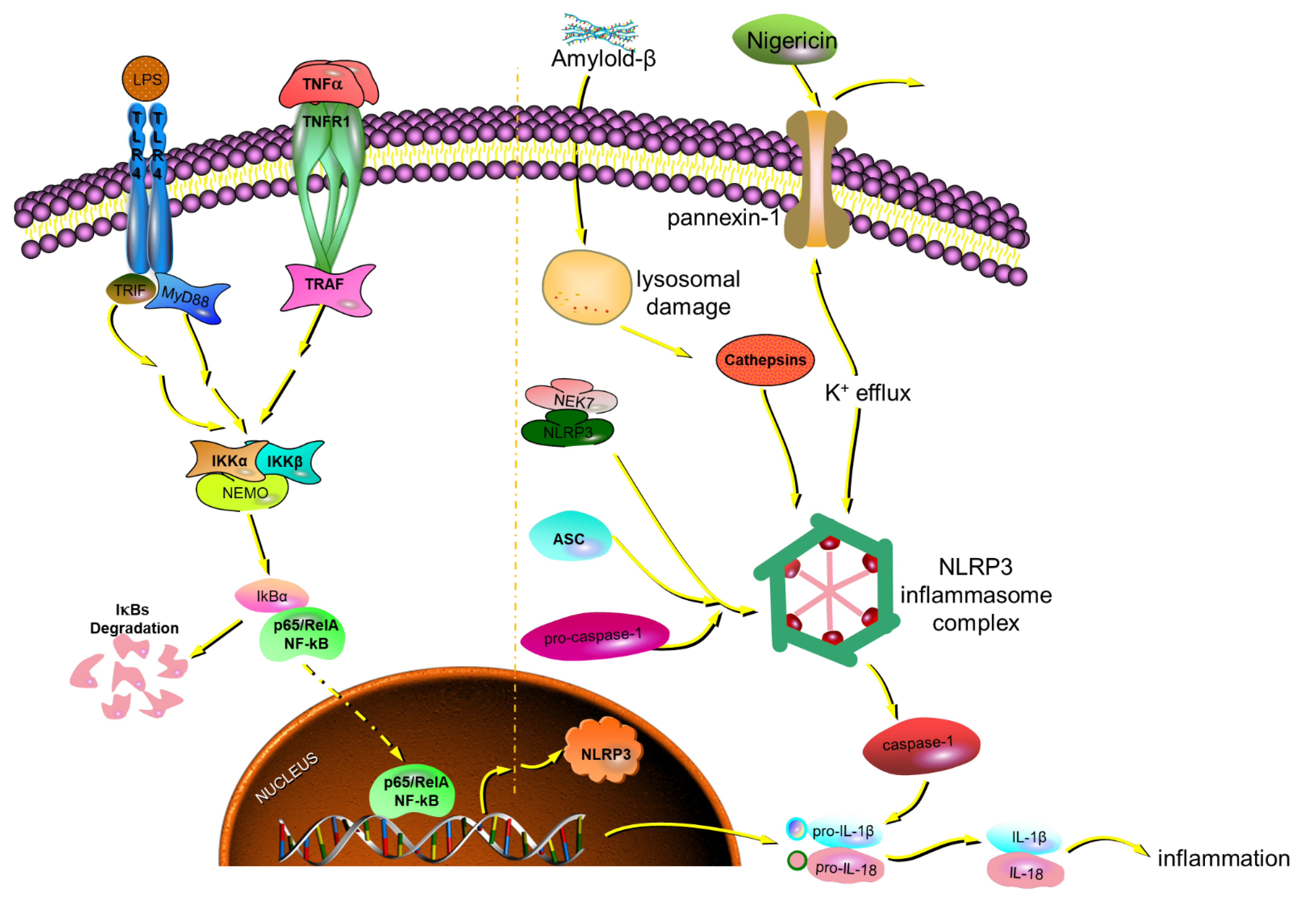
| No. | tR (min) | Precursor Ions and/or Adduct Ions | Exact Mass | Error (ppm) | Formula | Identification |
|---|---|---|---|---|---|---|
| 1 | 0.86 | 173.0076 [M − H]− | 173.0086 | −5.78 | C6H6O6 | cis-Aconitate acid |
| 2 | 0.92 | 191.0193 [M − H]− | 191.0192 | 0.52 | C6H8O7 | Isocitric acid |
| 3 | 0.98 | 254.9802 [M − H]− | 254.9811 | −3.53 | C6H8O9S | Ascorbic acid-2-sulfate |
| 4 | 2.66 | 286.0014 [M + FA-H]− | 286.0021 | −2.62 | C9H7NO5S | Indole-3-carboxilic acid-O-sulphate |
| 5 | 2.81 | 158.0809 [M − H]− | 158.0817 | −5.06 | C7H13NO3 | Valerylglycine |
| 6 | 3.18 | 206.0445 [M − H]− | 206.0453 | −3.88 | C10H9NO4 | 4-(2-Aminophenyl)-2,4-dioxobutanoic acid |
| 7 | 3.19 | 178.0498 [M − H]− | 178.0504 | −3.37 | C9H9NO3 | Hippuric acid |
| 8 | 3.31 | 212.0017 [M − H]− | 212.0018 | −0.47 | C8H7NO4S | Indoxyl sulfate |
| 9 | 3.49 | 222.0788 [M − H]− | 222.0767 | 9.46 | C11H13NO4 | N-Acetyl-L-tyrosine |
| 10 | 3.68 | 283.0818 [M − H]− | 283.0818 | 0.00 | C13H16O7 | p-Cresol glucuronide |
| 11 | 4.91 | 377.1959 [M − H]− | 377.1964 | −1.33 | C21H30O6 | 18-Hydroxycortisol |
| 12 | 8.59 | 407.2787 [M − H]− | 407.2798 | −2.70 | C24H40O5 | Cholic acid |
| 13 | 10.26 | 391.2840 [M − H]− | 391.2849 | −2.30 | C24H40O4 | Chenodeoxycholic acid |
| 14 | 2.21 | 206.0460 [M + H]+ | 206.0453 | 3.40 | C10H7NO4 | Xanthurenic acid |
| 15 | 2.83 | 162.0560 [M + H]+ | 162.0555 | 3.09 | C9H7NO2 | Indole-3-carboxylic acid |
| 16 | 2.84 | 338.0883 [M + H]+ | 338.0876 | 2.07 | C15H15NO8 | 3-Indole carboxylic acid glucuronide |
| 17 | 2.84 | 338.0883 [M + H]+ | 338.0876 | 2.07 | C15H15NO8 | 2,8-Dihydroxyquinoline-beta-D-glucuronide |
| 18 | 3.20 | 164.0705 [M + H]+ | 164.0711 | −3.66 | C9H9NO2 | 3-Methyldioxyindole |
| 19 | 3.81 | 162.0554 [M + H]+ | 162.0555 | −0.62 | C9H7NO2 | 2-Indolecarboxylic acid |
| 20 | 4.90 | 379.2115 [M + H]+ | 379.2121 | −1.48 | C21H30O6 | 18-Hydroxycortisol |
| 21 | 6.32 | 181.0870 [M + H]+ | 181.0864 | 3.31 | C10H12O3 | 3-Methoxybenzenepropanoic acid |
| 22 | 10.46 | 302.3061 [M + H]+ | 302.3059 | 0.66 | C18H39NO2 | Sphinganine |
| 23 | 10.86 | 328.3220 [M + H]+ | 328.3215 | 1.52 | C20H41NO2 | D-erythro-C20-Sphingosine |
| 24 | 11.89 | 330.3385 [M + H]+ | 330.3372 | 3.94 | C20H43NO2 | eicosasphinganine |
| Gene | Forward (5′→3′) | Reverse (3′→5′) | Length |
|---|---|---|---|
| iNOs | GAGCGAGTTGTGGATTGTC | CCAGGAAGTAGGTGAGGG | 133 bp |
| TNF-α | GTGAAGGGAATGGGTGTT | GGTCACTGTCCCAGCATC | 198 bp |
| IL-6 | CCACCAAGAACGATAGTCAA | TTTCCACGATTTCCCAGA | 392 bp |
| IL-1β | TGGGCTGGACTGTTTCTA | ATCAGAGGCAAGGAGGAA | 184 bp |
| NLRP3 | ACCTCCAAGACCACTACGG | CAGCCAGTGAACAGAGCC | 118 bp |
| ASC | TGCCAGGGTCACAGAAGT | CCAGGTCCATCACCAAGTA | 209 bp |
| Caspase-1 | CCCCAGGCAAGCCAAATC | TGAGGGTCCCAGTCAGTCC | 202 bp |
| GAPDH | ATGTACGTAGCCATCCAGGC | AGGAAGGAAGGCTGGAAGAG | 420 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, X.; Zhang, H.; Li, Q.; Ren, H.; Xu, X.; Li, X. Structural-Activity Relationship of Rare Ginsenosides from Red Ginseng in the Treatment of Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 8625. https://doi.org/10.3390/ijms24108625
Ye X, Zhang H, Li Q, Ren H, Xu X, Li X. Structural-Activity Relationship of Rare Ginsenosides from Red Ginseng in the Treatment of Alzheimer’s Disease. International Journal of Molecular Sciences. 2023; 24(10):8625. https://doi.org/10.3390/ijms24108625
Chicago/Turabian StyleYe, Xianwen, Haixia Zhang, Qian Li, Hongmin Ren, Xinfang Xu, and Xiangri Li. 2023. "Structural-Activity Relationship of Rare Ginsenosides from Red Ginseng in the Treatment of Alzheimer’s Disease" International Journal of Molecular Sciences 24, no. 10: 8625. https://doi.org/10.3390/ijms24108625
APA StyleYe, X., Zhang, H., Li, Q., Ren, H., Xu, X., & Li, X. (2023). Structural-Activity Relationship of Rare Ginsenosides from Red Ginseng in the Treatment of Alzheimer’s Disease. International Journal of Molecular Sciences, 24(10), 8625. https://doi.org/10.3390/ijms24108625







